

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146570.7 - phase: 0
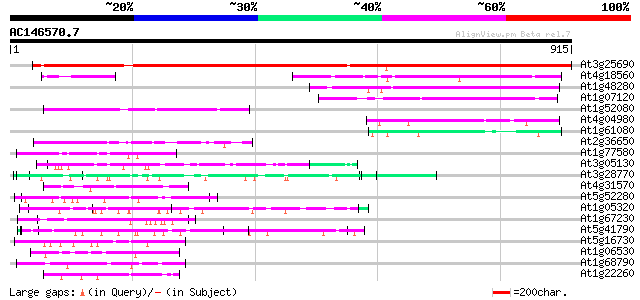
(915 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g25690 unknown protein 1031 0.0
At4g18560 pherophorin - like protein 312 6e-85
At1g48280 unknown protein 283 4e-76
At1g07120 hypothetical protein 242 6e-64
At1g52080 unknown protein 216 6e-56
At4g04980 94 5e-19
At1g61080 hypothetical protein 70 7e-12
At2g36650 hypothetical protein 68 2e-11
At1g77580 unknown protein 64 3e-10
At3g05130 hypothetical protein 60 4e-09
At3g28770 hypothetical protein 60 7e-09
At4g31570 putative protein 59 2e-08
At5g52280 hyaluronan mediated motility receptor-like protein 58 2e-08
At1g05320 unknown protein 57 5e-08
At1g67230 unknown protein 55 1e-07
At5g41790 myosin heavy chain-like protein 54 3e-07
At5g16730 putative protein 53 7e-07
At1g06530 hypothetical protein 53 7e-07
At1g68790 putative nuclear matrix constituent protein 1 (NMCP1) 53 9e-07
At1g22260 hypothetical protein 52 1e-06
>At3g25690 unknown protein
Length = 939
Score = 1031 bits (2665), Expect = 0.0
Identities = 554/908 (61%), Positives = 685/908 (75%), Gaps = 47/908 (5%)
Query: 37 DDSKKHGGNETEMAKNGSDSEIEWLRNVVEELEEREMKLQSELLEYYSLKEQVPVIEEFQ 96
+D K E EMA N D E+E L+ +V+ELEERE+KL+ ELLEYY LKEQ I E Q
Sbjct: 47 NDKNKERKYEVEMAYN--DGELERLKQLVKELEEREVKLEGELLEYYGLKEQESDIVELQ 104
Query: 97 RQLRIKSVEIDMLHMTIKSLQEENNKLQEELIHEASAKRELEVARNKIKELQRQIKIIAN 156
RQL+IK+VEIDML++TI SLQ E KLQEEL ++ELEVARNKIKELQRQI++ AN
Sbjct: 105 RQLKIKTVEIDMLNITINSLQAERKKLQEELSQNGIVRKELEVARNKIKELQRQIQLDAN 164
Query: 157 QTKGQLLLLKQKVSGLQAKEEEVVKKDAEIENNLKTVNDFEIEKKLKTVNDLEIEAVGLK 216
QTKGQLLLLKQ VS LQ KEEE + KD E+E +KLK V DLE++ + LK
Sbjct: 165 QTKGQLLLLKQHVSSLQMKEEEAMNKDTEVE------------RKLKAVQDLEVQVMELK 212
Query: 217 RRNKELQHEKRELTVKLNAAESRITELSSVTENEMIADAKSETGRLRHANEDLQKQVEGL 276
R+N+ELQHEKREL++KL++AE+RI LS++TE++ +A + E L+H NEDL KQVEGL
Sbjct: 213 RKNRELQHEKRELSIKLDSAEARIATLSNMTESDKVAKVREEVNNLKHNNEDLLKQVEGL 272
Query: 277 QMNRFSEVEELVYLRWVNACLRYELRNYKAPSGKSLARDLNNSFNPKSQEKAKQLMLEYA 336
QMNRFSEVEELVYLRWVNACLRYELRNY+ P+GK ARDL+ + +PKSQ KAK+LMLEYA
Sbjct: 273 QMNRFSEVEELVYLRWVNACLRYELRNYQTPAGKISARDLSKNLSPKSQAKAKRLMLEYA 332
Query: 337 GSERGHGDTDIESNFSHDHSSPGSEDLDNAYINSPTYKYSNLSKKTSLIQKLKKWNKN-N 395
GSERG GDTD+ESN+S SSPGS+D DNA ++S T ++S+ SKK LIQKLKKW K+ +
Sbjct: 333 GSERGQGDTDLESNYSQP-SSPGSDDFDNASMDSSTSRFSSFSKKPGLIQKLKKWGKSKD 391
Query: 396 DSSAFSSPARSLSIGSPCRVSTSY-RPRNTLESLMLRNAHDAVAITTFEQKEHKPTHSPE 454
DSS SSP+RS GSP R+S+S + R LESLM+RNA ++VAITTF Q + + +PE
Sbjct: 392 DSSVQSSPSRSFYGGSPGRLSSSMNKQRGPLESLMIRNAGESVAITTFGQVDQESPGTPE 451
Query: 455 TPFLPSLR---KVSSSDDILDSVSASFQLMSKSVDRSLDEKYPAYKDLHKLALAREKQLK 511
TP LP +R + SS + L+SV+ASF +MSKSVD LDEKYPAYKD HKLA+ REK +K
Sbjct: 452 TPNLPRIRTQQQASSPGEGLNSVAASFHVMSKSVDNVLDEKYPAYKDRHKLAVEREKHIK 511
Query: 512 EKAEKARGEKFGDNSNLNTTKAE-EERSTSLDSELTQVNEKACVSDGLNKQSEDVKDFDN 570
KA++AR E+FG N L A+ +E+ + S +T ++ S+ N+ +E +
Sbjct: 512 HKADQARAERFGGNVALPPKLAQLKEKRVVVPSVITATGDQ---SNESNESNEGKASENA 568
Query: 571 QTITEMKLFKIEKRPPRLPRPPPKPSDGAPVSNSLNEIPYAPS----------------- 613
T+T+MKL IEKRPPR+PRPPP+ + G +N + P P
Sbjct: 569 ATVTKMKLVDIEKRPPRVPRPPPRSAGGGKSTNLPSARPPLPGGGPPPPPPPPGGGPPPP 628
Query: 614 ----VPSPPPPPGSLPRGAVGDDKVKRAPELVEFYQSLMKREAKKDA--SLLTSSTSNAA 667
P PPPPPG+L RGA G +KV RAPELVEFYQSLMKRE+KK+ SL++S T N++
Sbjct: 629 PGGGPPPPPPPPGALGRGAGGGNKVHRAPELVEFYQSLMKRESKKEGAPSLISSGTGNSS 688
Query: 668 DTRSNVIAEIENRSSFLLAVKADVETQGDFVMSLATEVRAASFSKIEDVVAFVNWLDEEL 727
R+N+I EIENRS+FLLAVKADVETQGDFV SLATEVRA+SF+ IED++AFV+WLDEEL
Sbjct: 689 AARNNMIGEIENRSTFLLAVKADVETQGDFVQSLATEVRASSFTDIEDLLAFVSWLDEEL 748
Query: 728 SFLSDERAVLKHFDWPEGKSDALREASFEYQDLMKLEKQVSNFTDDPKLPCEDALQKMYS 787
SFL DERAVLKHFDWPEGK+DALREA+FEYQDLMKLEKQV++F DDP L CE AL+KMY
Sbjct: 749 SFLVDERAVLKHFDWPEGKADALREAAFEYQDLMKLEKQVTSFVDDPNLSCEPALKKMYK 808
Query: 788 LLEKLEQSVYALLRTRDFAISRYKEFGVPVNWLLDSGVVGKIKLSSVQLANKYMKRIASE 847
LLEK+EQSVYALLRTRD AISRYKEFG+PV+WL D+GVVGKIKLSSVQLA KYMKR+A E
Sbjct: 809 LLEKVEQSVYALLRTRDMAISRYKEFGIPVDWLSDTGVVGKIKLSSVQLAKKYMKRVAYE 868
Query: 848 IDTLSGPENEPTREFLILQGVRFSFRVHQFAGGFDTESMKAFEELRNNIHVQAGEYNNKP 907
+D++SG + +P REFL+LQGVRF+FRVHQFAGGFD ESMKAFEELR+ ++G+ NN
Sbjct: 869 LDSVSGSDKDPNREFLLLQGVRFAFRVHQFAGGFDAESMKAFEELRSRAKTESGDNNNNN 928
Query: 908 EMATKDDQ 915
+ +++
Sbjct: 929 NNNSNEEE 936
>At4g18560 pherophorin - like protein
Length = 637
Score = 312 bits (799), Expect = 6e-85
Identities = 206/473 (43%), Positives = 279/473 (58%), Gaps = 49/473 (10%)
Query: 461 LRKV--SSSDDILDSVSASFQ-LMSKSVDRSLDEKYPAYKDLHKLALAREKQLK-EKAEK 516
LRK+ S SDD SVS FQ LM S +L L L Q K+
Sbjct: 163 LRKLVSSESDDHALSVSQRFQGLMDVSAKSNLIRSLKRVGSLRNLPEPITNQENTNKSIS 222
Query: 517 ARGEKFGDNSNLNTTKAEEERSTSLDSELTQVNEKACVSDGLNKQSEDVKDFDNQTITEM 576
+ G+ GD + +E S S S ++ E + +S ++ K ++I+
Sbjct: 223 SSGDADGD-----IYRKDEIESYSRSSNSEELTESSSLSTVRSRVPRVPKPPPKRSISLG 277
Query: 577 KLFKIEKR---PPRLPRPPPKPSDGAPVSNSLNEIPYAPSV------PSPPPPPGSLPRG 627
E R PP+ PPP P P+ L + P PSV P PPPPP SL
Sbjct: 278 D--STENRADPPPQKSIPPPPPPPPPPL---LQQPPPPPSVSKAPPPPPPPPPPKSL--- 329
Query: 628 AVGDDKVKRAPELVEFYQSLMKREA---KKDASLLTSSTSNAADTRSN---VIAEIENRS 681
++ KV+R PE+VEFY SLM+R++ ++D++ ++ + A SN +I EIENRS
Sbjct: 330 SIASAKVRRVPEVVEFYHSLMRRDSTNSRRDSTGGGNAAAEAILANSNARDMIGEIENRS 389
Query: 682 SFLLAVKADVETQGDFVMSLATEVRAASFSKIEDVVAFVNWLDEELSFLS---------- 731
+LLA+K DVETQGDF+ L EV A+FS IEDVV FV WLD+ELS+L
Sbjct: 390 VYLLAIKTDVETQGDFIRFLIKEVGNAAFSDIEDVVPFVKWLDDELSYLFKVCKFVVVSL 449
Query: 732 --DERAVLKHFDWPEGKSDALREASFEYQDLMKLEKQVSNFTDDPKLPCEDALQKMYSLL 789
DERAVLKHF+WPE K+DALREA+F Y DL KL + S F +DP+ AL+KM +L
Sbjct: 450 KVDERAVLKHFEWPEQKADALREAAFCYFDLKKLISEASRFREDPRQSSSSALKKMQALF 509
Query: 790 EKLEQSVYALLRTRDFAISRYKEFGVPVNWLLDSGVVGKIKLSSVQLANKYMKRIASEID 849
EKLE VY+L R R+ A +++K F +PV+W+L++G+ +IKL+SV+LA KYMKR+++E++
Sbjct: 510 EKLEHGVYSLSRMRESAATKFKSFQIPVDWMLETGITSQIKLASVKLAMKYMKRVSAELE 569
Query: 850 TLSGPENEPTREFLILQGVRFSFRVHQFAGGFDTESMKAFEELRN---NIHVQ 899
+ G P E LI+QGVRF+FRVHQFAGGFD E+MKAFEELR+ + HVQ
Sbjct: 570 AIEG--GGPEEEELIVQGVRFAFRVHQFAGGFDAETMKAFEELRDKARSCHVQ 620
Score = 43.5 bits (101), Expect = 5e-04
Identities = 36/122 (29%), Positives = 56/122 (45%), Gaps = 21/122 (17%)
Query: 52 NGSDSEIEWLRNVVEELEEREMKLQSELLEYYSLKEQVPVIEEFQRQLRIKSVEIDMLHM 111
NG SE LR VEEL ERE L++E LE + K+ EID L
Sbjct: 84 NGVVSE---LRRQVEELREREALLKTENLE-----------------IADKNGEIDELRK 123
Query: 112 TIKSLQEENNKLQEELIHEASAKRELEVARNKIK-ELQRQIKIIANQTKGQLLLLKQKVS 170
L E+N +L+ E +RE E +++ E+ K++++++ L + Q+
Sbjct: 124 ETARLAEDNERLRREFDRSEEMRRECETREKEMEAEIVELRKLVSSESDDHALSVSQRFQ 183
Query: 171 GL 172
GL
Sbjct: 184 GL 185
Score = 32.3 bits (72), Expect = 1.2
Identities = 42/196 (21%), Positives = 79/196 (39%), Gaps = 22/196 (11%)
Query: 599 APVSNSLNEIPYAPSVPSPPPPPGSLPRGAVGDDKVKRAPELVEFYQSLMKREAKKDASL 658
+P + ++P +P PPPPP P K P F +S + A +
Sbjct: 15 SPSTKKTKDMPSPLPLPPPPPPPLKPPSSGSATTKPPINPSKPGFTRSFGVYFPRASAQV 74
Query: 659 LTSSTSNAADTRSNVIAEIENRSSFLLAVKADVETQGDFVMSLATEVRAASFSKIEDVVA 718
++ AA + + V++E+ + L +A ++T+ +I D
Sbjct: 75 HATA---AAASHNGVVSELRRQVEELREREALLKTEN---------------LEIADKNG 116
Query: 719 FVNWLDEELSFLS-DERAVLKHFDWPE--GKSDALREASFEYQDLMKLEKQVSNFTDDPK 775
++ L +E + L+ D + + FD E + RE E ++++L K VS+ +DD
Sbjct: 117 EIDELRKETARLAEDNERLRREFDRSEEMRRECETREKEME-AEIVELRKLVSSESDDHA 175
Query: 776 LPCEDALQKMYSLLEK 791
L Q + + K
Sbjct: 176 LSVSQRFQGLMDVSAK 191
>At1g48280 unknown protein
Length = 558
Score = 283 bits (723), Expect = 4e-76
Identities = 159/429 (37%), Positives = 254/429 (59%), Gaps = 28/429 (6%)
Query: 489 LDEKYPAYKDLHKLALAREKQLKEKAEKARGEKFGDNSNLNTTKAEEERSTSLDSELTQV 548
L+EK + L K + LK + E+AR NSN+ + S L S ++
Sbjct: 121 LEEKLVVNESLIKDLQLQVLNLKTELEEAR------NSNVELELNNRKLSQDLVSAEAKI 174
Query: 549 NEKACVSDGLNK-QSEDVKDFDNQTITEMKLFKIEKR--------------PPRLPRPPP 593
+ + + Q+ KD ++++ K++K P RLP PP
Sbjct: 175 SSLSSNDKPAKEHQNSRFKDIQRLIASKLEQPKVKKEVAVESSRLSPPSPSPSRLPPTPP 234
Query: 594 KPSDGAPVSNSL-----NEIPYAPSVPSPPPPPGSLPRGAVGDDKVKRAPELVEFYQSLM 648
P ++SL N P+AP P PPPPP PR + +++P + + +Q L
Sbjct: 235 LPKFLVSPASSLGKRDENSSPFAPPTPPPPPPPPP-PRPLAKAARAQKSPPVSQLFQLLN 293
Query: 649 KREAKKDASLLTSSTSNAADTRSN-VIAEIENRSSFLLAVKADVETQGDFVMSLATEVRA 707
K++ ++ S + + ++ N ++ EI+NRS+ L+A+KAD+ET+G+F+ L +V
Sbjct: 294 KQDNSRNLSQSVNGNKSQVNSAHNSIVGEIQNRSAHLIAIKADIETKGEFINDLIQKVLT 353
Query: 708 ASFSKIEDVVAFVNWLDEELSFLSDERAVLKHFDWPEGKSDALREASFEYQDLMKLEKQV 767
FS +EDV+ FV+WLD+EL+ L+DERAVLKHF WPE K+D L+EA+ EY++L KLEK++
Sbjct: 354 TCFSDMEDVMKFVDWLDKELATLADERAVLKHFKWPEKKADTLQEAAVEYRELKKLEKEL 413
Query: 768 SNFTDDPKLPCEDALQKMYSLLEKLEQSVYALLRTRDFAISRYKEFGVPVNWLLDSGVVG 827
S+++DDP + AL+KM +LL+K EQ + L+R R ++ Y++F +PV W+LDSG++
Sbjct: 414 SSYSDDPNIHYGVALKKMANLLDKSEQRIRRLVRLRGSSMRSYQDFKIPVEWMLDSGMIC 473
Query: 828 KIKLSSVQLANKYMKRIASEIDTLSGPENEPTREFLILQGVRFSFRVHQFAGGFDTESMK 887
KIK +S++LA YM R+A+E+ + + E T+E L+LQGVRF++R HQFAGG D E++
Sbjct: 474 KIKRASIKLAKTYMNRVANELQSARNLDRESTKEALLLQGVRFAYRTHQFAGGLDPETLC 533
Query: 888 AFEELRNNI 896
A EE++ +
Sbjct: 534 ALEEIKQRV 542
Score = 35.8 bits (81), Expect = 0.11
Identities = 26/80 (32%), Positives = 43/80 (53%), Gaps = 1/80 (1%)
Query: 166 KQKVSGLQAKEEEVVKKDAEIENNLKTVNDFEIEKKLKTVNDLEIEAVGLKRRNKELQHE 225
++ V A E+E K+ E+E L VN+ I+ V +L+ E + N EL+
Sbjct: 101 EETVMATAAAEDEKRKRMEELEEKL-VVNESLIKDLQLQVLNLKTELEEARNSNVELELN 159
Query: 226 KRELTVKLNAAESRITELSS 245
R+L+ L +AE++I+ LSS
Sbjct: 160 NRKLSQDLVSAEAKISSLSS 179
>At1g07120 hypothetical protein
Length = 392
Score = 242 bits (618), Expect = 6e-64
Identities = 149/393 (37%), Positives = 222/393 (55%), Gaps = 34/393 (8%)
Query: 504 LAREKQLKEKAEKARGEKFGDNSNLNTTKAEEERSTSLDSELTQVNEKACVSDGLN-KQS 562
L R +L+++ + R E + ++ K+ E S+ + Q + +DG N K
Sbjct: 22 LVRNDKLEKENHELRQEVARLRAQVSNLKSHENERKSMLWKKLQSSYDGSNTDGSNLKAP 81
Query: 563 EDVKDFDNQTITEMKLFKIEKRPPRLPRPPPKPSDGAPVSNSLNEIPYAPSVPSPPPPPG 622
E VK + + P PKP+ I + PPPPP
Sbjct: 82 ESVKS--------------NTKGQEVRNPNPKPT-----------IQGQSTATKPPPPPP 116
Query: 623 SLPRGAVGDDKVKRAPELVEFYQSLMKREAKKDASLLTSSTSNAADTRSNVIAEIENRSS 682
+ +G V+RAPE+VEFY++L KRE+ + + + A R N+I EIENRS
Sbjct: 117 LPSKRTLGKRSVRRAPEVVEFYRALTKRESHMGNKINQNGVLSPAFNR-NMIGEIENRSK 175
Query: 683 FLLAVKADVETQGDFVMSLATEVRAASFSKIEDVVAFVNWLDEELSFLSDERAVLKHFD- 741
+L +K+D + D + L ++V AA+F+ I +V FV W+DEELS L DERAVLKHF
Sbjct: 176 YLSDIKSDTDRHRDHIHILISKVEAATFTDISEVETFVKWIDEELSSLVDERAVLKHFPK 235
Query: 742 WPEGKSDALREASFEYQDLMKLEKQVSNFTDDPKLPCEDALQKMYSLLEKLEQSVYALLR 801
WPE K D+LREA+ Y+ L ++ +F D+PK ALQ++ SL ++LE+SV +
Sbjct: 236 WPERKVDSLREAACNYKRPKNLGNEILSFKDNPKDSLTQALQRIQSLQDRLEESVNNTEK 295
Query: 802 TRDFAISRYKEFGVPVNWLLDSGVVGKIKLSSVQLANKYMKRIASEIDTL-SGPENEPTR 860
RD RYK+F +P W+LD+G++G++K SS++LA +YMKRIA E+++ SG E
Sbjct: 296 MRDSTGKRYKDFQIPWEWMLDTGLIGQLKYSSLRLAQEYMKRIAKELESNGSGKEGN--- 352
Query: 861 EFLILQGVRFSFRVHQFAGGFDTESMKAFEELR 893
L+LQGVRF++ +HQFAGGFD E++ F EL+
Sbjct: 353 --LMLQGVRFAYTIHQFAGGFDGETLSIFHELK 383
>At1g52080 unknown protein
Length = 573
Score = 216 bits (549), Expect = 6e-56
Identities = 132/339 (38%), Positives = 200/339 (58%), Gaps = 20/339 (5%)
Query: 55 DSEIEWLRNVVEELEEREMKLQSELLEYYSLKEQVPVIEEFQRQLRIKSVEIDMLHMTIK 114
++EI LRN V L ERE L+ +LLEYYSLKEQ + E + +L++ +E + + IK
Sbjct: 138 ENEINRLRNTVRALRERERCLEDKLLEYYSLKEQQKIAMELRSRLKLNQMETKVFNFKIK 197
Query: 115 SLQEENNKLQEELIHEASAKRELEVARNKIKELQRQIKIIANQTKGQLLLLKQKVSGLQA 174
LQ EN KL+ E + EL++A+++++ L++++ I Q Q+L LKQ+V+ LQ
Sbjct: 198 KLQAENEKLKAECFEHSKVLLELDMAKSQVQVLKKKLNINTQQHVAQILSLKQRVARLQE 257
Query: 175 KEEEVVKKDAEIENNLKTVNDFEIEKKLKTVNDLEIEAVGLKRRNKELQHEKRELTVKLN 234
+E + V + D E +K ++ + DLE E L N LQ E EL+ KL
Sbjct: 258 EEIKAV------------LPDLEADKMMQRLRDLESEINELTDTNTRLQFENFELSEKLE 305
Query: 235 AAESRITELSSVTENEMIADAKSETGRLRHANEDLQKQVEGLQMNRFSEVEELVYLRWVN 294
+ + I S + E E I + + RLR NE+L+K VE LQ +R +++E+LVYLRW+N
Sbjct: 306 SVQ--IIANSKLEEPEEIETLREDCNRLRSENEELKKDVEQLQGDRCTDLEQLVYLRWIN 363
Query: 295 ACLRYELRNYKAPSGKSLARDLNNSFNPKSQEKAKQLMLEYAGSERGHGDTDIESNFSHD 354
ACLRYELR Y+ P+GK++ARDL+ + +P S+EKAKQL+LEYA SE +TD + S
Sbjct: 364 ACLRYELRTYQPPAGKTVARDLSTTLSPTSEEKAKQLILEYAHSE---DNTDYDRWSSSQ 420
Query: 355 HSSPGSED---LDNAYINSPTYKYSNLSKKTSLIQKLKK 390
S D LD++ +++ + + K L+ KL K
Sbjct: 421 EESSMITDSMFLDDSSVDTLFATKTKKTGKKKLMHKLMK 459
>At4g04980
Length = 681
Score = 93.6 bits (231), Expect = 5e-19
Identities = 84/344 (24%), Positives = 162/344 (46%), Gaps = 37/344 (10%)
Query: 582 EKRPPRLPRPPP--KPSDGAPV-----SNSLNEIPYAPSVPSPPPPPGSLPRGAVGDDKV 634
E + P P PPP K S+ ++S N APS+P+PP PPGS K+
Sbjct: 333 EHKAPAPPPPPPMSKASESGEFCQFSKTHSTNG-DNAPSMPAPPAPPGSGRSLKKATSKL 391
Query: 635 KRAPELVEFYQSLM------------KREAKKDASLLTSSTSNAADT-RSNVIAEIENRS 681
+R+ ++ Y +L K+ +K S+ S A + ++ +AE+ RS
Sbjct: 392 RRSAQIANLYWALKGKLEGRGVEGKTKKASKGQNSVAEKSPVKVARSGMADALAEMTKRS 451
Query: 682 SFLLAVKADVETQGDFVMSLATEVRAASFSKIEDVVAFVNWLDEELSFLSDERAVLKHFD 741
S+ ++ DV+ + L + + + +++++ F + ++ L L+DE VL F+
Sbjct: 452 SYFQQIEEDVQKYAKSIEELKSSIHSFQTKDMKELLEFHSKVESILEKLTDETQVLARFE 511
Query: 742 -WPEGKSDALREASFEYQDLMKLEKQVSNFTDDPKLPCEDALQKMYSLLEKLEQSVYALL 800
+PE K + +R A Y+ L + ++ N+ +P P D L K+ K + + +
Sbjct: 512 GFPEKKLEVIRTAGALYKKLDGILVELKNWKIEP--PLNDLLDKIERYFNKFKGEIETVE 569
Query: 801 RTRDFAISRYKEFGVPVNWLLDSGVVGKIKLSSVQLANKYM------KRIASEIDTLSGP 854
RT+D +K + + + D V+ ++K + V +++ M +R A+E + +G
Sbjct: 570 RTKDEDAKMFKRYNINI----DFEVLVQVKETMVDVSSNCMELALKERREANE-EAKNGE 624
Query: 855 ENEPTREFL--ILQGVRFSFRVHQFAGGFDTESMKAFEELRNNI 896
E++ E + + +F+F+V+ FAGG D + +L + I
Sbjct: 625 ESKMKEERAKRLWRAFQFAFKVYTFAGGHDERADCLTRQLAHEI 668
>At1g61080 hypothetical protein
Length = 907
Score = 69.7 bits (169), Expect = 7e-12
Identities = 79/349 (22%), Positives = 141/349 (39%), Gaps = 72/349 (20%)
Query: 585 PPRLP-----RPPPKPSDGAPVSNSLNEIPYAPSV------PSPPPPPGSLP--RGAVGD 631
PP +P PPP P A + + P P + PPPPPG+ R
Sbjct: 594 PPPMPLANGATPPPPPPPMAMANGAAGPPPPPPRMGMANGAAGPPPPPGAARSLRPKKAA 653
Query: 632 DKVKRAPELVEFYQSLMKREAKKDASLLTSSTSN-----------AADTRSNVIAEIENR 680
K+KR+ +L Y+ L + +D + T S S ++ +AEI +
Sbjct: 654 TKLKRSTQLGNLYRILKGKVEGRDPNAKTGSGSGRKAGAGSAPAGGKQGMADALAEITKK 713
Query: 681 SSFLLAVKADVETQGDFVMSLATEVRAASFSKIEDVVAFVNWLDEELSFLSDERAVLKHF 740
S++ L ++AD+ + L E+ + ++++F ++ L L+DE VL
Sbjct: 714 SAYFLQIQADIAKYMTSINELKIEITKFQTKDMTELLSFHRRVESVLENLTDESQVLARC 773
Query: 741 D-WPEGKSDALREASFEYQDLMKLEKQVSNFTDDPKLPCEDALQKMYSLLEKLEQSVYAL 799
+ +P+ K +A+R A Y L + ++ N +P L LL+K+E
Sbjct: 774 EGFPQKKLEAMRMAVALYTKLHGMITELQNMKIEPPL---------NQLLDKVE------ 818
Query: 800 LRTRDFAISRYKEFGVPVNWLLDSGVVGKIKLSSVQLANKYMKRIASE--IDTLSGPENE 857
RY KIK + V +++ M+ E + L P+ +
Sbjct: 819 ---------RY---------------FTKIKETMVDISSNCMELALKEKRDEKLVSPDAK 854
Query: 858 PTRE------FLILQGVRFSFRVHQFAGGFDTESMKAFEELRNNIHVQA 900
P+ + ++ + +F+F+V+ FAGG D + EL + I +
Sbjct: 855 PSLKKTVGSAKMLWRAFQFAFKVYTFAGGHDDRADSLTRELAHEIQTDS 903
Score = 34.3 bits (77), Expect = 0.33
Identities = 34/111 (30%), Positives = 43/111 (38%), Gaps = 22/111 (19%)
Query: 532 KAEEERSTSLDSELTQVNEKACVSDGLNKQSEDVKDFDNQTITEMKLFKIEKR------- 584
K E E S +LD E +N +Q +K ++ +E KL EK
Sbjct: 365 KMETEESVNLDEESVVLN---------GEQDTIMKISSLESTSESKLNHSEKYENSSQLF 415
Query: 585 -PPRLPRPPPKPSDGAPVSNSLNEIPYAP-----SVPSPPPPPGSLPRGAV 629
PP P PPP S S L P P ++ PPPPP P AV
Sbjct: 416 PPPPPPPPPPPLSFIKTASLPLPSPPPTPPIADIAISMPPPPPPPPPPPAV 466
Score = 31.6 bits (70), Expect = 2.1
Identities = 17/42 (40%), Positives = 20/42 (47%), Gaps = 3/42 (7%)
Query: 591 PPPKPSDGAPVSNSLNEIPYAPSVPSP---PPPPGSLPRGAV 629
PPP P P+ S P P +P+ PPPP PR AV
Sbjct: 493 PPPTPPAFKPLKGSAPPPPPPPPLPTTIAAPPPPPPPPRAAV 534
Score = 30.4 bits (67), Expect = 4.7
Identities = 16/40 (40%), Positives = 17/40 (42%), Gaps = 3/40 (7%)
Query: 585 PPRL---PRPPPKPSDGAPVSNSLNEIPYAPSVPSPPPPP 621
PPR P PPP P A P + P PPPPP
Sbjct: 529 PPRAAVAPPPPPPPPGTAAAPPPPPPPPGTQAAPPPPPPP 568
Score = 30.4 bits (67), Expect = 4.7
Identities = 19/46 (41%), Positives = 22/46 (47%), Gaps = 7/46 (15%)
Query: 585 PPRLPR-----PPPKPSDGAPVSNSLNEIP--YAPSVPSPPPPPGS 623
PP LP PPP P A V+ P A + P PPPPPG+
Sbjct: 513 PPPLPTTIAAPPPPPPPPRAAVAPPPPPPPPGTAAAPPPPPPPPGT 558
>At2g36650 hypothetical protein
Length = 374
Score = 68.2 bits (165), Expect = 2e-11
Identities = 80/362 (22%), Positives = 156/362 (42%), Gaps = 66/362 (18%)
Query: 39 SKKHGGNETEMAKNGSDSEIEWLRNVVEELEEREMKLQSELLEYYSLKEQVPVIEEFQRQ 98
S+++ E + N EI L++ EEL+ +E +++ + +LK+Q ++ E +
Sbjct: 59 SRENDEEEEAESPNQQKQEILSLKSRFEELQRKEYEMELHFERFCNLKDQEVMLIEHKSI 118
Query: 99 LRIKSVEIDMLHMTIKSLQEENNKLQEELIHEASAKRELEVARNKIKELQRQIKIIANQT 158
L ++ ++D + +++EE+ + Q +I E++ R++ L+ + K + ++
Sbjct: 119 LSLEKAQLDFFRKEVLAMEEEHKRGQALVIVYLKLVGEIQELRSENGLLEGKAKKLRRKS 178
Query: 159 KGQLLLLKQKVSGLQAKEEEVVKKDAEIENNLKTVNDFEIEKKLKTVNDLEIEAVGLKRR 218
K QL + + + E+E LK V+ E+E K V +LE + ++
Sbjct: 179 K-QLYRVVNESRKIIGVEKEF----------LKCVD--ELETKNNIVKELEGKVKDMEAY 225
Query: 219 NKELQHEKRELTVKLNAAESRITELSSVTENEMIADAKSETGRLRHANEDLQKQVEGLQM 278
LQ EK EL +K ++ S ++ SV + I + + E L+
Sbjct: 226 VDVLQEEKEELFMK--SSNSTSEKMVSVEDYRRIVE-----------------EYEELKK 266
Query: 279 NRFSEVEELVYLRWVNACLRYELRNYKAPSGKSLARDLNNSFNPKSQEKAKQLMLEYAGS 338
+ + V+E++ LRW NACLR+E+ G+ + F+P L G
Sbjct: 267 DYANGVKEVINLRWSNACLRHEVMRNGTNFGEIV-------FSPNGN-------LHELGL 312
Query: 339 ERGHGDTDIE----SNFSHDHSSPGSEDLDNAYINSPTYKYSNLSKKTSLIQKLKKWNKN 394
E D + +N H+ + E S++ L++KLKKW +
Sbjct: 313 EDDQVDNALALMRVANEQHEENHHQHES----------------SRRKRLMKKLKKWVEG 356
Query: 395 ND 396
N+
Sbjct: 357 NE 358
>At1g77580 unknown protein
Length = 779
Score = 64.3 bits (155), Expect = 3e-10
Identities = 74/289 (25%), Positives = 142/289 (48%), Gaps = 37/289 (12%)
Query: 12 DDEILSEFESLLSEQTDFPLLSEKTDDSKKHGGNETEMAKNGSDSEI---EWLRNVVEEL 68
++ + SE E L S + EK + K NE + + + I E L + +EL
Sbjct: 338 ENSLASEIEVLTSRIKELEEKLEKLEAEKHELENEVKCNREEAVVHIENSEVLTSRTKEL 397
Query: 69 EEREMKLQSELLEYYSLKEQVPVIEEFQRQLRIK---SVEIDMLHMTIKSLQEENNKLQE 125
EE+ KL++E E LK +V E + + ++ + EI++L K L+E+ KL+
Sbjct: 398 EEKLEKLEAEKEE---LKSEVKCNRE-KAVVHVENSLAAEIEVLTSRTKELEEQLEKLEA 453
Query: 126 ELIH---EASAKRELEVARNKIKELQRQIKIIANQTKGQLLLLKQKVSGLQAKEEEV--- 179
E + E RE VA+ + L +I+++ + K L++K+ L+ +++E+
Sbjct: 454 EKVELESEVKCNREEAVAQVE-NSLATEIEVLTCRIK----QLEEKLEKLEVEKDELKSE 508
Query: 180 VKKDAEIENNLK------TVNDFEIEKKLKTVN----DLEIEAVGLKRRNKELQHEKREL 229
VK + E+E+ L+ E+E KL+ + +L+I +K + +E Q +E+
Sbjct: 509 VKCNREVESTLRFELEAIACEKMELENKLEKLEVEKAELQISFDIIKDKYEESQVCLQEI 568
Query: 230 TVKLNAAESR---ITELSSVTENEMI---ADAKSETGRLRHANEDLQKQ 272
KL ++ + EL + E++ I ADAK+++ ++ ED++K+
Sbjct: 569 ETKLGEIQTEMKLVNELKAEVESQTIAMEADAKTKSAKIESLEEDMRKE 617
>At3g05130 hypothetical protein
Length = 634
Score = 60.5 bits (145), Expect = 4e-09
Identities = 106/478 (22%), Positives = 203/478 (42%), Gaps = 79/478 (16%)
Query: 45 NETEMAKNGSD---SEIEWLRNVVEELEEREM-------KLQSE----LLEYYSLKEQVP 90
+E ++ KNG D E+ L+ V LEE+E KL+SE + E +E++
Sbjct: 173 DERDLIKNGFDLQHEEVNRLKECVVRLEEKESNLEIVIGKLESENERLVKERKVREEEIE 232
Query: 91 VIEE----FQRQLRIKSVEIDMLHMTIKSLQEENNKLQEELIHEASAKRELEVARNKIKE 146
+++ ++ + K EID L IK L E N+++ I + ELE K+ +
Sbjct: 233 GVKKEKIGLEKIMEEKKNEIDGLKREIKVLLSEKNEMEIVKIEQKGVIEELE---RKLDK 289
Query: 147 LQRQIKIIANQTKGQLLLLKQKVSGLQAKEEEVVKKDA----EIE--NNLKTVNDFEIEK 200
L ++ + + K +L+ V GL+ +E ++K++ EI+ +T+ + E+E+
Sbjct: 290 LNETVRSLTKEEK----VLRDLVIGLEKNLDESMEKESGMMVEIDALGKERTIKESEVER 345
Query: 201 KL--KTVNDLEIEAVGLKRRNK-----ELQHEKRELTVKLNAAESRITELSSVTENEMIA 253
+ K + + ++E + ++ +K +L EK EL ++ + E ++ EL+ A
Sbjct: 346 LIGEKNLIEKQMEMLNVQSSDKGKLIDQLSREKVELEERIFSRERKLVELNRK------A 399
Query: 254 DAKSETGRLRHANEDLQKQVEGLQMNRFSEVEELVYLRWVNACLRYELRNYKAPSGKSLA 313
D + + N D Q ++ G + ++ NA + ELR +A K+L
Sbjct: 400 DELTHAVAVLQKNCDDQTKINGKLSCKVDQLS--------NALAQVELRREEA--DKALD 449
Query: 314 RDLNNSFNPKSQEKAKQLMLEYAGSERGHGDTDIESNFSHDHSSPGSEDLDNAYINSPTY 373
+ N + K++ + M+ E + +S FS DL++ S +
Sbjct: 450 EEKRNGEDLKAEVLKSEKMVAKTLEELEKVKIERKSLFS------AKNDLES---QSESL 500
Query: 374 KYSNLSKKTSLIQKLKKWN--KNNDSSAFSSPARSLSIGSPCRVSTSYRPRNTLESLMLR 431
K N+ + L++ K K SA RS+ + S L
Sbjct: 501 KSENVKLEKELVELRKAMEALKTELESAGMDAKRSMVMLKSAASMLS----------QLE 550
Query: 432 NAHDAVAITTFEQKEHKPTHSPETPFLPSLRKV-SSSDDILDSVSASFQLMSKSVDRS 488
N D + + EQK T P L S+ K + +DI++ + ++M +S + +
Sbjct: 551 NRED--RLISEEQKREIGT-EPYAMELESIEKAFKNKEDIIEEMKKEAEIMKQSTEEA 605
Score = 44.7 bits (104), Expect = 2e-04
Identities = 109/531 (20%), Positives = 205/531 (38%), Gaps = 104/531 (19%)
Query: 62 RNVVEELEEREMKLQSELLEY------------------YSLKEQVPVIEEF-QRQLRIK 102
RN ++ L + + +L++EL Y + LK ++ + F + Q R
Sbjct: 70 RNQIDSLVQAKDELETELARYCQEKTGLRDELDQVSDENFGLKFELDFVIVFVESQFREM 129
Query: 103 SVEIDMLHMTIKSLQEENNKLQEELIHEASAKRELEVAR-NKIKELQRQIKIIANQTKGQ 161
V +DML + E L+ E I E + K E+E + K+ + + IK + +
Sbjct: 130 CVGVDMLVKEKSDRESEIRVLKGEAI-ELTGKVEIEKEQLRKVCDERDLIKNGFDLQHEE 188
Query: 162 LLLLKQKVSGLQAKEE--EVVKKDAEIENNLKTVNDFEIEKKLKTVNDLEIEAVGLKRRN 219
+ LK+ V L+ KE E+V E EN + V + ++ ++ + ++ E +GL++
Sbjct: 189 VNRLKECVVRLEEKESNLEIVIGKLESENE-RLVKERKVREE--EIEGVKKEKIGLEKIM 245
Query: 220 KELQHE----KRELTVKLNAAESRITELSSVTENEMIADAKSETGRLRHANEDLQKQVEG 275
+E ++E KRE+ V L+ E E+ + + +I + + + +L L K+ +
Sbjct: 246 EEKKNEIDGLKREIKVLLS--EKNEMEIVKIEQKGVIEELERKLDKLNETVRSLTKEEKV 303
Query: 276 LQMNRFSEVEELVYLRWVNACLRYELRNYKAPSGKSLARDLNNSFNPKSQEKAKQLMLEY 335
L+ +LV L ++L+ S +E + ++
Sbjct: 304 LR--------DLVI---------------------GLEKNLDESME---KESGMMVEIDA 331
Query: 336 AGSERGHGDTDIESNFSHDHSSPGSEDLDNAYINSPTYKYSNLSKKTSLIQKLKKWNKNN 395
G ER ++++E + ++ N S K LI +L +
Sbjct: 332 LGKERTIKESEVERLIGEKNLIEKQMEMLNV----------QSSDKGKLIDQLSREKVEL 381
Query: 396 DSSAFSSPARSLSIGSPCRVSTSYRPRNTLESLMLRNAHDAVAITTFEQKEHKPTHSPET 455
+ FS + + + T +++ +N D I
Sbjct: 382 EERIFSRERKLVELNRKADELTH------AVAVLQKNCDDQTKING-------------- 421
Query: 456 PFLPSLRKVSSSDDILDSVSASFQLMSKSVDRSLDEKYPAYKDLHKLALAREKQLKEKAE 515
K+S D L + A +L + D++LDE+ +DL L EK + + E
Sbjct: 422 -------KLSCKVDQLSNALAQVELRREEADKALDEEKRNGEDLKAEVLKSEKMVAKTLE 474
Query: 516 KARGEKFGDNSNLNTTKAEEERSTSLDSELTQVNEKACVSDGLNKQSEDVK 566
+ K S + E +S SL SE ++ EK V L K E +K
Sbjct: 475 ELEKVKIERKSLFSAKNDLESQSESLKSENVKL-EKELVE--LRKAMEALK 522
Score = 38.5 bits (88), Expect = 0.017
Identities = 56/233 (24%), Positives = 105/233 (45%), Gaps = 24/233 (10%)
Query: 64 VVEELEEREMKLQSELLEYYSLKEQVPVIEEFQRQLRIKSVEIDMLHMTIKSLQEENNKL 123
+V+E +RE +++ E L +V + +E QLR E D++ EE N+L
Sbjct: 136 LVKEKSDRESEIRVLKGEAIELTGKVEIEKE---QLRKVCDERDLIKNGFDLQHEEVNRL 192
Query: 124 QEELIHEASAKRELEVARNKI----KELQRQIKIIANQTKGQLLLLKQKV---SGLQAKE 176
+E ++ + LE+ K+ + L ++ K+ + +G + K+K+ ++ K+
Sbjct: 193 KECVVRLEEKESNLEIVIGKLESENERLVKERKVREEEIEG---VKKEKIGLEKIMEEKK 249
Query: 177 EEVVKKDAEIENNLKTVNDFEIEK--KLKTVNDLEIEAVGLKRRNKELQHEK---RELTV 231
E+ EI+ L N+ EI K + + +LE + L + L E+ R+L +
Sbjct: 250 NEIDGLKREIKVLLSEKNEMEIVKIEQKGVIEELERKLDKLNETVRSLTKEEKVLRDLVI 309
Query: 232 KL--NAAESRITELSSVTENEMIADAK----SETGRLRHANEDLQKQVEGLQM 278
L N ES E + E + + + SE RL ++KQ+E L +
Sbjct: 310 GLEKNLDESMEKESGMMVEIDALGKERTIKESEVERLIGEKNLIEKQMEMLNV 362
>At3g28770 hypothetical protein
Length = 2081
Score = 59.7 bits (143), Expect = 7e-09
Identities = 111/576 (19%), Positives = 221/576 (38%), Gaps = 71/576 (12%)
Query: 12 DDEILSEFESLLSEQTDFPLLSEKTDDSKKHGGNETEMA--KNGSDSEIEWLRNVVEELE 69
D++ E E S+ + EK +K+ E + + K + + E ++ E+ E
Sbjct: 995 DNKEKKESEDSASKNREKKEYEEKKSKTKEEAKKEKKKSQDKKREEKDSEERKSKKEKEE 1054
Query: 70 EREMKLQSELLEYYSLKEQVPVIEEFQRQLRIKSVEIDMLHMTIKSLQEENNKLQEELIH 129
R++K + + E KE + K E H KS+++E +K +E+ H
Sbjct: 1055 SRDLKAKKKEEETKEKKES--------ENHKSKKKEDKKEHEDNKSMKKEEDK-KEKKKH 1105
Query: 130 EASAKRELEVARNKIKELQRQIKIIANQTKGQ------LLLLKQKVSGLQAKEEEVVKKD 183
E S R+ E + +++L+ Q + K + + L+K++ + KE E +
Sbjct: 1106 EESKSRKKEEDKKDMEKLEDQNSNKKKEDKNEKKKSQHVKLVKKESDKKEKKENEEKSET 1165
Query: 184 AEIENNLKTVNDFEIEKKLKTVNDLEIEAVGLKRRNKELQHEKRELTVKLNAAESRITEL 243
EIE++ N+ + +K+ K+ D + K++ KE++ + + KL E +
Sbjct: 1166 KEIESSKSQKNEVD-KKEKKSSKDQQ------KKKEKEMKESEEK---KLKKNEEDRKKQ 1215
Query: 244 SSVTENEMIADAKSETGRLRHANEDLQKQVEGLQMNRFSEVEELVYLRWVNACLRYELRN 303
+SV EN+ + K E + + ++ KQ G + + SE + E N
Sbjct: 1216 TSVEENKKQKETKKEKNKPKDDKKNTTKQSGGKKESMESESK--------------EAEN 1261
Query: 304 YKAPSGKSLARDLNNSFNPKSQEKAKQLMLEYAGSERGHGDTDIESNFSHDHSSPGSEDL 363
+ + A S E +++++ H D+ +S+ S + ++
Sbjct: 1262 QQKSQATTQA---------DSDESKNEILMQADSQADSHSDSQADSDESKNEILMQADSQ 1312
Query: 364 DNAYINSPTYKYSNLSKKTSLIQKLKKWNKNNDSSAFSSPARSLSIGSPCRVSTSYRPRN 423
N+ + S + QK K KN + + S +
Sbjct: 1313 ATTQRNNEEDRKKQTSVAENKKQKETKEEKNKPKDDKKNTTKQ---------SGGKKESM 1363
Query: 424 TLESLMLRNAHDAVAITTFEQKEHKPTHSPETPFLPSLRKVSSSDDILDSVSASFQLMSK 483
ES N + A T + E K E + S SD DS + +++ +
Sbjct: 1364 ESESKEAENQQKSQATTQADSDESK----NEILMQADSQADSHSDSQADSDESKNEILMQ 1419
Query: 484 SVDRSLDEKYPAYKDLHKLALAREKQLKEKAEKARGEKFGDNSNLNTT-----KAEEERS 538
+ ++ ++ + ++A K+ KE E+ K + NTT K E S
Sbjct: 1420 ADSQATTQRNNEEDRKKQTSVAENKKQKETKEEKNKPK---DDKKNTTEQSGGKKESMES 1476
Query: 539 TSLDSELTQVNEKACVSDGLNKQSEDVKDFDNQTIT 574
S ++E Q ++ + ++E + D+Q T
Sbjct: 1477 ESKEAENQQKSQATTQGESDESKNEILMQADSQADT 1512
Score = 50.1 bits (118), Expect = 6e-06
Identities = 108/584 (18%), Positives = 220/584 (37%), Gaps = 84/584 (14%)
Query: 33 SEKTDDSKKHGGNETEMAKNGSDSEIEWLRNVVEELEEREMKLQSELLEYYSLKEQVPVI 92
S+K +D K+H N++ M K E + +E + K +L + S K++
Sbjct: 1078 SKKKEDKKEHEDNKS-MKKEEDKKEKKKHEESKSRKKEEDKKDMEKLEDQNSNKKKEDKN 1136
Query: 93 EEFQRQLRIKSVEIDMLHMTIKSLQEENNKLQEELIHEASAKRELEVARNKIKEL-QRQI 151
E+ + Q +K +++E++K +++ E S +E+E ++++ E+ +++
Sbjct: 1137 EKKKSQ-------------HVKLVKKESDKKEKKENEEKSETKEIESSKSQKNEVDKKEK 1183
Query: 152 KIIANQTKGQLLLLKQKVSGLQAKEEEVVKKDAEIENNLKTVNDFEIEKKLKTVNDLEIE 211
K +Q K + +K+ K EE KK +E N K + + K K +
Sbjct: 1184 KSSKDQQKKKEKEMKESEEKKLKKNEEDRKKQTSVEENKKQKETKKEKNKPKDDKKNTTK 1243
Query: 212 AVGLKRRNKELQ------HEKRELTVKLNAAESRITEL----------------SSVTEN 249
G K+ + E + +K + T + ++ ES+ L S ++N
Sbjct: 1244 QSGGKKESMESESKEAENQQKSQATTQADSDESKNEILMQADSQADSHSDSQADSDESKN 1303
Query: 250 EMIADAKSETGRLRHANEDLQKQVEGLQMNRFSEVEELVYLRWVNACLRYELRNYKAPSG 309
E++ A S+ R+ ED +KQ + + E +E + + K S
Sbjct: 1304 EILMQADSQATTQRNNEEDRKKQTSVAENKKQKETKEEKNKPKDDKKNTTKQSGGKKESM 1363
Query: 310 KSLARDLNN------SFNPKSQEKAKQLMLEYAGSERGHGDTDIESNFSHDHSSPGSEDL 363
+S +++ N + S E +++++ H D+ +S+ S + ++
Sbjct: 1364 ESESKEAENQQKSQATTQADSDESKNEILMQADSQADSHSDSQADSDESKNEILMQADSQ 1423
Query: 364 DNAYINSPTYKYSNLSKKTSLIQKLKKWNKNNDSSAFSSPARSLSIGSPCRVSTSYRPRN 423
N+ + S + QK K KN + S +
Sbjct: 1424 ATTQRNNEEDRKKQTSVAENKKQKETKEEKNKPKDDKKNTTEQ---------SGGKKESM 1474
Query: 424 TLESLMLRNAHDAVAITTFEQKEHK-----------PTH------SPETPFLPSLRKVSS 466
ES N + A T E E K TH S E+ ++ S
Sbjct: 1475 ESESKEAENQQKSQATTQGESDESKNEILMQADSQADTHANSQGDSDESKNEILMQADSQ 1534
Query: 467 SDDILDSVSASFQLMSKSVDRSLDEKYPAYKDLHKLALAREKQLKEKAEKARGEKFGDNS 526
+D DS + +++ ++ D D + + + +++ + + Q K G++
Sbjct: 1535 ADSQTDSDESKNEILMQA-DSQADSQTDSDESKNEILMQADSQ----------AKIGESL 1583
Query: 527 NLNTTKAEEERSTSLDSELTQVNEKACVSDGLNKQSEDVKDFDN 570
N K +E+ + E N K G +++S+D K +N
Sbjct: 1584 EDNKVKGKEDNGDEVGKE----NSKTIEVKGRHEESKDGKTNEN 1623
Score = 46.2 bits (108), Expect = 8e-05
Identities = 124/617 (20%), Positives = 233/617 (37%), Gaps = 90/617 (14%)
Query: 6 NLANDFDDEILSEFESLLSEQTDFPLLSEKTDDSKKHGGNETEMA----KNGS-DSEIEW 60
NL N D + L + ES+ ++ + L EK + ++K N K G+ DS E
Sbjct: 601 NLENKEDKKELKDDESVGAKTNNETSLEEKREQTQKGHDNSINSKIVDNKGGNADSNKEK 660
Query: 61 LRNVVEELEEREMKLQSELLEYYSLKEQVPVIEEFQR-------QLRIKSVEIDMLHMTI 113
+V + + M+ + + +K+ E+ + + K +E
Sbjct: 661 EVHVGDSTNDNNMESKEDTKSEVEVKKNDGSSEKGEEGKENNKDSMEDKKLENKESQTDS 720
Query: 114 KSLQEENNKLQEELIHEASAKRELEV-ARNKIKELQRQIKIIANQTKGQLLLLKQKVSGL 172
K + ++K +E I+ +K + V A+ K KE + K N+ + + ++ V G
Sbjct: 721 KDDKSVDDKQEEAQIYGGESKDDKSVEAKGKKKESKENKKTKTNENR--VRNKEENVQGN 778
Query: 173 QAKEEEVVKKDAEIENNLKTVNDFEIEKKLKTVNDLEIEAVGLKRRNKELQHEKRELTVK 232
+ + E+V K + + + K+V + +K T N E + + ++ + K +V+
Sbjct: 779 KKESEKVEKGEKKESKDAKSVETKDNKKLSSTENRDEAKERSGEDNKEDKEESKDYQSVE 838
Query: 233 LNAAESRITELSSVTENEMIADAKSETGRLRHAN--EDLQKQVEGLQMNRFSEVEELV-Y 289
++V E D K + AN E ++K+ E +Q N S +E+ +
Sbjct: 839 AKEKNENGGVDTNVGNKEDSKDLKDDRSVEVKANKEESMKKKREEVQRNDKSSTKEVRDF 898
Query: 290 LRWVNACLRY---ELRNYKAPSGKSLARDLN-NSFNPKSQEKAKQLMLEYAGSERGHGDT 345
++ ++ E YK K ++ N ++ N S++K K D
Sbjct: 899 ANNMDIDVQKGSGESVKYKKDEKKEGNKEENKDTINTSSKQKGK--------------DK 944
Query: 346 DIESNFSHDHSSPGSEDLDNAYINSPTYKYSNLSKKTSLIQ--KLKKWNKNNDSSAFSSP 403
+ S + + E+ Y+N+ K + K+T+ + KLK+ NK+N S
Sbjct: 945 KKKKKESKNSNMKKKEEDKKEYVNNELKKQEDNKKETTKSENSKLKEENKDNKEKKESED 1004
Query: 404 ARSLSIGSPCRVSTSYRPRNTLESLMLRNAHDAVAITTFEQKEHKPTHSPETPFLPSLRK 463
+ S + R E +E+K+ K + K
Sbjct: 1005 SAS-------------KNREKKE---------------YEEKKSKTKEEAKK------EK 1030
Query: 464 VSSSDDILDSVSASFQLMSKSVDRSLDEKYPAYKDLHKLALAREKQLKEKAEKARGEKFG 523
S D + + + K + S D K A +E++ KEK E +
Sbjct: 1031 KKSQDKKREEKDSEERKSKKEKEESRDLK----------AKKKEEETKEKKESENHK--- 1077
Query: 524 DNSNLNTTKAEEERSTSLDSELTQVNEKACVSDGLNKQSEDVKD---FDNQTITEMKLFK 580
S K E E + S+ E + +K K+ ED KD ++Q + K K
Sbjct: 1078 --SKKKEDKKEHEDNKSMKKEEDKKEKKKHEESKSRKKEEDKKDMEKLEDQNSNKKKEDK 1135
Query: 581 IEKRPPRLPRPPPKPSD 597
EK+ + + K SD
Sbjct: 1136 NEKKKSQHVKLVKKESD 1152
Score = 45.8 bits (107), Expect = 1e-04
Identities = 116/612 (18%), Positives = 229/612 (36%), Gaps = 63/612 (10%)
Query: 119 ENNKLQEELIHEASAKRELEVARN-------KIKELQRQIKIIANQT-----KGQLLLLK 166
E++ +++ L + K E+E A+N K++E QR + N+T KG
Sbjct: 343 EDSDIEKNLESKEDVKSEVEAAKNAGSSMTGKLEEAQRNNGVSTNETMNSENKGSGESTN 402
Query: 167 QKVSGLQAKEEEVVKKDAEI--ENNLKTVNDFEIEKKLKTVNDLEIEAVGLKRRNKELQH 224
K+ +E+ K++ E ENN ++V +E K ++ E + K N+EL+
Sbjct: 403 DKMVNATTNDEDHKKENKEETHENNGESVKGENLENKAGNEESMKGENLENKVGNEELKG 462
Query: 225 EKRELTVKLNAAESRITELSSVTENEMIADAKSETGR-LRHANEDLQKQVEGLQMNRFSE 283
N + S NE+ + ++ G + E + + + +
Sbjct: 463 NASVEAKTNNESSKEEKREESQRSNEVYMNKETTKGENVNIQGESIGDSTKDNSLENKED 522
Query: 284 VEELVYLRWVNACLRYELRNYKAPSGKSLARDLNNSFNPKSQEKAKQ-LMLEYAGSERGH 342
V+ V + E R+ +A ++ + N N + E+ K +E ++ H
Sbjct: 523 VKPKVDANESDGNSTKE-RHQEAQVNNGVSTEDKNLDNIGADEQKKNDKSVEVTTNDGDH 581
Query: 343 GDTDIESNFSHDHSSPGSEDLDNAYINSPTYKYSNLSKKT----SLIQKLKKWNKNNDSS 398
E ++ S +E+L+N ++ KT SL +K ++ K +D+S
Sbjct: 582 TKEKREETQGNNGESVKNENLENKEDKKELKDDESVGAKTNNETSLEEKREQTQKGHDNS 641
Query: 399 ------------AFSSPARSLSIGSPCRVSTSYRPRNTLESLMLRNAHDAVAITTFEQKE 446
A S+ + + +G + +T + ++ +D + E KE
Sbjct: 642 INSKIVDNKGGNADSNKEKEVHVGDSTNDNNMESKEDTKSEVEVKK-NDGSSEKGEEGKE 700
Query: 447 HKPTHSPETPFLPSLRKVSSSDDILDSVSASFQLMSKSVDRSLDEKYPAYKDLHKLALAR 506
+ + +K+ + + DS D+S+D+K + +
Sbjct: 701 NNKDSMED-------KKLENKESQTDSKD----------DKSVDDKQ------EEAQIYG 737
Query: 507 EKQLKEKAEKARGEKFGDNSNLNTTKAEEERSTSLDSELTQVNEKAC--VSDGLNKQSED 564
+ +K+ +A+G+K N TK E R + + E Q N+K V G K+S+D
Sbjct: 738 GESKDDKSVEAKGKKKESKEN-KKTKTNENRVRNKE-ENVQGNKKESEKVEKGEKKESKD 795
Query: 565 VKDFDNQTITEMKLFKIEKRPPRLPRPPPKPSDGAPVSNSLNEIPYAPSVPSPPPPPGSL 624
K + T KL E R R + S + +
Sbjct: 796 AKSVE--TKDNKKLSSTENRDEAKERSGEDNKEDKEESKDYQSVEAKEKNENGGVDTNVG 853
Query: 625 PRGAVGDDKVKRAPELVEFYQSLMKREAKKDASLLTSSTSNAADTRSNVIAEIENRSSFL 684
+ D K R+ E+ + MK++ ++ SST D +N+ +++ S
Sbjct: 854 NKEDSKDLKDDRSVEVKANKEESMKKKREEVQRNDKSSTKEVRDFANNMDIDVQKGSGES 913
Query: 685 LAVKADVETQGD 696
+ K D + +G+
Sbjct: 914 VKYKKDEKKEGN 925
Score = 40.0 bits (92), Expect = 0.006
Identities = 97/506 (19%), Positives = 198/506 (38%), Gaps = 80/506 (15%)
Query: 113 IKSLQEENNKLQEELIHEASAKRELEVARNKIKELQRQIKIIANQTKGQLLLLKQKV-SG 171
++ L+E NN ++ E + + V N KE++ Q + I + + L K+ V S
Sbjct: 246 VEDLKEGNNVVENGETKENNGEN---VESNNEKEVEGQGESIGDSAIEKNLESKEDVKSE 302
Query: 172 LQAKEEE------------------VVKKDAEIENNLKTVNDFEIEKKLKTVNDLEIEAV 213
++A + + + + E+E +++ D +IEK L++ D++ E
Sbjct: 303 VEAAKNDGSSMTENLGEAQGNNGVSTIDNEKEVEGQGESIEDSDIEKNLESKEDVKSEVE 362
Query: 214 GLKRRNK----ELQHEKRELTVKLN---------AAESRITELSSVTENEMIADAKSETG 260
K +L+ +R V N + ES ++ + T N+ D K E
Sbjct: 363 AAKNAGSSMTGKLEEAQRNNGVSTNETMNSENKGSGESTNDKMVNATTND--EDHKKENK 420
Query: 261 RLRHANEDLQKQVEGLQMNRFSEVEELVYLRWVNACLRYELRNYKAPSGKSLARDLNNSF 320
H N + E L+ N+ E + N EL+ + K+ N S
Sbjct: 421 EETHENNGESVKGENLE-NKAGNEESMKGENLENKVGNEELKGNASVEAKT----NNESS 475
Query: 321 NPKSQEKAKQLMLEYAGSERGHGDTDIESNFSHDHSSPGSEDLDNAYINSPTYK---YSN 377
+ +E++++ Y E G+ N + S G DN+ N K +N
Sbjct: 476 KEEKREESQRSNEVYMNKETTKGE-----NVNIQGESIGDSTKDNSLENKEDVKPKVDAN 530
Query: 378 LSKKTSLIQKLKKWNKNNDSSAFSSPARSLSIGSPCRVSTSYRPRNTLESLMLRNAHDAV 437
S S ++ ++ NN S+ ++L + + +N + N D
Sbjct: 531 ESDGNSTKERHQEAQVNN---GVSTEDKNLD-----NIGADEQKKNDKSVEVTTNDGD-- 580
Query: 438 AITTFEQKEHKPTHSPETPFLPSLRKVSSSDDILDSVSASFQLMSKSVDRSLDEKYPAYK 497
T E++E ++ E+ +L ++ D S + +++ SL+EK +
Sbjct: 581 --HTKEKREETQGNNGESVKNENLENKEDKKELKDDESVGAKTNNET---SLEEKREQTQ 635
Query: 498 DLHKLALAREKQLKEKAEKARGEK-----FGDNSNLNTTKAEEERSTSLDSELTQVNEKA 552
H ++ K + K A K GD++N N +++E+ + ++ +
Sbjct: 636 KGHDNSI-NSKIVDNKGGNADSNKEKEVHVGDSTNDNNMESKEDTKSEVEVK-------- 686
Query: 553 CVSDGLNKQSEDVKDFDNQTITEMKL 578
+DG +++ E+ K+ + ++ + KL
Sbjct: 687 -KNDGSSEKGEEGKENNKDSMEDKKL 711
>At4g31570 putative protein
Length = 2712
Score = 58.5 bits (140), Expect = 2e-08
Identities = 62/250 (24%), Positives = 117/250 (46%), Gaps = 39/250 (15%)
Query: 56 SEIEWLRNVVEELEEREMKL---QSELLEYYSLKEQVPVIEEFQRQLRIKSVEIDMLHMT 112
S +E +R++V L +L +E LE KE +I QR+L K ++
Sbjct: 2246 SSMESVRSMVNRLSSAVKELVVANAETLERNE-KEMKVIIANLQRELHEKDIQ------- 2297
Query: 113 IKSLQEENNKLQEELIHE-----ASAK---RELEVARNKIKELQRQIKIIANQTKGQLLL 164
NN+ EL+ + A AK +L+ A +++++Q Q+ I+ +
Sbjct: 2298 -------NNRTCNELVGQVKEAQAGAKIFAEDLQSASARMRDMQDQLGILVRERDSMKER 2350
Query: 165 LKQKVSGLQAKEEEVVKKDAEIENNLKTVNDFEIEKKLKTVNDLEIEAVGLKRRNKELQH 224
+K+ ++G QA E+ +K + ++L D EIE ++ +++ E + LK R EL+
Sbjct: 2351 VKELLAG-QASHSELQEKVTSL-SDLLAAKDLEIEALMQALDEEESQMEDLKLRVTELEQ 2408
Query: 225 EKRELTVKLNAAES---RITELSSVTENEMIADAKSETGRLRHANEDLQKQVEGLQMNRF 281
E ++ + L AE+ +I++ S+T ++ L H +E+L ++E LQ
Sbjct: 2409 EVQQKNLDLQKAEASRGKISKKLSITVDKF--------DELHHLSENLLAEIEKLQQQVQ 2460
Query: 282 SEVEELVYLR 291
E+ +LR
Sbjct: 2461 DRDTEVSFLR 2470
Score = 42.4 bits (98), Expect = 0.001
Identities = 46/194 (23%), Positives = 88/194 (44%), Gaps = 18/194 (9%)
Query: 105 EIDMLHMTIKSLQEENNKLQEELIHEASAKRELEVARNKIKELQRQIKIIANQTKGQLLL 164
E D+ +SL EN L +++I ++ E ++E K+ KG+ L+
Sbjct: 1822 ERDLYMAKQQSLVAENEALDKKIIELQEFLKQEEQKSASVRE-----KLNVAVRKGKALV 1876
Query: 165 -----LKQKVSGLQAK----EEEVVKKDAEIENNLKTVNDFEIEKKLKTVNDLEIEAVGL 215
LKQ + + A+ + E++K+D ++ N K E+E V LE E L
Sbjct: 1877 QQRDSLKQTIEEVNAELGRLKSEIIKRDEKLLENEKKFR--ELESYSVRVESLESECQLL 1934
Query: 216 KRRNKELQHEKRELTVKLNAAESRITELSSV--TENEMIADAKSETGRLRHANEDLQKQV 273
K ++E ++ +E + +N ++ +S + T + + A+ E+ + R A E L ++
Sbjct: 1935 KIHSQETEYLLQERSGDINDPVMKLQRISQLFQTMSTTVTSAEQESRKSRRAAELLLAEL 1994
Query: 274 EGLQMNRFSEVEEL 287
+Q S E+L
Sbjct: 1995 NEVQETNDSLQEDL 2008
Score = 42.0 bits (97), Expect = 0.002
Identities = 58/220 (26%), Positives = 103/220 (46%), Gaps = 35/220 (15%)
Query: 13 DEILSEFESLLSEQTDFPLLSEKTDDSKKHGGNETE-MAKNGSDSEIEWLRNVVEELEER 71
+ +L +LSE+ + + +K N+ E M + DS LR +VE++E
Sbjct: 1203 ENLLEAVRKILSERLELQSVIDKLQSDLSSKSNDMEEMTQRSLDSTS--LRELVEKVEGL 1260
Query: 72 EMKLQSELLEYYSLKEQVPVIEEFQRQLRIKSVEIDML-HMTIKSLQEENNKL---QEEL 127
++L+S ++ + S QV E QL K +EI+ L ++ K L+ + N+L +E L
Sbjct: 1261 -LELESGVI-FESPSSQV---EFLVSQLVQKFIEIEELANLLRKQLEAKGNELMEIEESL 1315
Query: 128 IHE----ASAKRELEVARNKIKELQRQIKIIAN---QTKGQLLLLKQKVS-------GLQ 173
+H A + L A + ++ +++ +N Q++ +LL ++K+S GL
Sbjct: 1316 LHHKTKIAGLRESLTQAEESLVAVRSELQDKSNELEQSEQRLLSTREKLSIAVTKGKGLI 1375
Query: 174 AKEEEVVKKDAEI---------ENNLKTVNDFEIEKKLKT 204
+ + V + AE E N K E+EKKLKT
Sbjct: 1376 VQRDNVKQSLAEASAKLQKCSEELNSKDARLVEVEKKLKT 1415
Score = 37.4 bits (85), Expect = 0.039
Identities = 79/328 (24%), Positives = 123/328 (37%), Gaps = 47/328 (14%)
Query: 69 EEREMKLQSELLEYYSLKEQVPVIEEFQRQLRIKSVEIDMLHMTIKSLQEENNKLQEELI 128
EE + E + + L EQ + EF Q EI L + ++ E N+ L EEL
Sbjct: 432 EELFVSSTMEDILHVQLTEQSHLQIEFDHQHNQFVAEISQLRASYSAVTERNDSLAEEL- 490
Query: 129 HEASAKRELEVARNKIKELQRQIKIIANQTKGQLLLLKQKVSGLQAKEEEVVKKDAEIEN 188
E +K + N + QLL + +V AK E+ +E
Sbjct: 491 SECQSKLYAATSSN-------------TNLENQLLATEAQVEDFTAKMNEL---QLSLEK 534
Query: 189 NLKTVNDFEIEKKLKTVNDLEIEAVGLKRRNKELQHEKRELTVKLNAAESRITELSSVTE 248
+L ++ E K K +N L++E L + EK+EL + + I LSS
Sbjct: 535 SLLDLS----ETKEKFIN-LQVENDTLVAVISSMNDEKKELIEEKESKNYEIKHLSS--- 586
Query: 249 NEMIADAKSETGRLRHANEDLQKQVEGLQMNRFSEVEELVYLRWVNACLRYELRNYKAPS 308
+ + K+ L+ E + + L + VEE L L+ EL N K
Sbjct: 587 --ELCNCKNLAAILKAEVEQFENTIGPLTDEKIHLVEEKYSLLGEAEKLQEELANCKT-- 642
Query: 309 GKSLARDLNNSFNPKSQEKAKQLMLEYAGSERGHGDTDIESNFSHDHSSPGSEDLD-NAY 367
L N +E L + T E N H L+ +A+
Sbjct: 643 ----VVTLQEVENSNMKETLSLLTRQ---------QTMFEENNIHLREENEKAHLELSAH 689
Query: 368 INSPTY---KYSNLSKKTSLI-QKLKKW 391
+ S TY +YSNL + +L+ KL K+
Sbjct: 690 LISETYLLSEYSNLKEGYTLLNNKLLKF 717
Score = 36.2 bits (82), Expect = 0.087
Identities = 37/146 (25%), Positives = 60/146 (40%), Gaps = 14/146 (9%)
Query: 117 QEENNKLQEELIHEASAKRELEVARNKIKELQRQIKIIANQTKGQLLLLKQKVSGLQAKE 176
+ EN +L ++L EA + V ++ L Q+ + A + Q L + +
Sbjct: 897 ESENAQLTDDL-SEADQFVSVNVQIRNLRGLLDQLLLNARKAGIQFNQLNDDRTSTNQRL 955
Query: 177 EEVVKKDAEIENNLKTVNDFEIEKKLKTV-------------NDLEIEAVGLKRRNKELQ 223
EE+ + A ++++ + IE K+ +DLE+ LK RN +
Sbjct: 956 EELNVEFASHQDHINVLEADTIESKVSFEALKHYSYELQHKNHDLELLCDSLKLRNDNIS 1015
Query: 224 HEKRELTVKLNAAESRITELSSVTEN 249
E EL KLN RI EL EN
Sbjct: 1016 VENTELNKKLNYCSLRIDELEIQLEN 1041
Score = 33.1 bits (74), Expect = 0.73
Identities = 53/239 (22%), Positives = 97/239 (40%), Gaps = 63/239 (26%)
Query: 66 EELEEREMKLQSELL-------EYYSLKEQVPVIE----EFQRQLRIKSVEIDMLHMTIK 114
EE E+ ++L + L+ EY +LKE ++ +FQ + E D L +
Sbjct: 677 EENEKAHLELSAHLISETYLLSEYSNLKEGYTLLNNKLLKFQGEKEHLVEENDKLTQELL 736
Query: 115 SLQEENNKLQEELIHEASAKRELEVARNKIKE---------LQRQIKIIANQTKGQLLLL 165
+LQE + ++EE H RE +K+ E + + +++ N + L+
Sbjct: 737 TLQEHMSTVEEERTHLEVELREAIARLDKLAEENTSLTSSIMVEKARMVDNGSADVSGLI 796
Query: 166 KQKVSGLQAKEEEV-VKKDAE--IEN----NLKTVNDF---------EIEKKLKTVNDLE 209
Q++S + E+ V K + +EN NL+ V ++ +EK K V +LE
Sbjct: 797 NQEISEKLGRSSEIGVSKQSASFLENTQYTNLEEVREYTSEFSALMKNLEKGEKMVQNLE 856
Query: 210 ---------------------------IEAVGLKRRNKELQHEKRELTVKLNAAESRIT 241
I+A KR+ +E + E +LT L+ A+ ++
Sbjct: 857 EAIKQILTDSSVSKSSDKGATPAVSKLIQAFESKRKPEEPESENAQLTDDLSEADQFVS 915
>At5g52280 hyaluronan mediated motility receptor-like protein
Length = 853
Score = 58.2 bits (139), Expect = 2e-08
Identities = 80/368 (21%), Positives = 152/368 (40%), Gaps = 68/368 (18%)
Query: 20 ESLLSEQTDFPLLSEKTDDSKKHGGNETEMAKNGSDSEIEWLRNVVEEL----------- 68
E L + + L+ +++KK E + ++EI+ L+ +E+L
Sbjct: 391 EMLEQKNNEISSLNSLLEEAKKL---EEHKGMDSGNNEIDTLKQQIEDLDWELDSYKKKN 447
Query: 69 EEREMKLQSELLEYYSLKEQ------------------------VPVIEEFQRQLRI--- 101
EE+E+ L EY SLKE+ +I+E + Q+ I
Sbjct: 448 EEQEILLDELTQEYESLKEENYKNVSSKLEQQECSNAEDEYLDSKDIIDELKSQIEILEG 507
Query: 102 ----KSVEIDMLHMTIKSLQEENNKLQEELIHEASAKRE--LEVARNKIKELQRQIK--- 152
+S+E +T+ L+ + +L++EL +A A E + R K ++ QR IK
Sbjct: 508 KLKQQSLEYSECLITVNELESQVKELKKELEDQAQAYDEDIDTMMREKTEQEQRAIKAEE 567
Query: 153 ----------IIANQTKGQLLLLKQKVSGLQAKEEEVVKKDAEIENNLKTVNDFEIEKKL 202
I A + + + L ++ ++ E + KK NNL+ N E +
Sbjct: 568 NLRKTRWNNAITAERLQEKCKRLSLEMESKLSEHENLTKKTLAEANNLRLQNKTLEEMQE 627
Query: 203 KTVNDLEIEAVGLKRRNKELQHEKRELTVKLNAAESRITELSSVTENEMIADAKSE--TG 260
KT ++ E K + K ++ + + L++K+ ES + +L+ + + A ++E
Sbjct: 628 KTHTEITQE----KEQRKHVEEKNKALSMKVQMLESEVLKLTKLRDESSAAATETEKIIQ 683
Query: 261 RLRHANEDLQKQVEGLQMNRFSEVEELVYLRWVNACLRYELRNYKAP-SGKSLA-RDLNN 318
R ++ ++++ + + +EL + N LRN K G SL +L N
Sbjct: 684 EWRKERDEFERKLSLAKEVAKTAQKELTLTKSSNDDKETRLRNLKTEVEGLSLQYSELQN 743
Query: 319 SFNPKSQE 326
SF + E
Sbjct: 744 SFVQEKME 751
Score = 46.2 bits (108), Expect = 8e-05
Identities = 71/350 (20%), Positives = 142/350 (40%), Gaps = 33/350 (9%)
Query: 8 ANDFDDEILSEFESLLS-------EQTDFPLLSEKTDDSKKHGGNETEMAKNGSDSEIEW 60
+ D DE+ S+ E L E ++ + + + K E E D +I+
Sbjct: 491 SKDIIDELKSQIEILEGKLKQQSLEYSECLITVNELESQVKELKKELEDQAQAYDEDIDT 550
Query: 61 LRNVVEELEEREMKLQSELLEYYSLKEQVPVIEEFQRQLRIKSVEIDML-----HMTIKS 115
+ E E+R +K + L + + E Q + + S+E++ ++T K+
Sbjct: 551 MMREKTEQEQRAIKAEENLRK--TRWNNAITAERLQEKCKRLSLEMESKLSEHENLTKKT 608
Query: 116 LQEENN-KLQEELIHEASAKRELEVARNKIKELQRQIKIIANQTKGQLLLLKQ-KVSGLQ 173
L E NN +LQ + + E K E+ + K + + K A K Q+L + K++ L+
Sbjct: 609 LAEANNLRLQNKTLEEMQEKTHTEITQEKEQRKHVEEKNKALSMKVQMLESEVLKLTKLR 668
Query: 174 AKEEEVVKKDAEIENNLKTVNDFEIEKKLKTVNDL----EIEAVGLKRRNKELQHEKREL 229
+ + +I + D E E+KL ++ + E K N + + R L
Sbjct: 669 DESSAAATETEKIIQEWRKERD-EFERKLSLAKEVAKTAQKELTLTKSSNDDKETRLRNL 727
Query: 230 TVKLNAAESRITELSSVTENEMIADAKSETGRLRHANEDLQKQVEGLQMNRFSEVEELVY 289
++ + +EL +N + + + N++L+KQV L+++ + EE+
Sbjct: 728 KTEVEGLSLQYSEL----QNSFVQE--------KMENDELRKQVSNLKVDIRRKEEEMTK 775
Query: 290 LRWVNACLRYELRNYKAPSGKSLARDLNNSFNPKSQEKAKQLMLEYAGSE 339
+ R + +K + L+ +L N S + + +E SE
Sbjct: 776 ILDARMEARSQENGHKEENLSKLSDELAYCKNKNSSMERELKEMEERYSE 825
Score = 33.9 bits (76), Expect = 0.43
Identities = 62/247 (25%), Positives = 103/247 (41%), Gaps = 42/247 (17%)
Query: 183 DAEIENNLKTVNDFE-----IEKKLKTVNDLEIEAVGLKRRNKELQHEKRELTVKLNAAE 237
++ IE+ N F+ + + + L++E L+R+++ + EK+ L +
Sbjct: 239 ESYIESRNSPENSFQRGFSSVTESSDPIERLKMELEALRRQSELSELEKQSLRKQAIKES 298
Query: 238 SRITELS------------SVTENEMI--------ADAKSETGRLRHANEDLQKQVEGLQ 277
RI ELS ++ E E + ADA+S RLR +ED +E ++
Sbjct: 299 KRIQELSKEVSCLKGERDGAMEECEKLRLQNSRDEADAES---RLRCISEDSSNMIEEIR 355
Query: 278 MNRFSEVEELVYLRWVNACLRYEL-RNYKAPSGKSLA-RDLNNSFNPKSQE-KAKQLMLE 334
+EL + + + L+ +L R ++ S LA RDLN K+ E + +LE
Sbjct: 356 -------DELSCEKDLTSNLKLQLQRTQESNSNLILAVRDLNEMLEQKNNEISSLNSLLE 408
Query: 335 YAGSERGHGDTDIESNFSHDHSSPGSEDLD---NAYINSPTYKYSNLSKKTSLIQKLKKW 391
A H D N D EDLD ++Y + L + T + LK+
Sbjct: 409 EAKKLEEHKGMD-SGNNEIDTLKQQIEDLDWELDSYKKKNEEQEILLDELTQEYESLKEE 467
Query: 392 NKNNDSS 398
N N SS
Sbjct: 468 NYKNVSS 474
>At1g05320 unknown protein
Length = 841
Score = 57.0 bits (136), Expect = 5e-08
Identities = 63/291 (21%), Positives = 128/291 (43%), Gaps = 48/291 (16%)
Query: 16 LSEFESLLSEQTDFPLLSEKTDDSKKHGGNETEMAKNGSDSEIEWLRNVVEELEEREMKL 75
L E+++ E L + D++K E + N +S IE L +L E +KL
Sbjct: 448 LKSHENVIEEHKRQVLEASGVADTRKVEVEEALLKLNTLESTIEELEKENGDLAEVNIKL 507
Query: 76 QSELL-------------------EYYSLKEQVPVIEEFQRQLRIKSVEIDMLHMTIKSL 116
+L +Y KE IE+ +QL + E + L I SL
Sbjct: 508 NQKLANQGSETDDFQAKLSVLEAEKYQQAKELQITIEDLTKQL---TSERERLRSQISSL 564
Query: 117 QEENNKLQEELIHEASAKR------ELEVARNKIKELQRQIKIIANQTKGQLLLLKQKVS 170
+EE N++ E I++++ +L+V ++K ++ QI+ ++ + +L+ K
Sbjct: 565 EEEKNQVNE--IYQSTKNELVKLQAQLQVDKSKSDDMVSQIEKLSALV-AEKSVLESKFE 621
Query: 171 GLQAKEEEVVKKDAEIENNLKT----VNDFEI--EKKLKTVNDLEIEAVGLKRRNKELQH 224
++ +E V+K AE+ + L+ +D ++ EK ++ +L+ + + + L H
Sbjct: 622 QVEIHLKEEVEKVAELTSKLQEHKHKASDRDVLEEKAIQLHKELQASHTAISEQKEALSH 681
Query: 225 EKRELTVKLNAAESRI-----------TELSSVTENEMIADAKSETGRLRH 264
+ EL L ++ + ++L+ + + +ADAKS+ ++H
Sbjct: 682 KHSELEATLKKSQEELDAKKSVIVHLESKLNELEQKVKLADAKSKVSHIKH 732
Score = 43.9 bits (102), Expect = 4e-04
Identities = 126/619 (20%), Positives = 245/619 (39%), Gaps = 90/619 (14%)
Query: 31 LLSEKTDDSKKHGGNETEMAKNGSDSEIEWLRNVVEELEEREMKLQSELLEYYSLKEQVP 90
L +EK ++ +K +E E SD ++ + + L+ +L L E+V
Sbjct: 124 LEAEKLEELQKQSASELEEKLKISDERYSKTDALLSQALSQNSVLEQKLKSLEELSEKV- 182
Query: 91 VIEEFQRQLRIKSVEIDMLHMTIKSLQEENNKLQEELIHEASAKRELE------------ 138
E + L + E + ++ QE+ +KL+ L ++ ELE
Sbjct: 183 --SELKSALIVAEEEGKKSSIQMQEYQEKVSKLESSLNQSSARNSELEEDLRIALQKGAE 240
Query: 139 ------VARNKIKELQ-----RQIKI-IANQTKGQLLLLKQKVSGLQAKEEEVVKKDAEI 186
V+ + ELQ Q+K+ A + L ++ K S L+A ++K+ ++
Sbjct: 241 HEDIGNVSTKRSVELQGLFQTSQLKLEKAEEKLKDLEAIQVKNSSLEATLSVAMEKERDL 300
Query: 187 ENNLKTV------NDFEIEKKLKTVNDLEIEAVGLKRRNKELQHEKRELTVKLNAAESRI 240
NL V ++ +EK+ + +++ ++ L+ +K H + ++ + SR
Sbjct: 301 SENLNAVMEKLKSSEERLEKQAREIDEATTRSIELEALHK---HSELKVQKTMEDFSSRD 357
Query: 241 TELSSVTENEMIADAKSET--GRLRHA-NEDLQKQVEGLQMNRFSEVEELVYLRWVNACL 297
TE S+TE + K G+L A + L Q E +++ S EL L N L
Sbjct: 358 TEAKSLTEKSKDLEEKIRVYEGKLAEACGQSLSLQEE---LDQSSAENEL--LADTNNQL 412
Query: 298 R---YELRNYKAPSGKSLARDLNNSFNPKSQEKAKQLMLEYAGSERGHGDTDIESNFSHD 354
+ EL Y ++ LN + +AK L+ + E IE +
Sbjct: 413 KIKIQELEGYLDSEKETAIEKLN-----QKDTEAKDLITKLKSHE-----NVIEEHKRQV 462
Query: 355 HSSPGSEDLDNAYINSPTYKYSNLSKKTSLIQKLKKWNKN-------------NDSSAFS 401
+ G D + K + L S I++L+K N + N S
Sbjct: 463 LEASGVADTRKVEVEEALLKLNTLE---STIEELEKENGDLAEVNIKLNQKLANQGSETD 519
Query: 402 SPARSLSIGSPCRVSTSYRPRNTLESL--MLRNAHDAV--AITTFEQKEHK-----PTHS 452
LS+ + + + T+E L L + + + I++ E+++++ +
Sbjct: 520 DFQAKLSVLEAEKYQQAKELQITIEDLTKQLTSERERLRSQISSLEEEKNQVNEIYQSTK 579
Query: 453 PETPFLPSLRKV--SSSDDILDSVSASFQLMS-KSVDRSLDEKYPAY--KDLHKLALARE 507
E L + +V S SDD++ + L++ KSV S E+ + +++ K+A
Sbjct: 580 NELVKLQAQLQVDKSKSDDMVSQIEKLSALVAEKSVLESKFEQVEIHLKEEVEKVAELTS 639
Query: 508 KQLKEKAEKARGEKFGDNSNLNTTKAEEERSTSLDSELTQVNEKACVSDGLNKQSEDVKD 567
K L+E KA + + K + T++ + ++ K + K+S++ D
Sbjct: 640 K-LQEHKHKASDRDVLEEKAIQLHKELQASHTAISEQKEALSHKHSELEATLKKSQEELD 698
Query: 568 FDNQTIT--EMKLFKIEKR 584
I E KL ++E++
Sbjct: 699 AKKSVIVHLESKLNELEQK 717
Score = 43.9 bits (102), Expect = 4e-04
Identities = 94/459 (20%), Positives = 192/459 (41%), Gaps = 67/459 (14%)
Query: 136 ELEVARNKIKELQRQIKIIANQTKGQLLLLKQKVSGLQAK----EEEVVKKDAEIENNLK 191
ELE +R K+ EL+ +I+I A + + L KQ S L+ K +E K DA + L
Sbjct: 104 ELENSRKKMIELEDRIRISALEAEKLEELQKQSASELEEKLKISDERYSKTDALLSQALS 163
Query: 192 TVNDFEIEKKLKTVNDLEIEAVGLKRRNKELQHEKRELTVKLNAAESRITEL------SS 245
+ +E+KLK++ +L + LK + E ++ ++++ + ++++L SS
Sbjct: 164 --QNSVLEQKLKSLEELSEKVSELKSALIVAEEEGKKSSIQMQEYQEKVSKLESSLNQSS 221
Query: 246 VTENEMIADAK------SETGRLRHANEDLQKQVEGL----QMNRFSEVEELVYLRWVNA 295
+E+ D + +E + + + +++GL Q+ E+L L +
Sbjct: 222 ARNSELEEDLRIALQKGAEHEDIGNVSTKRSVELQGLFQTSQLKLEKAEEKLKDLEAI-- 279
Query: 296 CLRYELRNYKAPSGKSLA----RDLNNSFNPKSQE-KAKQLMLEYAGSERGHGDT-DIES 349
+++N + S+A RDL+ + N ++ K+ + LE E T IE
Sbjct: 280 ----QVKNSSLEATLSVAMEKERDLSENLNAVMEKLKSSEERLEKQAREIDEATTRSIEL 335
Query: 350 NFSHDHSSPGSEDLDNAYINSPTYKYSNLSKKTSLIQKLKKWNKNNDSSAFSSPARSLSI 409
H HS + + + T S K L +K++ + + + +SLS+
Sbjct: 336 EALHKHSELKVQKTMEDFSSRDTEAKSLTEKSKDLEEKIRVY----EGKLAEACGQSLSL 391
Query: 410 GSPCRVSTSYRPRNTLESLMLRNAHDAVAITTFEQKEHKPTHSPETPFLPSLRKVSSSDD 469
S++ E+ +L + ++ + I E + + + ET +K + + D
Sbjct: 392 QEELDQSSA-------ENELLADTNNQLKIKIQELEGYLDSEK-ETAIEKLNQKDTEAKD 443
Query: 470 ILDSVSASFQLMSKSVDRSLDEKYPAYKDLHKLALAREKQLKEKAEKARGEKFGDNSNLN 529
++ + KS + ++E + +A R+ +++E K LN
Sbjct: 444 LITKL--------KSHENVIEEHKRQVLEASGVADTRKVEVEEALLK-----------LN 484
Query: 530 TTKAEEERSTSLDSELTQVNEKACVSDGLNKQSEDVKDF 568
T ++ E + +L +VN K ++ L Q + DF
Sbjct: 485 TLESTIEELEKENGDLAEVNIK--LNQKLANQGSETDDF 521
>At1g67230 unknown protein
Length = 1132
Score = 55.5 bits (132), Expect = 1e-07
Identities = 74/326 (22%), Positives = 143/326 (43%), Gaps = 50/326 (15%)
Query: 14 EILSEFESLLSEQTDFPLLSEKTDDSKKHGGNETEMAKNGSDSEIEWLRNVVEELEEREM 73
E L E+E L E + S+ ++ NE++ ++ +ELEE +
Sbjct: 230 EDLREWERKLQEGEERVAKSQMIVKQREDRANESDKI----------IKQKGKELEEAQK 279
Query: 74 KLQSELLEYYSLKEQVPVIEEFQRQLRIKSVEIDMLHMTIKSLQEENNKLQEEL-IHEAS 132
K+ + L L++ V + L ++ E D+L +I++ E LQE+L E
Sbjct: 280 KIDAANLAVKKLEDDV---SSRIKDLALREQETDVLKKSIETKARELQALQEKLEAREKM 336
Query: 133 AKREL-EVARNKIKELQRQIKIIANQTKGQLL-LLKQKVSGLQAKEEEVVKKDAEIENNL 190
A ++L + + K+ QR+ ++ Q + + LK KV+ ++ +E E + ++
Sbjct: 337 AVQQLVDEHQAKLDSTQREFELEMEQKRKSIDDSLKSKVAEVEKREAEWKHMEEKVAKRE 396
Query: 191 KTVNDFEIEKKLKTVNDLEIEAVGLKRRNKELQHEKR-------------ELTVKLNAA- 236
+ + D ++EK + ND ++ G+ R K L+ E++ E+ + L A
Sbjct: 397 QAL-DRKLEKHKEKENDFDLRLKGISGREKALKSEEKALETEKKKLLEDKEIILNLKALV 455
Query: 237 -------ESRITELSS------VTENE------MIADAKSETGRLRHANEDLQKQVEGLQ 277
+++++E++ VTE E + + K + + R E LQK+ E L+
Sbjct: 456 EKVSGENQAQLSEINKEKDELRVTEEERSEYLRLQTELKEQIEKCRSQQELLQKEAEDLK 515
Query: 278 MNRFSEVEELVYLRWVNACLRYELRN 303
R S +E L A + EL+N
Sbjct: 516 AQRESFEKEWEELDERKAKIGNELKN 541
Score = 48.9 bits (115), Expect = 1e-05
Identities = 71/291 (24%), Positives = 124/291 (42%), Gaps = 50/291 (17%)
Query: 46 ETEMAKNGSDSEIEWLRNVVEELEEREMKLQSELLEYYSLKEQVPVIEEFQRQLRIKSVE 105
ET++ K +++ L+ + E+LE RE +L++ + K EF+ ++ K
Sbjct: 308 ETDVLKKSIETKARELQALQEKLEAREKMAVQQLVDEHQAKLD-STQREFELEMEQKRKS 366
Query: 106 IDMLHMTIKSLQEENNKLQEELIH--EASAKRELEVARN--KIKELQRQIKIIANQTKGQ 161
ID ++KS E K + E H E AKRE + R K KE + + G+
Sbjct: 367 IDD---SLKSKVAEVEKREAEWKHMEEKVAKREQALDRKLEKHKEKENDFDLRLKGISGR 423
Query: 162 LLLLKQKVSGLQAKEEEVVKKDAEIENNLKTVND----------FEIEKK---LKTVNDL 208
LK + L+ +++++++ D EI NLK + + EI K+ L+ +
Sbjct: 424 EKALKSEEKALETEKKKLLE-DKEIILNLKALVEKVSGENQAQLSEINKEKDELRVTEEE 482
Query: 209 EIEAVGLKRRNKE-----------LQHEKRELTVKLNAAESRITELSSV-----TENEMI 252
E + L+ KE LQ E +L + + E EL E + I
Sbjct: 483 RSEYLRLQTELKEQIEKCRSQQELLQKEAEDLKAQRESFEKEWEELDERKAKIGNELKNI 542
Query: 253 ADAKSETGRLRH------------ANEDLQKQVEGLQMNRFSEVEELVYLR 291
D K + R H ANE++++++E L++ + S E + Y R
Sbjct: 543 TDQKEKLERHIHLEEERLKKEKQAANENMERELETLEVAKASFAETMEYER 593
Score = 36.6 bits (83), Expect = 0.066
Identities = 87/524 (16%), Positives = 198/524 (37%), Gaps = 48/524 (9%)
Query: 46 ETEMAKNGSDSEIEWLRNVVEELEEREMKLQSELLEYYSLKEQVPVIEEFQRQLRIKSVE 105
E K +DS++ +V +EE+ ++++++L ++ ++ + + K+ E
Sbjct: 143 ENAEIKFTADSKLTEANALVRSVEEKSLEVEAKLR---AVDAKLAEVSRKSSDVERKAKE 199
Query: 106 IDMLHMTIKSLQEENNKLQEELIHEASAKRELEVARNKIKELQRQIKIIANQTKGQLLLL 165
++ +++ +E + + E EA+ ++ R ++E +R+++ + +++
Sbjct: 200 VEARESSLQ--RERFSYIAEREADEATLSKQ----REDLREWERKLQEGEERVAKSQMIV 253
Query: 166 KQKVSGLQAKEEEVVKKDAEIENNLKTVN---------DFEIEKKLKTVNDLEIEAVGLK 216
KQ+ ++ + +K E+E K ++ + ++ ++K + E E LK
Sbjct: 254 KQREDRANESDKIIKQKGKELEEAQKKIDAANLAVKKLEDDVSSRIKDLALREQETDVLK 313
Query: 217 RRNKELQHEKRELTVKLNAAESRITELSSVTENEMIADAKSETGRLRHANEDLQKQVEGL 276
+ + E + L KL A E + V E++ D+ L E +K ++
Sbjct: 314 KSIETKARELQALQEKLEAREKMAVQ-QLVDEHQAKLDSTQREFELE--MEQKRKSIDDS 370
Query: 277 QMNRFSEVEELVYLRWVNACLRYELRNYKAPSGKSLARDLNNSFNPKSQEKAKQLMLEYA 336
++ +EVE+ W + + A ++L R L K +EK L
Sbjct: 371 LKSKVAEVEKR-EAEWKH------MEEKVAKREQALDRKLE-----KHKEKENDFDLRLK 418
Query: 337 GSERGHGDTDIESNFSHDHSSPGSEDLDNAY-INSPTYKYSNLSKKTSLIQKLKKWNKNN 395
G E ED + + + K S ++ +L + NK
Sbjct: 419 GISGREKALKSEEKALETEKKKLLEDKEIILNLKALVEKVSGENQ-----AQLSEINKEK 473
Query: 396 DSSAFSSPARSLSIGSPCRVSTSYRPRNTLESLMLRNAHDAVAITTFEQKEHKPTHSPET 455
D + RS + + + + L+ + A D A +KE + +
Sbjct: 474 DELRVTEEERSEYLRLQTELKEQIEKCRSQQELLQKEAEDLKAQRESFEKEWEELDERKA 533
Query: 456 PFLPSLRKVSSSDDIL--------DSVSASFQLMSKSVDRSLDEKYPAYKDLHKLALARE 507
L+ ++ + L + + Q +++++R L+ A +
Sbjct: 534 KIGNELKNITDQKEKLERHIHLEEERLKKEKQAANENMERELETLEVAKASFAETMEYER 593
Query: 508 KQLKEKAEKARGEKFGDNSNLNTTKAEEERSTSLDSELTQVNEK 551
L +KAE R + D + K E + T L+ + ++ K
Sbjct: 594 SMLSKKAESERSQLLHD-IEMRKRKLESDMQTILEEKERELQAK 636
>At5g41790 myosin heavy chain-like protein
Length = 1305
Score = 54.3 bits (129), Expect = 3e-07
Identities = 124/620 (20%), Positives = 242/620 (39%), Gaps = 138/620 (22%)
Query: 14 EILSEFESLLSEQTDFPLLSEKTDDSKKHGGNETEMAKNGSDSEIEWLRNVVEELEEREM 73
E+ ++ ES + +D + ++ K ++ N ++E +N ++EL
Sbjct: 158 ELEAQLESSKQQVSDLSASLKAAEEENKAISSKNVETMN----KLEQTQNTIQELMAELG 213
Query: 74 KLQSELLEYYS-LKEQVPVIEEFQRQ--LRIKSVE---------IDMLHMTIKSLQEENN 121
KL+ E S L V V E QR + +K +E + L+ T+ + +EE
Sbjct: 214 KLKDSHREKESELSSLVEVHETHQRDSSIHVKELEEQVESSKKLVAELNQTLNNAEEEKK 273
Query: 122 KLQEELIHEASAKRELEVARNKIKEL----------------------------QRQIKI 153
L +++ A E++ A+N I+EL QR+
Sbjct: 274 VLSQKI---AELSNEIKEAQNTIQELVSESGQLKESHSVKDRDLFSLRDIHETHQRESST 330
Query: 154 IANQTKGQLLLLKQKVSGLQAKEEEVVKKDAEIENNLKTVNDFEIEKKLK----TVNDLE 209
++ + QL +Q++S L V KDAE EN + + EI KL+ T+ +L
Sbjct: 331 RVSELEAQLESSEQRISDLT-----VDLKDAEEENKAISSKNLEIMDKLEQAQNTIKELM 385
Query: 210 IEAVGLKRRNKELQHEKRELTVKLNAAESRITELSSVTEN-------------------- 249
E LK R+KE ++ EL+ + +A+ ++ ++ +N
Sbjct: 386 DELGELKDRHKE---KESELSSLVKSADQQVADMKQSLDNAEEEKKMLSQRILDISNEIQ 442
Query: 250 ---EMIADAKSETGRLRHANEDLQKQVEGLQ----------MNRFSEVE---ELVYLRWV 293
+ I + SE+ +L+ ++ ++++ GL+ R SE+E +L+ R V
Sbjct: 443 EAQKTIQEHMSESEQLKESHGVKERELTGLRDIHETHQRESSTRLSELETQLKLLEQRVV 502
Query: 294 NACLRYELRNYKAPSGKSLARDLNNSFNPKSQEKAKQLMLEYAGSERGHGDTDIE-SNFS 352
+ + S S+ ++ + ++Q K ++L+ E A S+ + E S+F
Sbjct: 503 DLSASLNAAEEEKKSLSSMILEITDELK-QAQSKVQELVTELAESKDTLTQKENELSSFV 561
Query: 353 HDHSSPGSEDLDNAYINSPTYKYSNLSKKT-SLIQKLKKWNKNNDSSAFSSPARSLSIGS 411
H A+ + + L + S +++K+ N+N +SS S I
Sbjct: 562 EVHE---------AHKRDSSSQVKELEARVESAEEQVKELNQNLNSSEEEKKILSQQISE 612
Query: 412 PCRVSTSYRPRNTLESLMLRNAHDAVAITTFEQKEHKPTHSPETPFLPSLRKVSSSDDIL 471
S+ ++ A + + E + K +H+ + L SLR DI
Sbjct: 613 M--------------SIKIKRAESTIQELSSESERLKGSHAEKDNELFSLR------DIH 652
Query: 472 DSVSASFQLMSKSVDRSLDEKYPAYKDLHKLALAREKQLKEKAEKARGEKFGDNSNLNTT 531
++ + ++ L+ R +L E + A E ++ ++ T
Sbjct: 653 ETHQRELSTQLRGLEAQLESSEH-----------RVLELSESLKAAEEESRTMSTKISET 701
Query: 532 KAEEERSTSLDSELTQVNEK 551
E ER+ + ELT + K
Sbjct: 702 SDELERTQIMVQELTADSSK 721
Score = 52.8 bits (125), Expect = 9e-07
Identities = 95/414 (22%), Positives = 180/414 (42%), Gaps = 54/414 (13%)
Query: 20 ESLLSEQTDFPLLSEKTDDSKKHGGNETEMAKNGSDSEIEWLRNVVEELEEREMKLQSEL 79
E L +++ LL+EK D + E E + E+E +R + +LE E+ ++ +
Sbjct: 724 EQLAEKESKLFLLTEK-DSKSQVQIKELEATVATLELELESVRARIIDLET-EIASKTTV 781
Query: 80 LEYYSL--KEQVPVIEEFQRQLRIKSVEIDMLHMTIKSLQEENNKLQEELIHEASAKR-E 136
+E +E V I E ++ + + E+ L ++ ++++ E L E R E
Sbjct: 782 VEQLEAQNREMVARISELEKTMEERGTELSALTQKLEDNDKQSSSSIETLTAEIDGLRAE 841
Query: 137 LEVARNKIKELQRQIKIIANQTKGQLLLLKQKVSGLQAK-----------EEEVVKKDAE 185
L+ + +E+++Q+ + + ++ L +V+GL+ + E ++ KK E
Sbjct: 842 LDSMSVQKEEVEKQMVCKSEEASVKIKRLDDEVNGLRQQVASLDSQRAELEIQLEKKSEE 901
Query: 186 IENNLKTVNDF--EIEKKLKT-------VN---------DLEIEAVGLKRR--NKELQHE 225
I L + + EI K+K +N +LE+E +G +R ++EL+ +
Sbjct: 902 ISEYLSQITNLKEEIINKVKVHESILEEINGLSEKIKGRELELETLGKQRSELDEELRTK 961
Query: 226 KRE---LTVKLNAAESRI---TELSSVTENEM--IADAKSETGRLRHANEDLQKQVEGLQ 277
K E + K+N A S I TEL + +NE+ + KSET A + +KQ +
Sbjct: 962 KEENVQMHDKINVASSEIMALTELINNLKNELDSLQVQKSET----EAELEREKQEKSEL 1017
Query: 278 MNRFSEVEELVYLRWVNACLRYELRNYKAPSGKSLARDLNNSFNPKSQE-KAKQLMLEYA 336
N+ ++V++ + V Y + L ++ + N + + K Q +LE
Sbjct: 1018 SNQITDVQKAL----VEQEAAYNTLEEEHKQINELFKETEATLNKVTVDYKEAQRLLEER 1073
Query: 337 GSERGHGDTDIESNFSHDHSSPGSEDLDNAYINSPTYKYSNLSKKTSLI-QKLK 389
G E D+ I + S ++ I + K SN+ K L QKL+
Sbjct: 1074 GKEVTSRDSTIGVHEETMESLRNELEMKGDEIETLMEKISNIEVKLRLSNQKLR 1127
Score = 50.4 bits (119), Expect = 4e-06
Identities = 117/608 (19%), Positives = 246/608 (40%), Gaps = 97/608 (15%)
Query: 18 EFESLLSEQ-TDFPLLSEKTDDSKKHGGNETEMAKNG---SDSEIEWLRNVVEELEEREM 73
E + +LS+Q ++ + ++ + + + +E+E K D+E+ LR++ E +RE+
Sbjct: 601 EEKKILSQQISEMSIKIKRAESTIQELSSESERLKGSHAEKDNELFSLRDI-HETHQREL 659
Query: 74 KLQSELLEYYSLKEQVPVIEEFQRQLRIKSVEIDMLHMTIKSLQEENNKLQEELIHEASA 133
Q LE + V+E L ++K+ +EE+ + ++ +
Sbjct: 660 STQLRGLEAQLESSEHRVLE---------------LSESLKAAEEESRTMSTKI---SET 701
Query: 134 KRELEVARNKIKELQRQIKIIANQTKGQLLLLKQKVSGLQAKEEEVVKKDAEIENNLKTV 193
ELE + ++EL +++ K QL + K+ L K+ + + E+E + T+
Sbjct: 702 SDELERTQIMVQELTAD----SSKLKEQLAEKESKLFLLTEKDSKSQVQIKELEATVATL 757
Query: 194 NDFEIEKKLKTVNDLEIEAVGLKRRNKELQHEKRELTVKLN----AAESRITELSSVTEN 249
+ E+E + DLE E ++L+ + RE+ +++ E R TELS++T+
Sbjct: 758 -ELELESVRARIIDLETEIASKTTVVEQLEAQNREMVARISELEKTMEERGTELSALTQK 816
Query: 250 EMIADAKSETGRLRHANEDLQKQVEGL--QMNRFSEVEELVYLRWVNACLRYELRNYKAP 307
D +S + + E L +++GL +++ S +E V + V K+
Sbjct: 817 LEDNDKQSSS-----SIETLTAEIDGLRAELDSMSVQKEEVEKQMV----------CKSE 861
Query: 308 SGKSLARDLNNSFNPKSQEKAKQLMLEYAGSERGHGDTDIESNFSHDHSSPGSE--DLDN 365
+ L++ N Q+ A S+R + +E S + S S+ +L
Sbjct: 862 EASVKIKRLDDEVNGLRQQVAS------LDSQRAELEIQLEKK-SEEISEYLSQITNLKE 914
Query: 366 AYINSPTYKYSNLSKKTSLIQKLK----------KWNKNNDSSAFSSPARSLSIGSPCRV 415
IN S L + L +K+K K D + ++ + V
Sbjct: 915 EIINKVKVHESILEEINGLSEKIKGRELELETLGKQRSELDEELRTKKEENVQMHDKINV 974
Query: 416 STS---------YRPRNTLESLMLRNAHDAVAITTFEQKEHKPTHSPETPFLPSLRKVSS 466
++S +N L+SL ++ + + E++E + T +L + +
Sbjct: 975 ASSEIMALTELINNLKNELDSLQVQKSETEAELER-EKQEKSELSNQITDVQKALVEQEA 1033
Query: 467 SDDILDSVSASFQLMSKSVDRSLDEKYPAYKDLHKLALAREKQL----------KEKAEK 516
+ + L+ + K + +L++ YK+ +L R K++ +E E
Sbjct: 1034 AYNTLEEEHKQINELFKETEATLNKVTVDYKEAQRLLEERGKEVTSRDSTIGVHEETMES 1093
Query: 517 ARGEKFGDNSNLNTTKAEEERSTSLDSELTQVNEKACVSDGLNKQ------SEDVKDFDN 570
R E + T E+ ++++ +L N+K V++ + + E+ K +
Sbjct: 1094 LRNELEMKGDEIETLM---EKISNIEVKLRLSNQKLRVTEQVLTEKEEAFRKEEAKHLEE 1150
Query: 571 QTITEMKL 578
Q + E L
Sbjct: 1151 QALLEKNL 1158
Score = 48.9 bits (115), Expect = 1e-05
Identities = 69/319 (21%), Positives = 134/319 (41%), Gaps = 22/319 (6%)
Query: 48 EMAKNGSDSEIEWLRNV-VEELEEREMKLQSELLEYYSLKEQVPVIEEFQRQLRIKSVEI 106
EM + + E E V + + ERE Q + LE + ++ ++ +F + L E
Sbjct: 38 EMKEKYKEKESEHSSLVELHKTHERESSSQVKELEAH-IESSEKLVADFTQSLNNAEEEK 96
Query: 107 DMLHMTIKSL----QEENNKLQEELIHEASAKRELEVARNKIKEL-------QRQIKIIA 155
+L I L QE N +QE + K V ++ L QR A
Sbjct: 97 KLLSQKIAELSNEIQEAQNTMQELMSESGQLKESHSVKERELFSLRDIHEIHQRDSSTRA 156
Query: 156 NQTKGQLLLLKQKVSGLQAKEEEVVKKDAEIEN-NLKTVNDFEIEKKLKTVNDLEIEAVG 214
++ + QL KQ+VS L A + +++ I + N++T+N ++E+ T+ +L E
Sbjct: 157 SELEAQLESSKQQVSDLSASLKAAEEENKAISSKNVETMN--KLEQTQNTIQELMAELGK 214
Query: 215 LKRRNKELQHEKRELTVKLNAAESRITELSSVTENEMIADAKSETGRLRHA--NEDLQKQ 272
LK ++E + E L V+++ R + + E + +K L N + +K+
Sbjct: 215 LKDSHREKESELSSL-VEVHETHQRDSSIHVKELEEQVESSKKLVAELNQTLNNAEEEKK 273
Query: 273 VEGLQMNRFS-EVEELVYLRWVNACLRYELRNYKAPSGKSL--ARDLNNSFNPKSQEKAK 329
V ++ S E++E +L+ + + L RD++ + +S +
Sbjct: 274 VLSQKIAELSNEIKEAQNTIQELVSESGQLKESHSVKDRDLFSLRDIHETHQRESSTRVS 333
Query: 330 QLMLEYAGSERGHGDTDIE 348
+L + SE+ D ++
Sbjct: 334 ELEAQLESSEQRISDLTVD 352
Score = 36.6 bits (83), Expect = 0.066
Identities = 64/290 (22%), Positives = 113/290 (38%), Gaps = 64/290 (22%)
Query: 4 IMNLANDFDDEILSEFES---LLSEQTDFPLLSEKTDDSKKHGGNETEMAKNGSDSEIEW 60
I NL N+ D + + E+ L E+ + LS + D +K E E A N + E +
Sbjct: 986 INNLKNELDSLQVQKSETEAELEREKQEKSELSNQITDVQK-ALVEQEAAYNTLEEEHKQ 1044
Query: 61 LRNVVEELEEREMKLQSELLEYYSLKEQ---------------VPVIEEFQRQLRIKSVE 105
+ + +E E K+ + E L E+ +E + +L +K E
Sbjct: 1045 INELFKETEATLNKVTVDYKEAQRLLEERGKEVTSRDSTIGVHEETMESLRNELEMKGDE 1104
Query: 106 IDMLHMTIKSLQ--------------------------EENNKLQEELIHEASAKRELEV 139
I+ L I +++ EE L+E+ + E + E
Sbjct: 1105 IETLMEKISNIEVKLRLSNQKLRVTEQVLTEKEEAFRKEEAKHLEEQALLEKNLTMTHET 1164
Query: 140 ARNKIKELQRQIKIIANQTKGQLLLLKQKVSGLQAKEEEVVKKDAEI----------ENN 189
R IKE+ ++ I + + + +K++ Q + E+ V + ++I N+
Sbjct: 1165 YRGMIKEIADKVNITVDGFQS----MSEKLTEKQGRYEKTVMEASKILWTATNWVIERNH 1220
Query: 190 LKTVNDFEIEKKLKTVNDLEIEAVGLKRRNKELQHEKRELTVKLNAAESR 239
K + EIEKK D EI+ +G K R E + E + T+ E R
Sbjct: 1221 EKEKMNKEIEKK-----DEEIKKLGGKVREDEKEKEMMKETLMGLGEEKR 1265
>At5g16730 putative protein
Length = 853
Score = 53.1 bits (126), Expect = 7e-07
Identities = 80/349 (22%), Positives = 148/349 (41%), Gaps = 76/349 (21%)
Query: 9 NDFDDEILSEFESLLSEQTDFPLLSEKTDDSKKHGGN-ETEMAKNGSD-----SEIEWLR 62
+D + EI E +++ +T E + S++ G+ E E++KN + SE+E ++
Sbjct: 362 HDTETEITDLKERIVTLETTVAKQKEDLEVSEQRLGSVEEEVSKNEKEVEKLKSELETVK 421
Query: 63 NV--------------VEELEEREMKLQSELLE-----------YYSLKEQVPVIEEFQR 97
V+ L E + KL S+L SL + + R
Sbjct: 422 EEKNRALKKEQDATSRVQRLSEEKSKLLSDLESSKEEEEKSKKAMESLASALHEVSSEGR 481
Query: 98 QLRIK---------SVEIDMLHMTIKSLQEENNKLQEELIHEA---------------SA 133
+L+ K +ID L + IK+ E+ + +E HE S+
Sbjct: 482 ELKEKLLSQGDHEYETQIDDLKLVIKATNEKYENMLDEARHEIDVLVSAVEQTKKHFESS 541
Query: 134 KRELEVAR----NKIKELQRQIKIIANQTKGQLLLLKQKVSGLQAKEEEVVKKDAEIENN 189
K++ E+ N +K+++ + + + LLK+ + + + KK+A+ +++
Sbjct: 542 KKDWEMKEANLVNYVKKMEEDVASMGKEMNRLDNLLKRT----EEEADAAWKKEAQTKDS 597
Query: 190 LKTVNDFEIEKKLKTVNDLEIEAVGLKRR--NKELQ-----HEKRELTVKLNAAESRITE 242
LK V + EI +T+ + + E++ LK +KE + HE +L K + + +I E
Sbjct: 598 LKEVEE-EIVYLQETLGEAKAESMKLKENLLDKETEFQNVIHENEDLKAKEDVSLKKIEE 656
Query: 243 LSSVTENEMIA--DAKSETGRLRHANED---LQKQVEGLQMNRFSEVEE 286
LS + E ++A + E G L + +D L K VE N VEE
Sbjct: 657 LSKLLEEAILAKKQPEEENGELSESEKDYDLLPKVVEFSSENGHRSVEE 705
Score = 41.6 bits (96), Expect = 0.002
Identities = 58/278 (20%), Positives = 117/278 (41%), Gaps = 16/278 (5%)
Query: 18 EFESLLSEQTDFPLLSEKTDDSKKHGGNETEMAKNGSDSEIEWLRNVVEELEEREMKLQS 77
+ E+ +++ LS + K + E A S L +V+++LE KL
Sbjct: 304 DLEAAKMAESNAHSLSNEWQSKAKELEEQLEEANKLERSASVSLESVMKQLEGSNDKLHD 363
Query: 78 ELLEYYSLKEQVPVIE----EFQRQLRIKSVEIDMLHMTIKSLQEENNKLQEELIHEASA 133
E LKE++ +E + + L + + + + ++E KL+ EL
Sbjct: 364 TETEITDLKERIVTLETTVAKQKEDLEVSEQRLGSVEEEVSKNEKEVEKLKSELETVKEE 423
Query: 134 K-RELEVARNKIKELQR----QIKIIAN--QTKGQLLLLKQKVSGLQAKEEEVVKKDAEI 186
K R L+ ++ +QR + K++++ +K + K+ + L + EV + E+
Sbjct: 424 KNRALKKEQDATSRVQRLSEEKSKLLSDLESSKEEEEKSKKAMESLASALHEVSSEGREL 483
Query: 187 ENNLKTVNDFEIEKKLKTVNDLEIEAVGLKRRNKELQHEKRELTVKLNAAESRITELSSV 246
+ L + D E E ++ + L I+A K N L + E+ V ++A E S
Sbjct: 484 KEKLLSQGDHEYETQIDDLK-LVIKATNEKYENM-LDEARHEIDVLVSAVEQTKKHFESS 541
Query: 247 TENEMIADAKSETGRLRHANEDLQKQVEGLQMNRFSEV 284
++ + +A + ++ ED+ G +MNR +
Sbjct: 542 KKDWEMKEA-NLVNYVKKMEEDVASM--GKEMNRLDNL 576
Score = 38.1 bits (87), Expect = 0.023
Identities = 55/276 (19%), Positives = 118/276 (41%), Gaps = 31/276 (11%)
Query: 22 LLSEQTDFPLLSEKTDDSKKHGGNETEMAKNGSDS------EIEWLRNVVEELEEREMKL 75
L SE T L + T + K ++ EM D ++E R E++E+EM +
Sbjct: 241 LSSELTRLKALLDSTRE--KTAISDNEMVAKLEDEIVVLKRDLESARGFEAEVKEKEMIV 298
Query: 76 QSELLEYYSLKEQVPVIEEFQRQLRIKSVEIDMLHMTIKSLQEENNKLQEELIHEASAKR 135
+ ++ + K + + K+ E++ + L+E N + + S +
Sbjct: 299 EKLNVDLEAAKMAESNAHSLSNEWQSKAKELE------EQLEEANKLERSASVSLESVMK 352
Query: 136 ELEVARNKIKELQRQIKIIANQTKGQLLLLKQKVSGLQAKEEEVVKKDAEIENNLKTVND 195
+LE + +K+ + + +I + + + ++ L+ E+ + + E+ N K V
Sbjct: 353 QLEGSNDKLHDTETEITDLKERIVTLETTVAKQKEDLEVSEQRLGSVEEEVSKNEKEVE- 411
Query: 196 FEIEKKLKTVNDLEIEAVGLKR----RNKELQHEKRELTVKLNAAESRITELSSVTENEM 251
+++ +L+TV + + A+ ++ R + L EK +L L + S+ E S E
Sbjct: 412 -KLKSELETVKEEKNRALKKEQDATSRVQRLSEEKSKLLSDLES--SKEEEEKSKKAMES 468
Query: 252 IADAKSET---GR------LRHANEDLQKQVEGLQM 278
+A A E GR L + + + Q++ L++
Sbjct: 469 LASALHEVSSEGRELKEKLLSQGDHEYETQIDDLKL 504
Score = 30.4 bits (67), Expect = 4.7
Identities = 45/176 (25%), Positives = 69/176 (38%), Gaps = 21/176 (11%)
Query: 581 IEKRPPRLPRPPPKPSDGAPVSNSLNEIPYAPSVPSPPPPPGSLPRGAVGD---DKVKRA 637
+E+R P+LP PP K S + E P + S + DK K
Sbjct: 67 VERRSPKLPTPPEK-SQARVAAVKGTESPQTTTRLSQIKEDLKKANERISSLEKDKAKAL 125
Query: 638 PELVEFYQSLMKREAKKDASLLTSSTSNAADTRSNVIAEIENRSSFLLAVKADVETQGDF 697
EL ++AKK+A +T +A + +V E + AV+A +E +
Sbjct: 126 DEL---------KQAKKEAEQVTLKLDDALKAQKHV--EENSEIEKFQAVEAGIEAVQNN 174
Query: 698 VMSLATEVRAASFSKIEDVVAFVNWLDEELSFLSDERAVLKHFDWPEGKSDALREA 753
L E+ D A V + +EL +++E A FD KS AL +A
Sbjct: 175 EEELKKELETVKNQHASDSAALVA-VRQELEKINEELAAA--FD---AKSKALSQA 224
>At1g06530 hypothetical protein
Length = 323
Score = 53.1 bits (126), Expect = 7e-07
Identities = 61/261 (23%), Positives = 125/261 (47%), Gaps = 34/261 (13%)
Query: 34 EKTDDSKKHGGNETEMAKNGSDSE-----IEWLRNVVEELEEREMKLQSELLEYYSLKEQ 88
+ T+ ++K G E++ + D++ IE L +EEL E K + ++ E ++ +
Sbjct: 30 KSTELNQKIGDLESQNQELARDNDAINRKIESLTAEIEELRGAESKAKRKMGE---MERE 86
Query: 89 VPVIEEFQRQLRIKSVEIDMLHMTIKSLQEENNKLQEELIHEASAKRELEVARNKIKELQ 148
+ +E ++ L + + L+ E +LQ ELI +A+ E E A + ++L+
Sbjct: 87 IDKSDEERKVL-------EAIASRASELETEVARLQHELI---TARTEGEEATAEAEKLR 136
Query: 149 RQIKIIANQTKGQLLLLKQKVSGLQAKEEEVVKKDAEIENNLKTVNDFEIEKKLK----- 203
+I +Q G + L+++V+GL+ +EE K+ E+E+ L + E+++K K
Sbjct: 137 SEI----SQKGGGIEELEKEVAGLRTVKEENEKRMKELESKLGALEVKELDEKNKKFRAE 192
Query: 204 -----TVNDLEIEAVGLKRRNKELQHE--KRELTVKLNAAESRITELSSVTENEMIADAK 256
+++ E E LK + K L+ + K + ++ E + E S + + +
Sbjct: 193 EEMREKIDNKEKEVHDLKEKIKSLESDVAKGKTELQKWITEKMVVEDSLKDSEKKVVALE 252
Query: 257 SETGRLRHANEDLQKQVEGLQ 277
SE L+ +D +K + GL+
Sbjct: 253 SEIVELQKQLDDAEKMINGLK 273
>At1g68790 putative nuclear matrix constituent protein 1 (NMCP1)
Length = 1085
Score = 52.8 bits (125), Expect = 9e-07
Identities = 68/292 (23%), Positives = 134/292 (45%), Gaps = 35/292 (11%)
Query: 12 DDEILSEFESLLSEQTDFPLLSEKTDDSKKHGGNETEMAKNGSDSEIEWLRNVVEELEER 71
+++ LSE + ++ + + + +E+T + K+ + + + SE+ E+ E
Sbjct: 253 EEDRLSEVKRSINHREERVMENERTIEKKEKILENLQQKISVAKSEL------TEKEESI 306
Query: 72 EMKLQSELL---EYYSLKEQVPVIE----EFQRQL-RIKSVEIDMLHMTIKSLQEENNKL 123
++KL L ++ ++K +V + E EF+ L + +EI L K++ + +
Sbjct: 307 KIKLNDISLKEKDFEAMKAKVDIKEKELHEFEENLIEREQMEIGKLLDDQKAVLDSRRRE 366
Query: 124 QEELIHEASAKRELEVARNKIKELQRQIKIIANQTKGQLLLLKQKVSGLQAKEEEVVKKD 183
E + + + E+ K + Q Q++I + K L ++ + L+ KEE V KK+
Sbjct: 367 FEMELEQMRRSLDEELEGKKAEIEQLQVEISHKEEK-----LAKREAALEKKEEGVKKKE 421
Query: 184 AEIENNLKTVNDFE-----IEKKLKTVNDLEIEAVGLKRRNKELQHEKRELTVKLNAAES 238
+++ LKTV + E EKKL N+ +E K ++L+ E E+ + ES
Sbjct: 422 KDLDARLKTVKEKEKALKAEEKKLHMENERLLED---KECLRKLKDEIEEIGTETTKQES 478
Query: 239 RITELSSVTENEMIADAKSETGRLRHANEDLQKQVEGLQMNR---FSEVEEL 287
RI E E+E + K E +L++Q++ ++ E EEL
Sbjct: 479 RIRE-----EHESLRITKEERVEFLRLQSELKQQIDKVKQEEELLLKEREEL 525
Score = 35.0 bits (79), Expect = 0.19
Identities = 40/232 (17%), Positives = 100/232 (42%), Gaps = 7/232 (3%)
Query: 65 VEELEEREMKLQSELLEYYSLKEQVPVIEEFQRQLRIKSVEIDMLHMTIKS-LQEENNKL 123
++ ++E+E L++E + + E++ +E R+L+ + EI +S ++EE+ L
Sbjct: 428 LKTVKEKEKALKAEEKKLHMENERLLEDKECLRKLKDEIEEIGTETTKQESRIREEHESL 487
Query: 124 Q---EELIHEASAKRELEVARNKIKELQRQIKIIANQTKGQLLLLKQKVSGLQAKEEEVV 180
+ EE + + EL+ +K+K+ + + + K +++ L K +
Sbjct: 488 RITKEERVEFLRLQSELKQQIDKVKQEEELLLKEREELKQDKERFEKEWEALDKKRANIT 547
Query: 181 KKDAEIENNLKTVNDFEIEKKLKTVNDLEIEAVGLKRRNKELQHEKRELTVKLNAAESRI 240
++ E+ + + + +I +K + + LKR ++ +K + E +
Sbjct: 548 REQNEVAEENEKLRNLQISEKHRLKREEMTSRDNLKRELDGVKMQKESFEADMEDLEMQK 607
Query: 241 TELSSVTENEMIADAKSETGRLRHANEDLQKQVEGLQMNR---FSEVEELVY 289
L + + A + R R + Q++++ + + E+EE+ Y
Sbjct: 608 RNLDMEFQRQEEAGERDFNERARTYEKRSQEELDNINYTKKLAQREMEEMQY 659
Score = 33.5 bits (75), Expect = 0.56
Identities = 28/133 (21%), Positives = 62/133 (46%), Gaps = 12/133 (9%)
Query: 470 ILDSVSASFQLMSKSVDRSLDEKYPAYK-DLHKLAL----------AREKQLKEKAEKAR 518
+LDS F++ + + RSLDE+ K ++ +L + RE L++K E +
Sbjct: 359 VLDSRRREFEMELEQMRRSLDEELEGKKAEIEQLQVEISHKEEKLAKREAALEKKEEGVK 418
Query: 519 GEKFGDNSNLNTTKAEEERSTSLDSELTQVNEKACV-SDGLNKQSEDVKDFDNQTITEMK 577
++ ++ L T K +E+ + + +L NE+ + L K +++++ +T +
Sbjct: 419 KKEKDLDARLKTVKEKEKALKAEEKKLHMENERLLEDKECLRKLKDEIEEIGTETTKQES 478
Query: 578 LFKIEKRPPRLPR 590
+ E R+ +
Sbjct: 479 RIREEHESLRITK 491
>At1g22260 hypothetical protein
Length = 851
Score = 52.4 bits (124), Expect = 1e-06
Identities = 61/258 (23%), Positives = 116/258 (44%), Gaps = 45/258 (17%)
Query: 55 DSEIEWLRNVVEELEEREMKLQSELLEYY----SLKEQVPVIEEFQRQLRIKSVEIDMLH 110
+ E++ L ++ E+LE+ + +Q + S +++V ++E + L + E+D +
Sbjct: 245 EDEVKDLVSIQEKLEKEKTSVQLSADNCFEKLVSSEQEVKKLDELVQYLVAELTELDKKN 304
Query: 111 MTIKS--------------------------LQEENNKLQEELIHEASAKRELEVARN-- 142
+T K Q + LQ EL A+ K LE A N
Sbjct: 305 LTFKEKFDKLSGLYDTHIMLLQKDRDLALDRAQRSFDNLQGELFRVAATKEALESAGNEL 364
Query: 143 --KIKELQRQIKIIANQTKGQLLLLKQKVSGLQAKEEEVVKKDAEIENNLKTVNDFEIEK 200
KI ELQ + + +Q G Q + L+++ + +V K A+ E+ + + + E+E
Sbjct: 365 NEKIVELQNDKESLISQLSGLRCSTSQTIDKLESEAKGLVSKHADAESAISQLKE-EMET 423
Query: 201 KLKTVNDLEIEAVGLKRRNKELQHEKRELTVKLNA-AESRITELSSVTENEMIADAKSET 259
L++V E + L + L+ E +E KL A A+ ++ EL ++ + +SE+
Sbjct: 424 LLESVKTSEDKKQELSLKLSSLEMESKEKCEKLQADAQRQVEELETLQK-------ESES 476
Query: 260 GRLRHANEDLQKQVEGLQ 277
+L+ + L K+V LQ
Sbjct: 477 HQLQ--ADLLAKEVNQLQ 492
Score = 37.7 bits (86), Expect = 0.030
Identities = 45/203 (22%), Positives = 79/203 (38%), Gaps = 28/203 (13%)
Query: 67 ELEEREMKLQSELLEYYSLKEQVPVIEEFQRQLRIKSVEIDMLHMTIKSLQEEN----NK 122
+LE KL+ + Y+L+E++ +LR++ E + L ++S ++
Sbjct: 61 DLELANCKLKKSMEHVYALEEKLQNAFNENAKLRVRKKEDEKLWRGLESKFSSTKTLCDQ 120
Query: 123 LQEELIHEAS----AKRELEVARNKIKELQRQIKIIANQTKGQLLLLKQKVSGLQAKEEE 178
L E L H AS A+++ K I + Q + L L A +EE
Sbjct: 121 LTETLQHLASQVQDAEKDKGFFETKFSTSSEAIDSLNQQMRDMSL-------RLDAAKEE 173
Query: 179 VVKKDAEIENNLKTVNDFE-------------IEKKLKTVNDLEIEAVGLKRRNKELQHE 225
+ +D E+E E IEKK + LE A K + L +
Sbjct: 174 ITSRDKELEELKLEKQQKEMFYQTERCGTASLIEKKDAVITKLEASAAERKLNIENLNSQ 233
Query: 226 KRELTVKLNAAESRITELSSVTE 248
++ ++L E + +L S+ E
Sbjct: 234 LEKVHLELTTKEDEVKDLVSIQE 256
Score = 32.3 bits (72), Expect = 1.2
Identities = 26/104 (25%), Positives = 45/104 (43%), Gaps = 12/104 (11%)
Query: 463 KVSSSDDILDSVSASFQLMSKSVDRSLDEKYPAYKDLHKLALAREKQLKE-KAEKARGEK 521
K S+S + +DS++ + MS +D + +E +R+K+L+E K EK + E
Sbjct: 145 KFSTSSEAIDSLNQQMRDMSLRLDAAKEE-----------ITSRDKELEELKLEKQQKEM 193
Query: 522 FGDNSNLNTTKAEEERSTSLDSELTQVNEKACVSDGLNKQSEDV 565
F T E++ + E+ + LN Q E V
Sbjct: 194 FYQTERCGTASLIEKKDAVITKLEASAAERKLNIENLNSQLEKV 237
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.310 0.128 0.345
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 20,465,184
Number of Sequences: 26719
Number of extensions: 971545
Number of successful extensions: 8210
Number of sequences better than 10.0: 498
Number of HSP's better than 10.0 without gapping: 99
Number of HSP's successfully gapped in prelim test: 410
Number of HSP's that attempted gapping in prelim test: 5482
Number of HSP's gapped (non-prelim): 2000
length of query: 915
length of database: 11,318,596
effective HSP length: 108
effective length of query: 807
effective length of database: 8,432,944
effective search space: 6805385808
effective search space used: 6805385808
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 65 (29.6 bits)
Medicago: description of AC146570.7


