

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146570.6 + phase: 0
(101 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
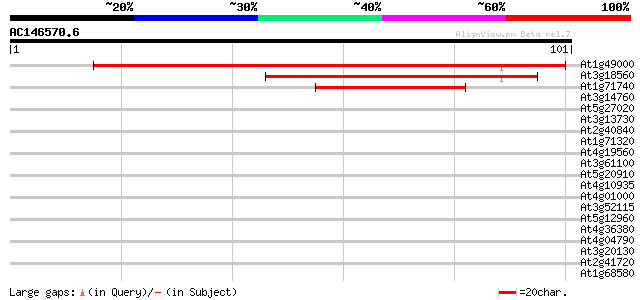
At1g49000 hypothetical protein; similar to ESTs gb|R64892.1, gb|... 65 6e-12
At3g18560 unknown protein 59 4e-10
At1g71740 hypothetical protein 39 3e-04
At3g14760 unknown protein 37 0.002
At5g27020 putative protein 36 0.003
At3g13730 CYP90D 29 0.45
At2g40840 4-alpha-glucanotransferase 27 1.7
At1g71320 hypothetical protein 26 2.9
At4g19560 putative protein 26 3.8
At3g61100 unknown protein 26 3.8
At5g20910 ABI3-interacting protein 2 25 5.0
At4g10935 unknown protein 25 5.0
At4g01000 putative protein 25 5.0
At3g52115 gamma response I protein 25 5.0
At5g12960 unknown protein 25 6.5
At4g36380 cytochrome P450 (ROTUNDIFOLIA3) 25 6.5
At4g04790 hypothetical protein 25 6.5
At3g20130 cytochrome P450, putative 25 6.5
At2g41720 salt-inducible protein-like 25 6.5
At1g68580 unknown protein 25 8.5
>At1g49000 hypothetical protein; similar to ESTs gb|R64892.1,
gb|T04316.1
Length = 156
Score = 65.1 bits (157), Expect = 6e-12
Identities = 33/86 (38%), Positives = 52/86 (60%), Gaps = 1/86 (1%)
Query: 16 SLKTKKLEELSTSSKNIIKKIYKAIHCKILKMKKQEDDKCLWKKTILMGEKCQPLEFPGA 75
S+ + + +EL T+ N + + + K E++ LW++ ILMG KC+PL+F G
Sbjct: 67 SMSSPRPKELFTTLSNKAMTMVRRKNPPEEKATAMEEEHGLWQREILMGGKCEPLDFSGV 126
Query: 76 IFYDSEGNQLSE-PPRTPRSSPSSSF 100
I+YDS G L+E PPR+PR +P S+
Sbjct: 127 IYYDSNGRLLNEVPPRSPRGTPLPSY 152
>At3g18560 unknown protein
Length = 195
Score = 58.9 bits (141), Expect = 4e-10
Identities = 24/50 (48%), Positives = 39/50 (78%), Gaps = 1/50 (2%)
Query: 47 MKKQEDDKCLWKKTILMGEKCQPLEFPGAIFYDSEGNQLSE-PPRTPRSS 95
M++ E++ +W++ ILMG KC+PL++ G I+YD G+QL + PPR+PR+S
Sbjct: 124 MEEDEEEYGVWQREILMGGKCEPLDYSGVIYYDCSGHQLKQVPPRSPRAS 173
>At1g71740 hypothetical protein
Length = 130
Score = 39.3 bits (90), Expect = 3e-04
Identities = 16/27 (59%), Positives = 20/27 (73%)
Query: 56 LWKKTILMGEKCQPLEFPGAIFYDSEG 82
LW+K ILMG KCQ +F G I YD++G
Sbjct: 88 LWQKNILMGGKCQLPDFSGVILYDADG 114
>At3g14760 unknown protein
Length = 168
Score = 36.6 bits (83), Expect = 0.002
Identities = 17/43 (39%), Positives = 23/43 (52%)
Query: 56 LWKKTILMGEKCQPLEFPGAIFYDSEGNQLSEPPRTPRSSPSS 98
+W++ ILMGEKC+ F G I YD G+ P + SS
Sbjct: 116 VWQRPILMGEKCELPRFSGLILYDECGHPRHHPQLQEKVKQSS 158
>At5g27020 putative protein
Length = 164
Score = 36.2 bits (82), Expect = 0.003
Identities = 18/39 (46%), Positives = 26/39 (66%), Gaps = 4/39 (10%)
Query: 50 QEDDKCLWKKTILMGEKCQPLEFPGAIFYDSEGNQLSEP 88
+E++ C KK ILMGE+C+P+ G + YD +G L EP
Sbjct: 130 EEEELC--KKRILMGERCKPMN--GVLQYDGDGILLPEP 164
>At3g13730 CYP90D
Length = 491
Score = 28.9 bits (63), Expect = 0.45
Identities = 14/52 (26%), Positives = 28/52 (52%)
Query: 10 IISYIISLKTKKLEELSTSSKNIIKKIYKAIHCKILKMKKQEDDKCLWKKTI 61
++S I+ +L + KN++K++ + I KI K K +E+D + K +
Sbjct: 227 LMSLPINFPGTQLHRSLQAKKNMVKQVERIIEGKIRKTKNKEEDDVIAKDVV 278
>At2g40840 4-alpha-glucanotransferase
Length = 858
Score = 26.9 bits (58), Expect = 1.7
Identities = 14/44 (31%), Positives = 24/44 (53%), Gaps = 8/44 (18%)
Query: 19 TKKLEELSTSSKNIIKKIYKAIHCKILKMKKQEDDKCLWKKTIL 62
T ++L SKN++K++Y + ++QED LW+K L
Sbjct: 609 TSSFQDLDDHSKNVLKRLYYDYY-----FQRQED---LWRKNAL 644
>At1g71320 hypothetical protein
Length = 392
Score = 26.2 bits (56), Expect = 2.9
Identities = 11/36 (30%), Positives = 25/36 (68%), Gaps = 2/36 (5%)
Query: 5 KINKAIISYIISLKTKKLEELSTSSKNIIKKIYKAI 40
+++++++ Y IS+K ++ L+ K+I+KK YK +
Sbjct: 171 QLSQSVLDYDISVK--RMPSLAGFGKDIVKKSYKVV 204
>At4g19560 putative protein
Length = 474
Score = 25.8 bits (55), Expect = 3.8
Identities = 15/56 (26%), Positives = 31/56 (54%)
Query: 11 ISYIISLKTKKLEELSTSSKNIIKKIYKAIHCKILKMKKQEDDKCLWKKTILMGEK 66
I+ + L K+EE + +++I Y+ IH K L ++++ K+ +L+GE+
Sbjct: 104 IATVCMLLAGKVEETPVTLEDVIIASYERIHKKDLAGAQRKEVYDQQKELVLIGEE 159
>At3g61100 unknown protein
Length = 166
Score = 25.8 bits (55), Expect = 3.8
Identities = 14/65 (21%), Positives = 29/65 (44%), Gaps = 3/65 (4%)
Query: 1 MIINKINKAIISYIISLKTKKLEELSTSSKNIIKKIYKAIHCKILKMKKQEDDKCLWKKT 60
M+I+++N +Y+ K K L + K K +CK+ + + + +W+
Sbjct: 100 MVISRVNSKFATYLADWKAKDLYIFGAEPE---KAPGKCSNCKVTSLDELFTQQWVWESL 156
Query: 61 ILMGE 65
L G+
Sbjct: 157 SLGGD 161
>At5g20910 ABI3-interacting protein 2
Length = 310
Score = 25.4 bits (54), Expect = 5.0
Identities = 13/44 (29%), Positives = 27/44 (60%), Gaps = 1/44 (2%)
Query: 28 SSKNIIKKIYKAIHCK-ILKMKKQEDDKCLWKKTILMGEKCQPL 70
+SK +++K+ I + +LK E + C+ K+ +++G+K Q L
Sbjct: 203 ASKEVVEKLPVIIFTEELLKKFGAEAECCICKENLVIGDKMQEL 246
>At4g10935 unknown protein
Length = 984
Score = 25.4 bits (54), Expect = 5.0
Identities = 16/61 (26%), Positives = 30/61 (48%), Gaps = 1/61 (1%)
Query: 12 SYIISLKTKKLE-ELSTSSKNIIKKIYKAIHCKILKMKKQEDDKCLWKKTILMGEKCQPL 70
S +++K KKL + +N+ KKIY ++ K ++ + WK + G K + +
Sbjct: 314 SSALAVKAKKLMLQKGKVRENLTKKIYADLNGKRKSAWHRDCEVEFWKHRCIQGRKPEKI 373
Query: 71 E 71
E
Sbjct: 374 E 374
>At4g01000 putative protein
Length = 415
Score = 25.4 bits (54), Expect = 5.0
Identities = 13/50 (26%), Positives = 23/50 (46%)
Query: 16 SLKTKKLEELSTSSKNIIKKIYKAIHCKILKMKKQEDDKCLWKKTILMGE 65
+L+ K LE L S + + + K + K+E DKC+ + + E
Sbjct: 135 NLEKKALEYLKKQSNKVKQGVGNGATQKYVNKYKEESDKCILAVDLALNE 184
>At3g52115 gamma response I protein
Length = 588
Score = 25.4 bits (54), Expect = 5.0
Identities = 21/65 (32%), Positives = 30/65 (45%), Gaps = 5/65 (7%)
Query: 17 LKTKKLEELSTSSKNIIKKIYKAIHCKILKMKKQEDDKCLWKKTILMGEKCQPLEFPGAI 76
L+ K L+ELS N+I K LKMK + + L KKT++ + LE+
Sbjct: 146 LQVKGLDELSEDGINMI-----VSEVKSLKMKTEFLQEELSKKTLVTENLLKKLEYLSTE 200
Query: 77 FYDSE 81
D E
Sbjct: 201 AADGE 205
>At5g12960 unknown protein
Length = 865
Score = 25.0 bits (53), Expect = 6.5
Identities = 11/33 (33%), Positives = 17/33 (51%)
Query: 68 QPLEFPGAIFYDSEGNQLSEPPRTPRSSPSSSF 100
+P +FPG I + + L+ +P SSSF
Sbjct: 733 EPFDFPGMIVKQATDSSLTVQASSPSDKGSSSF 765
>At4g36380 cytochrome P450 (ROTUNDIFOLIA3)
Length = 524
Score = 25.0 bits (53), Expect = 6.5
Identities = 16/51 (31%), Positives = 24/51 (46%), Gaps = 3/51 (5%)
Query: 33 IKKIYKAIHCKILKMKKQEDDKCLWK---KTILMGEKCQPLEFPGAIFYDS 80
IKK+ I K+L +D + K + + G C P++FPG Y S
Sbjct: 210 IKKMTFEILVKVLMSTSPGEDMNILKLEFEEFIKGLICIPIKFPGTRLYKS 260
>At4g04790 hypothetical protein
Length = 731
Score = 25.0 bits (53), Expect = 6.5
Identities = 9/31 (29%), Positives = 20/31 (64%)
Query: 1 MIINKINKAIISYIISLKTKKLEELSTSSKN 31
M+++K+NK+++S + + + E S +S N
Sbjct: 1 MVVSKVNKSLLSSVFKSRIRPTGESSIASGN 31
>At3g20130 cytochrome P450, putative
Length = 515
Score = 25.0 bits (53), Expect = 6.5
Identities = 15/45 (33%), Positives = 28/45 (61%), Gaps = 3/45 (6%)
Query: 18 KTKKLEELSTSSKNIIKKIYKAIHCKILKMKKQEDDKCLWKKTIL 62
+T+KL L T S ++KK++ A+ +L+ + Q+ L+KK I+
Sbjct: 209 ETEKLRGLVTESIGLMKKMFLAV---LLRRQLQKLGISLFKKDIM 250
>At2g41720 salt-inducible protein-like
Length = 822
Score = 25.0 bits (53), Expect = 6.5
Identities = 24/82 (29%), Positives = 35/82 (42%), Gaps = 18/82 (21%)
Query: 7 NKAIISYIISLKTKKLEELSTSSKNIIKKIYKAIHCKILKMKKQEDDKCLWKKTILMGEK 66
N AI SYI + + +K L S ++ KK + D + TIL+
Sbjct: 499 NSAIGSYINAAELEKAIALYQS----------------MRKKKVKADSVTF--TILISGS 540
Query: 67 CQPLEFPGAIFYDSEGNQLSEP 88
C+ ++P AI Y E LS P
Sbjct: 541 CRMSKYPEAISYLKEMEDLSIP 562
>At1g68580 unknown protein
Length = 648
Score = 24.6 bits (52), Expect = 8.5
Identities = 10/38 (26%), Positives = 18/38 (47%)
Query: 44 ILKMKKQEDDKCLWKKTILMGEKCQPLEFPGAIFYDSE 81
+LK +DD LW +I+ GE + P + + +
Sbjct: 64 VLKKLNSKDDVALWLDSIVSGEIPHVADVPATVMTEKD 101
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.315 0.132 0.375
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,418,387
Number of Sequences: 26719
Number of extensions: 95131
Number of successful extensions: 262
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 12
Number of HSP's successfully gapped in prelim test: 9
Number of HSP's that attempted gapping in prelim test: 249
Number of HSP's gapped (non-prelim): 21
length of query: 101
length of database: 11,318,596
effective HSP length: 77
effective length of query: 24
effective length of database: 9,261,233
effective search space: 222269592
effective search space used: 222269592
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 52 (24.6 bits)
Medicago: description of AC146570.6


