

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146570.4 - phase: 0
(338 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
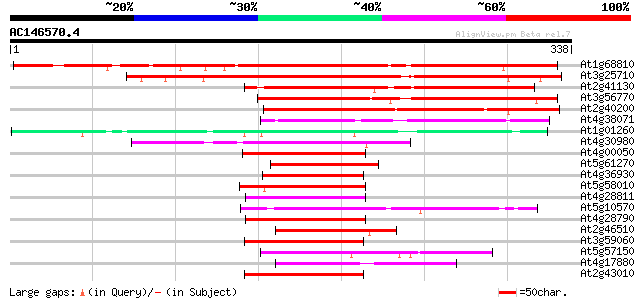
Score E
Sequences producing significant alignments: (bits) Value
At1g68810 putative bHLH transcription factor (bHLH030) 315 3e-86
At3g25710 putative bHLH transcription factor (bHLH032) 261 3e-70
At2g41130 putative bHLH transcription factor (bHLH106) 153 1e-37
At3g56770 putative HLH DNA binding protein 149 3e-36
At2g40200 putative bHLH transcription factor (bHLH051) 123 1e-28
At4g38071 putative bHLH transcription factor (bHLH131) 100 1e-21
At1g01260 transcription factor MYC7E, putative bHLH transcriptio... 68 8e-12
At4g30980 putative bHLH transcription factor (bHLH069) 67 2e-11
At4g00050 bHLH like transcription factor (bHLH016) 63 3e-10
At5g61270 bHLH transcription factor like protein 62 6e-10
At4g36930 putative bHLH transcription factor (AtbHLH024) / SPATU... 59 5e-09
At5g58010 putative bHLH transcription factor (bHLH082) 58 6e-09
At4g28811 AtbHLH119 58 8e-09
At5g10570 putative bHLH transcription factor (bHLH061) 57 1e-08
At4g28790 bHLH transcription factor like protein (bHLH023) 57 1e-08
At2g46510 putative bHLH transcription factor 57 1e-08
At3g59060 putative bHLH transcription factor (bHLH065) 57 1e-08
At5g57150 unknown protein 57 2e-08
At4g17880 putative transcription factor BHLH4 57 2e-08
At2g43010 putative transcription factor BHLH9 57 2e-08
>At1g68810 putative bHLH transcription factor (bHLH030)
Length = 368
Score = 315 bits (806), Expect = 3e-86
Identities = 196/358 (54%), Positives = 239/358 (66%), Gaps = 46/358 (12%)
Query: 3 QEDQGQCSSQTINNFETYQQEQFLLQQQMMRQQNSDHHIFGGGRSMFPTAGGDHN----- 57
+E++ + SS+ +NN + YQ + F Q S HH S T G N
Sbjct: 8 EEEEEEDSSEAMNNIQNYQNDLFF------HQLISHHHHHHHDPSQSETLGASGNVGSGF 61
Query: 58 ------QVSPILQQTWSMQQLHHHHHPFNPHDPFVIPQQQASPYASLFNR---RVPSLQF 108
VSPI WS+ PF+ P AS Y S FNR LQF
Sbjct: 62 TIFSQDSVSPI----WSLPPPTSIQPPFDQFPP--PSSSPASFYGSFFNRSRAHHQGLQF 115
Query: 109 AYDHHAGS--------EHLRIISDTLQH--GSGSGPFGGFQGEVGKMSAQEIMEAKALAA 158
Y+ G+ E LRI+S+ L +GSGPFG Q E+GKM+AQEIM+AKALAA
Sbjct: 116 GYEGFGGATSAAHHHHEQLRILSEALGPVVQAGSGPFG-LQAELGKMTAQEIMDAKALAA 174
Query: 159 SKSHSEAERRRRERINNHLAKLRSLLPSTTKTDKASLLAEVIQHVKELKRQTSLIAETSP 218
SKSHSEAERRRRERINNHLAKLRS+LP+TTKTDKASLLAEVIQHVKELKR+TS+I+ET+
Sbjct: 175 SKSHSEAERRRRERINNHLAKLRSILPNTTKTDKASLLAEVIQHVKELKRETSVISETNL 234
Query: 219 VPTECDELTVDAAADDEDYGSNGNKFIIKASLCCDDRSDLLPELIKTLKALRLRTLKADI 278
VPTE DELTV A ++E+ G +F+IKASLCC+DRSDLLP++IKTLKA+RL+TLKA+I
Sbjct: 235 VPTESDELTV-AFTEEEETGD--GRFVIKASLCCEDRSDLLPDMIKTLKAMRLKTLKAEI 291
Query: 279 TTLGGRVKNVLFITGEED-----DHEYCISSIQEALKAVMEKS-VGDESASGSVKRQR 330
TT+GGRVKNVLF+TGEE + EYCI +I+EALKAVMEKS V + S+SG+ KRQR
Sbjct: 292 TTVGGRVKNVLFVTGEESSGEEVEEEYCIGTIEEALKAVMEKSNVEESSSSGNAKRQR 349
>At3g25710 putative bHLH transcription factor (bHLH032)
Length = 344
Score = 261 bits (668), Expect = 3e-70
Identities = 160/320 (50%), Positives = 199/320 (62%), Gaps = 63/320 (19%)
Query: 71 QLHHHHHP---------FNPHDPFVIPQQQA--SPYASLFNRRVPSLQFAYDHHAG---- 115
Q H HHHP DP P S FNRR S ++D++ G
Sbjct: 22 QFHLHHHPQILPWSSTSLPSFDPLHFPSNPTRYSDPVHYFNRRASSSSSSFDYNDGFVSP 81
Query: 116 -------SEHLRIISDTLQHGSGSGPFGGFQGEV-GKMSAQEIMEAKALAASKSHSEAER 167
HLRI+S+ L G GF GE+ GK+SAQE+M+AKALAASKSHSEAER
Sbjct: 82 PPSMDHPQNHLRILSEALGPIMRRGSSFGFDGEIMGKLSAQEVMDAKALAASKSHSEAER 141
Query: 168 RRRERINNHLAKLRSLLPSTTKTDKASLLAEVIQHVKELKRQTSLIAETSPVPTECDELT 227
RRRERIN HLAKLRS+LP+TTKTDKASLLAEVIQH+KELKRQTS I +T VPTECD+LT
Sbjct: 142 RRRERINTHLAKLRSILPNTTKTDKASLLAEVIQHMKELKRQTSQITDTYQVPTECDDLT 201
Query: 228 VDAAADDEDYGSNGNKFIIKASLCCDDRSDLLPELIKTLKALRLRTLKADITTLGGRVKN 287
VD++ +DE+ GN +I+AS CC DR+DL+ ++I LK+LRLRTLKA+I T+GGRVKN
Sbjct: 202 VDSSYNDEE----GN-LVIRASFCCQDRTDLMHDVINALKSLRLRTLKAEIATVGGRVKN 256
Query: 288 VLFITGEEDDHE------------------------YCISSIQEALKAVMEKSVG----- 318
+LF++ E DD E +SSI+EALKAV+EK V
Sbjct: 257 ILFLSREYDDEEDHDSYRRNFDGDDVEDYDEERMMNNRVSSIEEALKAVIEKCVHNNDES 316
Query: 319 ------DESASGSVKRQRTN 332
++S+SG +KRQRT+
Sbjct: 317 NDNNNLEKSSSGGIKRQRTS 336
>At2g41130 putative bHLH transcription factor (bHLH106)
Length = 253
Score = 153 bits (387), Expect = 1e-37
Identities = 83/178 (46%), Positives = 131/178 (72%), Gaps = 11/178 (6%)
Query: 142 VGKMSAQEIMEAKALAASKSHSEAERRRRERINNHLAKLRSLLPSTTKTDKASLLAEVIQ 201
+G+ AQ+ +ALAA ++H EAERRRRERIN+HL KLR++L +KTDKA+LLA+V+Q
Sbjct: 55 IGETMAQD----RALAALRNHKEAERRRRERINSHLNKLRNVLSCNSKTDKATLLAKVVQ 110
Query: 202 HVKELKRQTSLIAETSP--VPTECDELTVDAAADDEDYGSNGNKFIIKASLCCDDRSDLL 259
V+ELK+QT +++ +P+E DE++V DY ++G+ I KASLCC+DRSDLL
Sbjct: 111 RVRELKQQTLETSDSDQTLLPSETDEISV---LHFGDYSNDGH-IIFKASLCCEDRSDLL 166
Query: 260 PELIKTLKALRLRTLKADITTLGGRVKNVLFITGEEDDHEY-CISSIQEALKAVMEKS 316
P+L++ LK+L ++TL+A++ T+GGR ++VL + +++ H + +Q ALK+++E+S
Sbjct: 167 PDLMEILKSLNMKTLRAEMVTIGGRTRSVLVVAADKEMHGVESVHFLQNALKSLLERS 224
>At3g56770 putative HLH DNA binding protein
Length = 230
Score = 149 bits (375), Expect = 3e-36
Identities = 83/191 (43%), Positives = 128/191 (66%), Gaps = 17/191 (8%)
Query: 150 IMEAKALAASKSHSEAERRRRERINNHLAKLRSLLPSTTKTDKASLLAEVIQHVKELKRQ 209
+ E KALA+ ++H EAER+RR RIN+HL KLR LL +KTDK++LLA+V+Q VKELK+Q
Sbjct: 37 VYEDKALASLRNHKEAERKRRARINSHLNKLRKLLSCNSKTDKSTLLAKVVQRVKELKQQ 96
Query: 210 TSLIAETSPVPTECDELTV----DAAADDEDYGSNGNKFIIKASLCCDDRSDLLPELIKT 265
T I + + +P+E DE++V D + D+ + I K S CC+DR +LL +L++T
Sbjct: 97 TLEITDET-IPSETDEISVLNIEDCSRGDD------RRIIFKVSFCCEDRPELLKDLMET 149
Query: 266 LKALRLRTLKADITTLGGRVKNVLFITGEEDDH-EYCISSIQEALKAVMEKS-----VGD 319
LK+L++ TL AD+TT+GGR +NVL + +++ H ++ +Q ALK+++E+S VG
Sbjct: 150 LKSLQMETLFADMTTVGGRTRNVLVVAADKEHHGVQSVNFLQNALKSLLERSSKSVMVGH 209
Query: 320 ESASGSVKRQR 330
G + +R
Sbjct: 210 GGGGGEERLKR 220
>At2g40200 putative bHLH transcription factor (bHLH051)
Length = 254
Score = 123 bits (309), Expect = 1e-28
Identities = 70/183 (38%), Positives = 115/183 (62%), Gaps = 7/183 (3%)
Query: 154 KALAASKSHSEAERRRRERINNHLAKLRSLLPSTTKTDKASLLAEVIQHVKELKRQTSLI 213
KA + S+SH AE+RRR+RIN+HL LR L+P++ K DKA+LLA VI+ VKELK++ +
Sbjct: 59 KAESLSRSHRLAEKRRRDRINSHLTALRKLVPNSDKLDKAALLATVIEQVKELKQKAAES 118
Query: 214 AETSPVPTECDELTVDAAADDEDYGSNGNKFIIKASLCCDDRSDLLPELIKTLKALRLRT 273
+PTE DE+TV D+ SN N I KAS CC+D+ + + E+I+ L L+L T
Sbjct: 119 PIFQDLPTEADEVTVQPET-ISDFESNTNTIIFKASFCCEDQPEAISEIIRVLTKLQLET 177
Query: 274 LKADITTLGGRVKNVLFITGEEDDHE-----YCISSIQEALKAVMEKSVGDESASGSVKR 328
++A+I ++GGR++ + FI + + +E +++++L + + + + + SV R
Sbjct: 178 IQAEIISVGGRMR-INFILKDSNCNETTNIAASAKALKQSLCSALNRITSSSTTTSSVCR 236
Query: 329 QRT 331
R+
Sbjct: 237 IRS 239
>At4g38071 putative bHLH transcription factor (bHLH131)
Length = 256
Score = 100 bits (249), Expect = 1e-21
Identities = 58/174 (33%), Positives = 101/174 (57%), Gaps = 14/174 (8%)
Query: 152 EAKALAASKSHSEAERRRRERINNHLAKLRSLLPSTTKTDKASLLAEVIQHVKELKRQTS 211
E+K +AA K HS+AERRRR RIN+ A LR++LP+ K DKAS+L E +++ ELK+
Sbjct: 87 ESKEVAAKK-HSDAERRRRLRINSQFATLRTILPNLVKQDKASVLGETVRYFNELKK--- 142
Query: 212 LIAETSPVPTECDELTVDAAADDEDYGSNGNKFIIKASLCCDDRSDLLPELIKTLKALRL 271
++ + P+ D L +D N N+ + + C DR L+ E+ +++KA++
Sbjct: 143 MVQDIPTTPSLEDNLRLDHC--------NNNRDLARVVFSCSDREGLMSEVAESMKAVKA 194
Query: 272 RTLKADITTLGGRVKNVLFITGEEDDHEYCISSIQEALKAVMEKSVGDESASGS 325
+ ++A+I T+GGR K LF+ G + + ++++LK V+ E+ + +
Sbjct: 195 KAVRAEIMTVGGRTKCALFVQGVNGNEG--LVKLKKSLKLVVNGKSSSEAKNNN 246
>At1g01260 transcription factor MYC7E, putative bHLH transcription
factor (AtbHLH013)
Length = 590
Score = 67.8 bits (164), Expect = 8e-12
Identities = 79/357 (22%), Positives = 134/357 (37%), Gaps = 57/357 (15%)
Query: 2 IQEDQGQCSSQTINNFETYQQEQFLLQQQMMRQQNSDHHIF------------------- 42
I +++ + + ++N Q Q QQQ Q H F
Sbjct: 256 IDDNRTKIFGKDLHNSGFLQHHQHHQQQQQQPPQQQQHRQFREKLTVRKMDDRAPKRLDA 315
Query: 43 ---GGGRSMFPTAGGDHNQVSPILQQTWSMQQLHHHHHPFNPHDPFVIPQQQASPYASLF 99
G R MF G ++N +L TW Q ++ P N + + + P
Sbjct: 316 YPNNGNRFMFSNPGTNNNT---LLSPTWV--QPENYTRPINVKEVPSTDEFKFLPLQQSS 370
Query: 100 NRRVPSLQFAYDHHAGSEHLRIISDTLQHGSGSGPFGGFQG--EVGKMSAQEI--MEAKA 155
R +P Q D A S S+ G G G + G E G ++ A
Sbjct: 371 QRLLPPAQMQIDFSAASSRA---SENNSDGEGGGEWADAVGADESGNNRPRKRGRRPANG 427
Query: 156 LAASKSHSEAERRRRERINNHLAKLRSLLPSTTKTDKASLLAEVIQHVKEL-------KR 208
A + +H EAER+RRE++N LRS++P+ +K DKASLL + + ++ EL +
Sbjct: 428 RAEALNHVEAERQRREKLNQRFYALRSVVPNISKMDKASLLGDAVSYINELHAKLKVMEA 487
Query: 209 QTSLIAETSPVPTECD-ELTVDAAADDEDYGSNGNKFIIKASLCCDDRSDLLPELIKTLK 267
+ + +S P D ++ V + +D + + C S + +
Sbjct: 488 ERERLGYSSNPPISLDSDINVQTSGED-----------VTVRINCPLESHPASRIFHAFE 536
Query: 268 ALRLRTLKADITTLGGRVKNVLFITGEEDDHEYCISSIQEALKAVMEKSVGDESASG 324
++ + +++ V + + EE E IS AL SV ++SG
Sbjct: 537 ESKVEVINSNLEVSQDTVLHTFVVKSEELTKEKLIS----ALSREQTNSVQSRTSSG 589
>At4g30980 putative bHLH transcription factor (bHLH069)
Length = 310
Score = 66.6 bits (161), Expect = 2e-11
Identities = 50/171 (29%), Positives = 80/171 (46%), Gaps = 11/171 (6%)
Query: 74 HHHHPFNPHDPFVIPQQQASPYASLFNR-RVPSLQFAYDHHAGSEHLRIISDTLQHGSGS 132
HHH + + +IP P +LFN SL F +G + + T + S
Sbjct: 60 HHHDADSRNQITMIPLSHNHPNDALFNGFSTGSLPFHLPQGSGGQ-----TQTQSQATAS 114
Query: 133 GPFGGFQGEVGKMSAQEIMEAKALAASKSHSEAERRRRERINNHLAKLRSLLPSTTKTDK 192
GG + + + A+ A+ HS AER RRERI + L+ L+P+ KTDK
Sbjct: 115 ATTGG---ATAQPQTKPKVRARRGQATDPHSIAERLRRERIAERMKSLQELVPNGNKTDK 171
Query: 193 ASLLAEVIQHVKELKRQTSLI--AETSPVPTECDELTVDAAADDEDYGSNG 241
AS+L E+I +VK L+ Q ++ + + +++ DA E+ S+G
Sbjct: 172 ASMLDEIIDYVKFLQLQVKVLSMSRLGGAASASSQISEDAGGSHENTSSSG 222
>At4g00050 bHLH like transcription factor (bHLH016)
Length = 399
Score = 62.8 bits (151), Expect = 3e-10
Identities = 28/74 (37%), Positives = 52/74 (69%)
Query: 141 EVGKMSAQEIMEAKALAASKSHSEAERRRRERINNHLAKLRSLLPSTTKTDKASLLAEVI 200
E K + + K A+ H+++ER+RR++IN + L+ L+P+++KTDKAS+L EVI
Sbjct: 197 EEKKAGGKSSVSTKRSRAAAIHNQSERKRRDKINQRMKTLQKLVPNSSKTDKASMLDEVI 256
Query: 201 QHVKELKRQTSLIA 214
+++K+L+ Q S+++
Sbjct: 257 EYLKQLQAQVSMMS 270
>At5g61270 bHLH transcription factor like protein
Length = 366
Score = 61.6 bits (148), Expect = 6e-10
Identities = 27/65 (41%), Positives = 46/65 (70%)
Query: 158 ASKSHSEAERRRRERINNHLAKLRSLLPSTTKTDKASLLAEVIQHVKELKRQTSLIAETS 217
A+ H+E+ERRRR+RIN + L+ LLP+ +K DK S+L +VI+H+K+L+ Q ++ +
Sbjct: 167 AAAIHNESERRRRDRINQRMRTLQKLLPTASKADKVSILDDVIEHLKQLQAQVQFMSLRA 226
Query: 218 PVPTE 222
+P +
Sbjct: 227 NLPQQ 231
>At4g36930 putative bHLH transcription factor (AtbHLH024) / SPATULA
(SPT)
Length = 373
Score = 58.5 bits (140), Expect = 5e-09
Identities = 26/61 (42%), Positives = 45/61 (73%)
Query: 153 AKALAASKSHSEAERRRRERINNHLAKLRSLLPSTTKTDKASLLAEVIQHVKELKRQTSL 212
+K A++ H+ +E+RRR RIN + L+SL+P++ KTDKAS+L E I+++K+L+ Q +
Sbjct: 193 SKRCRAAEVHNLSEKRRRSRINEKMKALQSLIPNSNKTDKASMLDEAIEYLKQLQLQVQM 252
Query: 213 I 213
+
Sbjct: 253 L 253
>At5g58010 putative bHLH transcription factor (bHLH082)
Length = 274
Score = 58.2 bits (139), Expect = 6e-09
Identities = 33/81 (40%), Positives = 50/81 (60%), Gaps = 5/81 (6%)
Query: 139 QGEVGKMSAQEIME-----AKALAASKSHSEAERRRRERINNHLAKLRSLLPSTTKTDKA 193
QG V SA + + A+ A+ HS AER RRERI + L+ L+P+T KTDKA
Sbjct: 59 QGTVSTTSAPVVRQKPRVRARRGQATDPHSIAERLRRERIAERMKSLQELVPNTNKTDKA 118
Query: 194 SLLAEVIQHVKELKRQTSLIA 214
S+L E+I++V+ L+ Q +++
Sbjct: 119 SMLDEIIEYVRFLQLQVKVLS 139
>At4g28811 AtbHLH119
Length = 470
Score = 57.8 bits (138), Expect = 8e-09
Identities = 30/72 (41%), Positives = 43/72 (59%)
Query: 143 GKMSAQEIMEAKALAASKSHSEAERRRRERINNHLAKLRSLLPSTTKTDKASLLAEVIQH 202
G A K A+ H+ +ERRRRERIN + L+ LLP KTDK S+L +VI++
Sbjct: 266 GTEEAHGSTSRKRSRAADMHNLSERRRRERINERMKTLQELLPRCRKTDKVSMLEDVIEY 325
Query: 203 VKELKRQTSLIA 214
VK L+ Q +++
Sbjct: 326 VKSLQLQIQMMS 337
>At5g10570 putative bHLH transcription factor (bHLH061)
Length = 315
Score = 57.4 bits (137), Expect = 1e-08
Identities = 49/187 (26%), Positives = 86/187 (45%), Gaps = 19/187 (10%)
Query: 140 GEVGKMSAQEIMEAKALAASKSHSEAERRRRERINNHLAKLRSLLPSTTKTDKASLLAEV 199
GE K + + +E + + AERRRR+R+N+ L+ LRS++P TK D+ S+L +
Sbjct: 134 GETNKKRSNKKLEGQP----SKNLMAERRRRKRLNDRLSLLRSIVPKITKMDRTSILGDA 189
Query: 200 IQHVKELKRQTSLIAETSPVPTECDELTVDAAADDEDYGSNGNKFII-------KASLCC 252
I ++KEL + + + E L+ +E N KF + +CC
Sbjct: 190 IDYMKELLDKINKLQEDEQELGSNSHLS--TLITNESMVRNSLKFEVDQREVNTHIDICC 247
Query: 253 DDRSDLLPELIKTLKALRLRTLKADITTLGG-RVKNVLFITGEEDDHEYCISSIQEALKA 311
+ L+ + TL+ L L + I+ ++ F GE+ Y ++S EA K
Sbjct: 248 PTKPGLVVSTVSTLETLGLEIEQCVISCFSDFSLQASCFEVGEQ---RYMVTS--EATKQ 302
Query: 312 VMEKSVG 318
+ ++ G
Sbjct: 303 ALIRNAG 309
>At4g28790 bHLH transcription factor like protein (bHLH023)
Length = 413
Score = 57.4 bits (137), Expect = 1e-08
Identities = 28/72 (38%), Positives = 46/72 (63%)
Query: 143 GKMSAQEIMEAKALAASKSHSEAERRRRERINNHLAKLRSLLPSTTKTDKASLLAEVIQH 202
G A++ +K A+ H +ERRRR++IN + L+ LLP TKTD++S+L +VI++
Sbjct: 263 GTEEARDSTSSKRSRAAIMHKLSERRRRQKINEMMKALQELLPRCTKTDRSSMLDDVIEY 322
Query: 203 VKELKRQTSLIA 214
VK L+ Q + +
Sbjct: 323 VKSLQSQIQMFS 334
>At2g46510 putative bHLH transcription factor
Length = 662
Score = 57.4 bits (137), Expect = 1e-08
Identities = 28/77 (36%), Positives = 50/77 (64%), Gaps = 4/77 (5%)
Query: 161 SHSEAERRRRERINNHLAKLRSLLPSTTKTDKASLLAEVIQHVKELKRQTSLIAE----T 216
+H EAER+RRE++N LRS++P+ +K DKASLL + I ++KEL+ + ++ + T
Sbjct: 395 NHVEAERQRREKLNQRFYALRSVVPNISKMDKASLLGDAISYIKELQEKVKIMEDERVGT 454
Query: 217 SPVPTECDELTVDAAAD 233
+E + +TV+ + +
Sbjct: 455 DKSLSESNTITVEESPE 471
>At3g59060 putative bHLH transcription factor (bHLH065)
Length = 442
Score = 57.0 bits (136), Expect = 1e-08
Identities = 27/72 (37%), Positives = 46/72 (63%)
Query: 142 VGKMSAQEIMEAKALAASKSHSEAERRRRERINNHLAKLRSLLPSTTKTDKASLLAEVIQ 201
+G S+Q + A++ H+ +ERRRR+RIN + L+ L+P ++TDKAS+L E I
Sbjct: 241 MGNKSSQRSGSTRRSRAAEVHNLSERRRRDRINERMKALQELIPHCSRTDKASILDEAID 300
Query: 202 HVKELKRQTSLI 213
++K L+ Q ++
Sbjct: 301 YLKSLQMQLQVM 312
>At5g57150 unknown protein
Length = 226
Score = 56.6 bits (135), Expect = 2e-08
Identities = 41/169 (24%), Positives = 81/169 (47%), Gaps = 30/169 (17%)
Query: 152 EAKALAASKSHSEAERRRRERINNHLAKLRSLLPSTTKTDKASLLAEVIQHVK----ELK 207
+ A + + + +ER RR+++N L LRS++P+ TK DKAS++ + I +++ E K
Sbjct: 46 DGAASSPASKNIVSERNRRQKLNQRLFALRSVVPNITKMDKASIIKDAISYIEGLQYEEK 105
Query: 208 RQTSLIAETSPVPTECDELTVDAAAD-----------DEDYGSN--------------GN 242
+ + I E P + D D D GS+ G
Sbjct: 106 KLEAEIRELESTPKSSLSFSKDFDRDLLVPVTSKKMKQLDSGSSTSLIEVLELKVTFMGE 165
Query: 243 KFIIKASLCCDDRSDLLPELIKTLKALRLRTLKADITTLGGRVKNVLFI 291
+ ++ S+ C+ R+D + +L + ++L L+ L +++T+ G + + +FI
Sbjct: 166 RTMV-VSVTCNKRTDTMVKLCEVFESLNLKILTSNLTSFSGMIFHTVFI 213
>At4g17880 putative transcription factor BHLH4
Length = 589
Score = 56.6 bits (135), Expect = 2e-08
Identities = 33/109 (30%), Positives = 56/109 (51%), Gaps = 8/109 (7%)
Query: 161 SHSEAERRRRERINNHLAKLRSLLPSTTKTDKASLLAEVIQHVKELKRQTSLIAETSPVP 220
+H EAER+RRE++N LR+++P+ +K DKASLL + I ++ ELK +
Sbjct: 416 NHVEAERQRREKLNQRFYSLRAVVPNVSKMDKASLLGDAISYISELKSKLQ--------K 467
Query: 221 TECDELTVDAAADDEDYGSNGNKFIIKASLCCDDRSDLLPELIKTLKAL 269
E D+ + D + + K +K C + S +L E+ +K +
Sbjct: 468 AESDKEELQKQIDVMNKEAGNAKSSVKDRKCLNQESSVLIEMEVDVKII 516
>At2g43010 putative transcription factor BHLH9
Length = 430
Score = 56.6 bits (135), Expect = 2e-08
Identities = 28/72 (38%), Positives = 45/72 (61%)
Query: 142 VGKMSAQEIMEAKALAASKSHSEAERRRRERINNHLAKLRSLLPSTTKTDKASLLAEVIQ 201
+G S Q + A++ H+ +ERRRR+RIN + L+ L+P +KTDKAS+L E I
Sbjct: 242 IGNKSNQRSGSNRRSRAAEVHNLSERRRRDRINERMKALQELIPHCSKTDKASILDEAID 301
Query: 202 HVKELKRQTSLI 213
++K L+ Q ++
Sbjct: 302 YLKSLQLQLQVM 313
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.315 0.130 0.370
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,400,921
Number of Sequences: 26719
Number of extensions: 314620
Number of successful extensions: 1719
Number of sequences better than 10.0: 185
Number of HSP's better than 10.0 without gapping: 139
Number of HSP's successfully gapped in prelim test: 47
Number of HSP's that attempted gapping in prelim test: 1492
Number of HSP's gapped (non-prelim): 227
length of query: 338
length of database: 11,318,596
effective HSP length: 100
effective length of query: 238
effective length of database: 8,646,696
effective search space: 2057913648
effective search space used: 2057913648
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 60 (27.7 bits)
Medicago: description of AC146570.4


