

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146569.7 + phase: 0
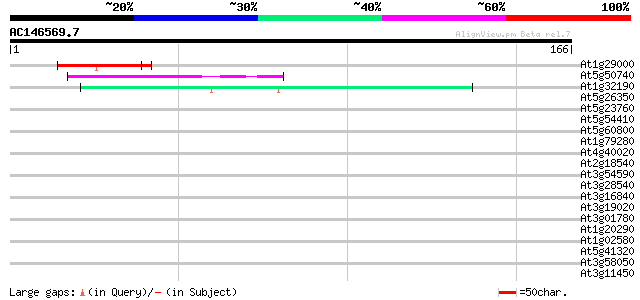
(166 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g29000 hypothetical protein 44 3e-05
At5g50740 putative protein 40 4e-04
At1g32190 unknown protein 40 6e-04
At5g26350 putative protein 37 0.005
At5g23760 unknown protein 36 0.008
At5g54410 unknown protein 35 0.014
At5g60800 unknown protein 35 0.019
At1g79280 hypothetical protein 35 0.019
At4g40020 putative protein 35 0.024
At2g18540 putative vicilin storage protein (globulin-like) 35 0.024
At3g54590 extensin precursor -like protein 34 0.032
At3g28540 hypothetical protein 34 0.032
At3g16840 ATP-dependent RNA helicase 34 0.032
At3g19020 hypothetical protein 34 0.041
At3g01780 unknown protein 34 0.041
At1g20290 hypothetical protein 34 0.041
At1g02580 MEDEA (MEA) 34 0.041
At5g41320 unknown protein 33 0.054
At3g58050 putative protein 33 0.054
At3g11450 putative cell division related protein 33 0.054
>At1g29000 hypothetical protein
Length = 287
Score = 44.3 bits (103), Expect = 3e-05
Identities = 21/25 (84%), Positives = 22/25 (88%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKK 39
EE KKK E+EKKKEEEKKKEEE KK
Sbjct: 198 EEDKKKKEDEKKKEEEKKKEEENKK 222
Score = 41.6 bits (96), Expect = 2e-04
Identities = 23/30 (76%), Positives = 24/30 (79%), Gaps = 2/30 (6%)
Query: 15 EEVKKKTEEE--KKKEEEKKKEEEKKKMME 42
EE KKK EEE KKKE+EKKKEEEKKK E
Sbjct: 190 EEDKKKKEEEDKKKKEDEKKKEEEKKKEEE 219
Score = 38.9 bits (89), Expect = 0.001
Identities = 21/30 (70%), Positives = 22/30 (73%), Gaps = 2/30 (6%)
Query: 15 EEVKKKTEEEKKKEEE--KKKEEEKKKMME 42
E + KTEEEKKKEEE KKKEEE KK E
Sbjct: 176 EIISSKTEEEKKKEEEDKKKKEEEDKKKKE 205
Score = 38.9 bits (89), Expect = 0.001
Identities = 18/25 (72%), Positives = 21/25 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKK 39
E+ KKK EE+KK+EE KKKE EKKK
Sbjct: 205 EDEKKKEEEKKKEEENKKKEGEKKK 229
Score = 37.7 bits (86), Expect = 0.003
Identities = 20/30 (66%), Positives = 24/30 (79%), Gaps = 2/30 (6%)
Query: 15 EEVKKKTEEEKKKEEE--KKKEEEKKKMME 42
EE KK+ E++KKKEEE KKKE+EKKK E
Sbjct: 184 EEKKKEEEDKKKKEEEDKKKKEDEKKKEEE 213
Score = 37.4 bits (85), Expect = 0.004
Identities = 18/28 (64%), Positives = 22/28 (78%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
+E +KK EEEKKKEEE KK+E +KK E
Sbjct: 204 KEDEKKKEEEKKKEEENKKKEGEKKKEE 231
Score = 36.6 bits (83), Expect = 0.006
Identities = 18/23 (78%), Positives = 18/23 (78%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEK 37
EE KKK EE KKKE EKKKEE K
Sbjct: 211 EEEKKKEEENKKKEGEKKKEEVK 233
>At5g50740 putative protein
Length = 162
Score = 40.4 bits (93), Expect = 4e-04
Identities = 24/64 (37%), Positives = 34/64 (52%), Gaps = 8/64 (12%)
Query: 18 KKKTEEEKKKEEEKKKEEEKKKMMEACRTVLFGSCNSCCKTNCNGKCEKCTCGSAKCDGI 77
+KK+EE + K E+KK EEEKKK + +F C C K K +C G +G+
Sbjct: 3 EKKSEEPQAKSEDKKPEEEKKKEPQEIVLKIFMHCEGCAK-----KIHRCLKG---FEGV 54
Query: 78 QCVT 81
+ VT
Sbjct: 55 EDVT 58
Score = 32.0 bits (71), Expect = 0.16
Identities = 18/54 (33%), Positives = 30/54 (55%), Gaps = 1/54 (1%)
Query: 3 KKKFNCVTILS-VEEVKKKTEEEKKKEEEKKKEEEKKKMMEACRTVLFGSCNSC 55
+K V ++S + E K ++E +KKE+EK K EEKK+ + + C +C
Sbjct: 84 RKSHRQVELISPIPEPKPVSDEPEKKEKEKPKPEEKKEEVVTVVLRVHMHCEAC 137
>At1g32190 unknown protein
Length = 422
Score = 40.0 bits (92), Expect = 6e-04
Identities = 30/127 (23%), Positives = 41/127 (31%), Gaps = 11/127 (8%)
Query: 22 EEEKKKEEEKKKEEEKKKMMEACRTVLFGSCNSCCKT----------NCNGKCEKCTCGS 71
E K K +E ++ E+ G C C +C C C C
Sbjct: 271 ENTTTKSRLKTIWQEIRRRDESTGCCCSGLCRPSCSCPKPRCPKPSCSCGCGCGDCGCFK 330
Query: 72 AKCDGIQ-CVTICFKCENPKSCCECKPIIKSCCNNKKCDDGCTPKKPPSPKVQQCPQWCT 130
C ++ C + C K SCC SC KC K P + C C
Sbjct: 331 CSCPTLKGCFSCCKKPSCVSSCCCPTFKCSSCFGKPKCPKCSCWKCLKCPDTECCRSSCC 390
Query: 131 CSRCYGY 137
CS C+ +
Sbjct: 391 CSGCFSW 397
>At5g26350 putative protein
Length = 126
Score = 37.0 bits (84), Expect = 0.005
Identities = 19/30 (63%), Positives = 25/30 (83%), Gaps = 1/30 (3%)
Query: 14 VEEVKKKTEEEKKK-EEEKKKEEEKKKMME 42
+EE KKK E+EKK+ EEEKK+ EE+KK +E
Sbjct: 60 LEEEKKKLEKEKKQLEEEKKQLEEEKKQLE 89
Score = 35.8 bits (81), Expect = 0.011
Identities = 16/28 (57%), Positives = 22/28 (78%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
E++KK EE+ K EEEKKK E++KK +E
Sbjct: 48 EDMKKHKEEKNKLEEEKKKLEKEKKQLE 75
Score = 34.7 bits (78), Expect = 0.024
Identities = 18/29 (62%), Positives = 23/29 (79%), Gaps = 1/29 (3%)
Query: 15 EEVKKKTEEEKKK-EEEKKKEEEKKKMME 42
+E K K EEEKKK E+EKK+ EE+KK +E
Sbjct: 54 KEEKNKLEEEKKKLEKEKKQLEEEKKQLE 82
Score = 29.3 bits (64), Expect = 1.0
Identities = 14/27 (51%), Positives = 21/27 (76%)
Query: 14 VEEVKKKTEEEKKKEEEKKKEEEKKKM 40
+E+ KK+ EEEKK+ EE+KK+ E + M
Sbjct: 67 LEKEKKQLEEEKKQLEEEKKQLEFEVM 93
Score = 28.9 bits (63), Expect = 1.3
Identities = 14/27 (51%), Positives = 19/27 (69%)
Query: 13 SVEEVKKKTEEEKKKEEEKKKEEEKKK 39
+ E KK +EEK K EE+KK+ EK+K
Sbjct: 45 ATNEDMKKHKEEKNKLEEEKKKLEKEK 71
Score = 26.2 bits (56), Expect = 8.6
Identities = 17/47 (36%), Positives = 25/47 (53%), Gaps = 8/47 (17%)
Query: 14 VEEVKKKTEEEKKK-EEEKKKEEEKKKMME-------ACRTVLFGSC 52
+EE KK+ EEEKK+ E E E++K++ C TV+ C
Sbjct: 74 LEEEKKQLEEEKKQLEFEVMGANEREKVLRQLIVLSWGCFTVVIAMC 120
>At5g23760 unknown protein
Length = 103
Score = 36.2 bits (82), Expect = 0.008
Identities = 16/36 (44%), Positives = 28/36 (77%)
Query: 4 KKFNCVTILSVEEVKKKTEEEKKKEEEKKKEEEKKK 39
KK V ++SV K++ +EEKK+E++++K+EEKK+
Sbjct: 59 KKVGKVDLISVGPAKEEKKEEKKEEKKEEKKEEKKE 94
>At5g54410 unknown protein
Length = 219
Score = 35.4 bits (80), Expect = 0.014
Identities = 17/44 (38%), Positives = 25/44 (56%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMMEACRTVLFGSCNSCCKT 58
EE KK EEKKK+ +++E E K++ R L + N CC +
Sbjct: 141 EEKKKDPTEEKKKDPAEEEELEIKRISNDARFCLHDNYNKCCNS 184
Score = 29.6 bits (65), Expect = 0.78
Identities = 17/28 (60%), Positives = 19/28 (67%), Gaps = 3/28 (10%)
Query: 15 EEVKKKTEEEKK---KEEEKKKEEEKKK 39
EE K TEE+KK +EEEK EEKKK
Sbjct: 110 EEEKDLTEEKKKDPTEEEEKDPTEEKKK 137
Score = 29.3 bits (64), Expect = 1.0
Identities = 19/31 (61%), Positives = 22/31 (70%), Gaps = 3/31 (9%)
Query: 12 LSVEEVKKKTEEEKK-KEEEKKKE--EEKKK 39
L+ E+ K TEEE+K EEKKKE EEKKK
Sbjct: 115 LTEEKKKDPTEEEEKDPTEEKKKEPAEEKKK 145
Score = 27.7 bits (60), Expect = 3.0
Identities = 16/28 (57%), Positives = 18/28 (64%), Gaps = 2/28 (7%)
Query: 14 VEEVKKKTEEEKKK--EEEKKKEEEKKK 39
VE KKT+ EK + EEEK EEKKK
Sbjct: 94 VENKLKKTQPEKDRAEEEEKDLTEEKKK 121
Score = 26.2 bits (56), Expect = 8.6
Identities = 10/25 (40%), Positives = 18/25 (72%)
Query: 13 SVEEVKKKTEEEKKKEEEKKKEEEK 37
++E ++KT E+K+ E EK+K+ K
Sbjct: 56 AMETFRRKTNEQKRLENEKRKQALK 80
>At5g60800 unknown protein
Length = 283
Score = 35.0 bits (79), Expect = 0.019
Identities = 23/64 (35%), Positives = 32/64 (49%), Gaps = 2/64 (3%)
Query: 15 EEVKKKTEEEKKKEEEKKK-EEEKKKMMEACRTVLFGSCNSCCKTNCNGKCEKCTCGSAK 73
+E K K +E+KKK EEKKK + KK E T N C+ C GK +K +
Sbjct: 101 KENKNKNDEDKKKSEEKKKPDNNDKKPKETPVTTAVLKLNFHCQ-GCIGKIQKTVTKTKG 159
Query: 74 CDGI 77
+G+
Sbjct: 160 VNGL 163
Score = 32.0 bits (71), Expect = 0.16
Identities = 19/38 (50%), Positives = 26/38 (68%), Gaps = 4/38 (10%)
Query: 3 KKKFNCVTILSVEEVKKKTEEEKKK-EEEKKKEEEKKK 39
KKK V ++S + K+K +E K K +E+KKK EEKKK
Sbjct: 85 KKK---VDLVSPQPKKEKEKENKNKNDEDKKKSEEKKK 119
Score = 30.4 bits (67), Expect = 0.46
Identities = 15/41 (36%), Positives = 25/41 (60%)
Query: 2 LKKKFNCVTILSVEEVKKKTEEEKKKEEEKKKEEEKKKMME 42
L++K T V+ V + ++EK+KE + K +E+KKK E
Sbjct: 76 LREKLEEKTKKKVDLVSPQPKKEKEKENKNKNDEDKKKSEE 116
>At1g79280 hypothetical protein
Length = 2111
Score = 35.0 bits (79), Expect = 0.019
Identities = 15/28 (53%), Positives = 22/28 (78%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
+EV+KKTE+ KKK+EE KE ++K +E
Sbjct: 1532 DEVRKKTEDLKKKDEELTKERSERKSVE 1559
>At4g40020 putative protein
Length = 615
Score = 34.7 bits (78), Expect = 0.024
Identities = 15/26 (57%), Positives = 23/26 (87%)
Query: 14 VEEVKKKTEEEKKKEEEKKKEEEKKK 39
VE V+KK EE++KKEE+K+ ++EKK+
Sbjct: 391 VEVVEKKIEEKEKKEEKKENKKEKKE 416
Score = 30.4 bits (67), Expect = 0.46
Identities = 14/26 (53%), Positives = 21/26 (79%)
Query: 14 VEEVKKKTEEEKKKEEEKKKEEEKKK 39
V EV +K EEK+K+EEKK+ +++KK
Sbjct: 390 VVEVVEKKIEEKEKKEEKKENKKEKK 415
Score = 30.4 bits (67), Expect = 0.46
Identities = 12/29 (41%), Positives = 23/29 (78%)
Query: 14 VEEVKKKTEEEKKKEEEKKKEEEKKKMME 42
+EE +KK E+++ K+E+K+ ++EKK+ E
Sbjct: 398 IEEKEKKEEKKENKKEKKESKKEKKEHSE 426
Score = 29.6 bits (65), Expect = 0.78
Identities = 11/22 (50%), Positives = 20/22 (90%)
Query: 18 KKKTEEEKKKEEEKKKEEEKKK 39
KK++++EKK+ EKK+++EKK+
Sbjct: 414 KKESKKEKKEHSEKKEDKEKKE 435
Score = 28.5 bits (62), Expect = 1.7
Identities = 11/25 (44%), Positives = 20/25 (80%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKK 39
+E KK+ ++EKK+ +++KKE +KK
Sbjct: 404 KEEKKENKKEKKESKKEKKEHSEKK 428
Score = 28.1 bits (61), Expect = 2.3
Identities = 11/25 (44%), Positives = 19/25 (76%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKK 39
E K+K E +K+K+E +K+E+K+K
Sbjct: 409 ENKKEKKESKKEKKEHSEKKEDKEK 433
Score = 26.9 bits (58), Expect = 5.1
Identities = 10/22 (45%), Positives = 18/22 (81%)
Query: 18 KKKTEEEKKKEEEKKKEEEKKK 39
+KK +++KKE +KKE+++KK
Sbjct: 413 EKKESKKEKKEHSEKKEDKEKK 434
Score = 26.2 bits (56), Expect = 8.6
Identities = 12/22 (54%), Positives = 17/22 (76%)
Query: 18 KKKTEEEKKKEEEKKKEEEKKK 39
K+ E +KK EEK+K+EEKK+
Sbjct: 388 KEVVEVVEKKIEEKEKKEEKKE 409
>At2g18540 putative vicilin storage protein (globulin-like)
Length = 699
Score = 34.7 bits (78), Expect = 0.024
Identities = 14/28 (50%), Positives = 21/28 (75%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +K+ EE K++EEE K+ EE++K E
Sbjct: 475 EEARKREEERKREEEEAKRREEERKKRE 502
Score = 34.3 bits (77), Expect = 0.032
Identities = 17/30 (56%), Positives = 23/30 (76%), Gaps = 2/30 (6%)
Query: 15 EEVKKKTEEEKKKEEE--KKKEEEKKKMME 42
EE +K EEE+K+EEE K++EEE+KK E
Sbjct: 474 EEEARKREEERKREEEEAKRREEERKKREE 503
Score = 33.5 bits (75), Expect = 0.054
Identities = 14/28 (50%), Positives = 21/28 (75%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
+E K++ EE +K+EEE+K+EEE K E
Sbjct: 545 QERKRREEEARKREEERKREEEMAKRRE 572
Score = 33.5 bits (75), Expect = 0.054
Identities = 14/28 (50%), Positives = 21/28 (75%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
E K++ EE +K+EEE+K+EEE+ K E
Sbjct: 468 ERKKREEEEARKREEERKREEEEAKRRE 495
Score = 33.1 bits (74), Expect = 0.071
Identities = 16/35 (45%), Positives = 24/35 (67%), Gaps = 4/35 (11%)
Query: 15 EEVKKKTEEEKKKEEE----KKKEEEKKKMMEACR 45
EE K++ EE KK+EEE +K+EEE++K E +
Sbjct: 489 EEAKRREEERKKREEEAEQARKREEEREKEEEMAK 523
Score = 32.3 bits (72), Expect = 0.12
Identities = 17/32 (53%), Positives = 22/32 (68%), Gaps = 4/32 (12%)
Query: 15 EEVKKKTEEE----KKKEEEKKKEEEKKKMME 42
EE +KK EEE +K+EEE++KEEE K E
Sbjct: 495 EEERKKREEEAEQARKREEEREKEEEMAKKRE 526
Score = 32.0 bits (71), Expect = 0.16
Identities = 13/29 (44%), Positives = 21/29 (71%)
Query: 14 VEEVKKKTEEEKKKEEEKKKEEEKKKMME 42
+E +K+ EE +K+EE K++EEE+ K E
Sbjct: 435 IERRRKEEEEARKREEAKRREEEEAKRRE 463
Score = 31.6 bits (70), Expect = 0.21
Identities = 13/23 (56%), Positives = 19/23 (82%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEK 37
EE K++ EE K++EEE+KK EE+
Sbjct: 482 EERKREEEEAKRREEERKKREEE 504
Score = 31.2 bits (69), Expect = 0.27
Identities = 13/29 (44%), Positives = 21/29 (71%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMMEA 43
EE K++ EEE K+ EE++ E +K++ EA
Sbjct: 449 EEAKRREEEEAKRREEEETERKKREEEEA 477
Score = 30.8 bits (68), Expect = 0.35
Identities = 12/29 (41%), Positives = 21/29 (72%)
Query: 14 VEEVKKKTEEEKKKEEEKKKEEEKKKMME 42
VE +++ +E K++EEE +K EE++K E
Sbjct: 537 VERKRREEQERKRREEEARKREEERKREE 565
Score = 30.4 bits (67), Expect = 0.46
Identities = 15/32 (46%), Positives = 22/32 (67%), Gaps = 4/32 (12%)
Query: 15 EEVKKKTEEE----KKKEEEKKKEEEKKKMME 42
EE K++ EEE K++EEE +K EE++K E
Sbjct: 457 EEAKRREEEETERKKREEEEARKREEERKREE 488
Score = 30.4 bits (67), Expect = 0.46
Identities = 13/27 (48%), Positives = 19/27 (70%)
Query: 16 EVKKKTEEEKKKEEEKKKEEEKKKMME 42
E K++ EE ++EEE+K+EEE K E
Sbjct: 648 ERKRREEEAMRREEERKREEEAAKRAE 674
Score = 30.0 bits (66), Expect = 0.60
Identities = 12/28 (42%), Positives = 20/28 (70%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
E+ +K+ EE +K+EE KK EE+++ E
Sbjct: 506 EQARKREEEREKEEEMAKKREEERQRKE 533
Score = 29.6 bits (65), Expect = 0.78
Identities = 14/35 (40%), Positives = 24/35 (68%), Gaps = 4/35 (11%)
Query: 15 EEVKKKTEEE----KKKEEEKKKEEEKKKMMEACR 45
EEV++K EE +++EE +K+EEE+K+ E +
Sbjct: 535 EEVERKRREEQERKRREEEARKREEERKREEEMAK 569
Score = 29.6 bits (65), Expect = 0.78
Identities = 15/30 (50%), Positives = 21/30 (70%), Gaps = 2/30 (6%)
Query: 15 EEVKKKTEEEKKKEEE--KKKEEEKKKMME 42
EE + EEE+K+EEE K+ EEE++K E
Sbjct: 653 EEEAMRREEERKREEEAAKRAEEERRKKEE 682
Score = 29.6 bits (65), Expect = 0.78
Identities = 13/27 (48%), Positives = 22/27 (81%), Gaps = 2/27 (7%)
Query: 15 EEVKKKTEEEKKKEEE--KKKEEEKKK 39
EE +K EEE+K+EEE K++E+E+++
Sbjct: 551 EEEARKREEERKREEEMAKRREQERQR 577
Score = 29.3 bits (64), Expect = 1.0
Identities = 15/29 (51%), Positives = 21/29 (71%), Gaps = 2/29 (6%)
Query: 16 EVKKKTEEE--KKKEEEKKKEEEKKKMME 42
E KK+ EEE K++EE K++EEE K+ E
Sbjct: 468 ERKKREEEEARKREEERKREEEEAKRREE 496
Score = 28.5 bits (62), Expect = 1.7
Identities = 14/27 (51%), Positives = 20/27 (73%), Gaps = 2/27 (7%)
Query: 15 EEVKKKTEEEKKK--EEEKKKEEEKKK 39
EE +K+ EE K+ EE +KKEEE++K
Sbjct: 660 EEERKREEEAAKRAEEERRKKEEEEEK 686
Score = 28.5 bits (62), Expect = 1.7
Identities = 17/37 (45%), Positives = 24/37 (63%), Gaps = 6/37 (16%)
Query: 15 EEVKKKTEEE----KKKEEE--KKKEEEKKKMMEACR 45
EE K+ EEE KK+EEE +K+EEE+K+ E +
Sbjct: 456 EEEAKRREEEETERKKREEEEARKREEERKREEEEAK 492
Score = 28.5 bits (62), Expect = 1.7
Identities = 14/37 (37%), Positives = 24/37 (64%), Gaps = 5/37 (13%)
Query: 14 VEEVKKKTEEE-----KKKEEEKKKEEEKKKMMEACR 45
+EE K++ EEE K++EE +K+EE K++ E +
Sbjct: 424 IEERKRREEEEIERRRKEEEEARKREEAKRREEEEAK 460
Score = 28.1 bits (61), Expect = 2.3
Identities = 13/25 (52%), Positives = 19/25 (76%), Gaps = 3/25 (12%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKK 39
EE K+ EEE++K+EE EEEK++
Sbjct: 667 EEAAKRAEEERRKKEE---EEEKRR 688
Score = 28.1 bits (61), Expect = 2.3
Identities = 13/33 (39%), Positives = 23/33 (69%), Gaps = 5/33 (15%)
Query: 15 EEVKKKTEEEKKKE-----EEKKKEEEKKKMME 42
EE+ KK EEE++++ E K++EE+++K E
Sbjct: 519 EEMAKKREEERQRKEREEVERKRREEQERKRRE 551
Score = 28.1 bits (61), Expect = 2.3
Identities = 10/24 (41%), Positives = 19/24 (78%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKK 38
EE +K+ E ++++EEE K+ EE++
Sbjct: 443 EEARKREEAKRREEEEAKRREEEE 466
Score = 28.1 bits (61), Expect = 2.3
Identities = 14/33 (42%), Positives = 22/33 (66%), Gaps = 5/33 (15%)
Query: 15 EEVKKKTEEEKKKE-----EEKKKEEEKKKMME 42
EE+ K+ E+E++K+ E KK+EEE +K E
Sbjct: 597 EEMAKRREQERQKKEREEMERKKREEEARKREE 629
Score = 28.1 bits (61), Expect = 2.3
Identities = 17/47 (36%), Positives = 27/47 (57%), Gaps = 7/47 (14%)
Query: 6 FNCVTILSVE--EVKKKTEEEKKKEEE-----KKKEEEKKKMMEACR 45
F C + E ++ ++ EE K++EEE +K+EEE +K EA R
Sbjct: 407 FECASCAEGELSKLMREIEERKRREEEEIERRRKEEEEARKREEAKR 453
Score = 27.7 bits (60), Expect = 3.0
Identities = 16/37 (43%), Positives = 25/37 (67%), Gaps = 6/37 (16%)
Query: 15 EEVKKKTEEE--KKKEEEKKKEE----EKKKMMEACR 45
EE ++K EEE K++E+E++K+E E+KK E R
Sbjct: 589 EEQERKREEEMAKRREQERQKKEREEMERKKREEEAR 625
Score = 27.7 bits (60), Expect = 3.0
Identities = 14/29 (48%), Positives = 24/29 (82%), Gaps = 4/29 (13%)
Query: 15 EEVKKKT--EEEKKKEEE--KKKEEEKKK 39
EEV++K E+E+K+EEE K++E+E++K
Sbjct: 581 EEVERKIREEQERKREEEMAKRREQERQK 609
Score = 27.3 bits (59), Expect = 3.9
Identities = 14/31 (45%), Positives = 21/31 (67%), Gaps = 3/31 (9%)
Query: 15 EEVKKKTEE---EKKKEEEKKKEEEKKKMME 42
E KK+ EE +K++EE +K+EEE K+ E
Sbjct: 606 ERQKKEREEMERKKREEEARKREEEMAKIRE 636
Score = 26.6 bits (57), Expect = 6.6
Identities = 13/26 (50%), Positives = 20/26 (76%), Gaps = 2/26 (7%)
Query: 16 EVKKKTEEEKKKEEE--KKKEEEKKK 39
E KK+ EE +K+EEE K +EEE+++
Sbjct: 616 ERKKREEEARKREEEMAKIREEERQR 641
Score = 26.6 bits (57), Expect = 6.6
Identities = 12/30 (40%), Positives = 18/30 (60%)
Query: 16 EVKKKTEEEKKKEEEKKKEEEKKKMMEACR 45
E K+ EEE+ K E+++ E KK+ E R
Sbjct: 449 EEAKRREEEEAKRREEEETERKKREEEEAR 478
Score = 26.2 bits (56), Expect = 8.6
Identities = 16/46 (34%), Positives = 27/46 (57%), Gaps = 3/46 (6%)
Query: 3 KKKFNCVTILSVEEVKKKTE--EEKKKEEE-KKKEEEKKKMMEACR 45
K++ I E +K+ E E K++EEE ++EEE+K+ EA +
Sbjct: 626 KREEEMAKIREEERQRKEREDVERKRREEEAMRREEERKREEEAAK 671
>At3g54590 extensin precursor -like protein
Length = 743
Score = 34.3 bits (77), Expect = 0.032
Identities = 20/58 (34%), Positives = 25/58 (42%), Gaps = 9/58 (15%)
Query: 115 KKPPSPKVQQCPQ---WCTCSRCYGYAPYQPYCNPCP------PPNYMVCYDPNPEPC 163
K PP P V P + + Y +P PY +P P PP+ VC P P PC
Sbjct: 619 KSPPPPYVYSSPPPPYYSPSPKVYYKSPPPPYYSPSPKVYYKSPPHPHVCVCPPPPPC 676
Score = 30.8 bits (68), Expect = 0.35
Identities = 19/50 (38%), Positives = 23/50 (46%), Gaps = 6/50 (12%)
Query: 115 KKPPSPKVQQCPQWCTCS---RCYGYAPYQPYCNPCPPPNYMVCYDPNPE 161
K PP P V P T S + Y +P PY PPP Y Y P+P+
Sbjct: 394 KSPPPPYVYSSPPPPTYSPSPKVYYKSPPPPYVYSSPPPPY---YSPSPK 440
Score = 30.0 bits (66), Expect = 0.60
Identities = 17/50 (34%), Positives = 22/50 (44%), Gaps = 6/50 (12%)
Query: 115 KKPPSPKVQQCPQ---WCTCSRCYGYAPYQPYCNPCPPPNYMVCYDPNPE 161
K PP P V P + + Y +P PY PPP Y Y P+P+
Sbjct: 444 KSPPPPYVYSSPPPPYYSPSPKVYYKSPPPPYVYSSPPPPY---YSPSPK 490
Score = 30.0 bits (66), Expect = 0.60
Identities = 17/50 (34%), Positives = 22/50 (44%), Gaps = 6/50 (12%)
Query: 115 KKPPSPKVQQCPQ---WCTCSRCYGYAPYQPYCNPCPPPNYMVCYDPNPE 161
K PP P V P + + Y +P PY PPP Y Y P+P+
Sbjct: 569 KSPPPPYVYNSPPPPYYSPSPKVYYKSPPPPYVYSSPPPPY---YSPSPK 615
Score = 30.0 bits (66), Expect = 0.60
Identities = 17/50 (34%), Positives = 22/50 (44%), Gaps = 6/50 (12%)
Query: 115 KKPPSPKVQQCPQ---WCTCSRCYGYAPYQPYCNPCPPPNYMVCYDPNPE 161
K PP P V P + + Y +P PY PPP Y Y P+P+
Sbjct: 469 KSPPPPYVYSSPPPPYYSPSPKVYYKSPPPPYVYSSPPPPY---YSPSPK 515
Score = 30.0 bits (66), Expect = 0.60
Identities = 17/50 (34%), Positives = 22/50 (44%), Gaps = 6/50 (12%)
Query: 115 KKPPSPKVQQCPQ---WCTCSRCYGYAPYQPYCNPCPPPNYMVCYDPNPE 161
K PP P V P + + Y +P PY PPP Y Y P+P+
Sbjct: 419 KSPPPPYVYSSPPPPYYSPSPKVYYKSPPPPYVYSSPPPPY---YSPSPK 465
Score = 30.0 bits (66), Expect = 0.60
Identities = 17/50 (34%), Positives = 22/50 (44%), Gaps = 6/50 (12%)
Query: 115 KKPPSPKVQQCPQ---WCTCSRCYGYAPYQPYCNPCPPPNYMVCYDPNPE 161
K PP P V P + + Y +P PY PPP Y Y P+P+
Sbjct: 494 KSPPPPYVYSSPPPPYYSPSPKVYYKSPPPPYVYSSPPPPY---YSPSPK 540
Score = 30.0 bits (66), Expect = 0.60
Identities = 17/50 (34%), Positives = 22/50 (44%), Gaps = 6/50 (12%)
Query: 115 KKPPSPKVQQCPQ---WCTCSRCYGYAPYQPYCNPCPPPNYMVCYDPNPE 161
K PP P V P + + Y +P PY PPP Y Y P+P+
Sbjct: 594 KSPPPPYVYSSPPPPYYSPSPKVYYKSPPPPYVYSSPPPPY---YSPSPK 640
Score = 28.1 bits (61), Expect = 2.3
Identities = 16/50 (32%), Positives = 22/50 (44%), Gaps = 6/50 (12%)
Query: 115 KKPPSPKVQQCPQ---WCTCSRCYGYAPYQPYCNPCPPPNYMVCYDPNPE 161
K PP P V P + + + +P PY PPP Y Y P+P+
Sbjct: 519 KSPPPPYVYSSPPPPYYSPSPKVHYKSPPPPYVYSSPPPPY---YSPSPK 565
>At3g28540 hypothetical protein
Length = 510
Score = 34.3 bits (77), Expect = 0.032
Identities = 16/31 (51%), Positives = 22/31 (70%)
Query: 7 NCVTILSVEEVKKKTEEEKKKEEEKKKEEEK 37
+C L+ + KKK E+E ++EEEKKKE EK
Sbjct: 300 DCSLDLTGQRKKKKEEDEDEEEEEKKKEAEK 330
Score = 29.3 bits (64), Expect = 1.0
Identities = 12/21 (57%), Positives = 18/21 (85%)
Query: 18 KKKTEEEKKKEEEKKKEEEKK 38
KKK EE++ +EEE+KK+E +K
Sbjct: 310 KKKKEEDEDEEEEEKKKEAEK 330
Score = 27.7 bits (60), Expect = 3.0
Identities = 10/25 (40%), Positives = 21/25 (84%)
Query: 18 KKKTEEEKKKEEEKKKEEEKKKMME 42
+KK +EE + EEE++K++E +K+++
Sbjct: 309 RKKKKEEDEDEEEEEKKKEAEKLLK 333
Score = 27.3 bits (59), Expect = 3.9
Identities = 12/23 (52%), Positives = 17/23 (73%)
Query: 21 TEEEKKKEEEKKKEEEKKKMMEA 43
T + KKK+EE + EEE++K EA
Sbjct: 306 TGQRKKKKEEDEDEEEEEKKKEA 328
Score = 26.9 bits (58), Expect = 5.1
Identities = 16/46 (34%), Positives = 23/46 (49%), Gaps = 5/46 (10%)
Query: 15 EEVKKKTEEEKKKEEEK-----KKEEEKKKMMEACRTVLFGSCNSC 55
EE + + EEEKKKE EK + E E K + + G ++C
Sbjct: 314 EEDEDEEEEEKKKEAEKLLKRERGERESKVTLSGLLNAIDGLWSAC 359
>At3g16840 ATP-dependent RNA helicase
Length = 832
Score = 34.3 bits (77), Expect = 0.032
Identities = 14/25 (56%), Positives = 21/25 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKK 39
EE +KK E++ K+ +EKKKE++KKK
Sbjct: 128 EETRKKKEKKAKRNKEKKKEKKKKK 152
Score = 28.9 bits (63), Expect = 1.3
Identities = 15/31 (48%), Positives = 22/31 (70%), Gaps = 3/31 (9%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMMEACR 45
+E K K +EKKKE++KKK+ KK+ EA +
Sbjct: 134 KEKKAKRNKEKKKEKKKKKQ---KKINEAAK 161
>At3g19020 hypothetical protein
Length = 951
Score = 33.9 bits (76), Expect = 0.041
Identities = 21/70 (30%), Positives = 31/70 (44%), Gaps = 3/70 (4%)
Query: 61 NGKCEKCTCGSAKCDGIQCVTICFKCE-NPKSCCECKPIIKSC--CNNKKCDDGCTPKKP 117
NG+ + C GS++ + C + N KS EC P++ C+ KC G
Sbjct: 342 NGEAQSCVPGSSQEKQFDDTSNCLQNRPNQKSAKECLPVVSRPVDCSKDKCAGGGGGGSN 401
Query: 118 PSPKVQQCPQ 127
PSPK P+
Sbjct: 402 PSPKPTPTPK 411
>At3g01780 unknown protein
Length = 1192
Score = 33.9 bits (76), Expect = 0.041
Identities = 14/25 (56%), Positives = 23/25 (92%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKK 39
EEVK+K E+E+ K++E+KK++EK+K
Sbjct: 1131 EEVKEKKEKEEGKDKEEKKKKEKEK 1155
Score = 30.8 bits (68), Expect = 0.35
Identities = 14/28 (50%), Positives = 21/28 (75%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
E+ ++ +E+K+KEE K KEE+KKK E
Sbjct: 1127 EDDDEEVKEKKEKEEGKDKEEKKKKEKE 1154
Score = 27.3 bits (59), Expect = 3.9
Identities = 13/25 (52%), Positives = 18/25 (72%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKK 39
EE + +EKK++EE K +EEKKK
Sbjct: 1126 EEDDDEEVKEKKEKEEGKDKEEKKK 1150
Score = 26.9 bits (58), Expect = 5.1
Identities = 15/37 (40%), Positives = 21/37 (56%), Gaps = 2/37 (5%)
Query: 2 LKKKFNCVTILSVEEVKK--KTEEEKKKEEEKKKEEE 36
LK + +L + KK K EEE + EEE++ EEE
Sbjct: 1091 LKISMERIALLKAAQPKKTSKIEEESENEEEEEGEEE 1127
Score = 26.2 bits (56), Expect = 8.6
Identities = 12/28 (42%), Positives = 19/28 (67%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE E ++KKE+E+ K++E+KK E
Sbjct: 1125 EEEDDDEEVKEKKEKEEGKDKEEKKKKE 1152
>At1g20290 hypothetical protein
Length = 497
Score = 33.9 bits (76), Expect = 0.041
Identities = 14/27 (51%), Positives = 24/27 (88%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMM 41
EE +++ EEE+++EEE+++EEE+KK M
Sbjct: 459 EEEEEEEEEEEEEEEEEEEEEEEKKYM 485
Score = 33.5 bits (75), Expect = 0.054
Identities = 14/27 (51%), Positives = 24/27 (88%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMM 41
EE +++ EEE+++EEE+++EEEKK M+
Sbjct: 460 EEEEEEEEEEEEEEEEEEEEEEKKYMI 486
Score = 33.5 bits (75), Expect = 0.054
Identities = 14/33 (42%), Positives = 27/33 (81%)
Query: 10 TILSVEEVKKKTEEEKKKEEEKKKEEEKKKMME 42
TI+ EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 359 TIVQAEEEEEEEEEEEEEEEEEEEEEEEEEEEE 391
Score = 30.8 bits (68), Expect = 0.35
Identities = 12/25 (48%), Positives = 23/25 (92%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKK 39
EE +++ EEE+++EEE+++EEE++K
Sbjct: 458 EEEEEEEEEEEEEEEEEEEEEEEEK 482
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 416 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 443
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 370 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 397
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 369 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 396
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 380 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 407
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 371 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 398
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 382 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 409
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 383 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 410
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 396 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 423
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 375 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 402
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 403 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 430
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 397 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 424
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 378 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 405
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 433 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 460
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 398 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 425
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 399 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 426
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 450 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 477
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 412 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 439
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 448 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 475
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 365 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 392
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 413 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 440
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 418 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 445
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 417 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 444
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 393 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 420
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 410 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 437
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 389 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 416
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 391 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 418
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 392 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 419
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 423 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 450
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 424 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 451
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 394 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 421
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 395 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 422
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 440 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 467
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 437 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 464
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 438 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 465
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 439 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 466
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 376 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 403
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 368 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 395
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 373 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 400
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 377 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 404
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 405 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 432
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 404 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 431
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 379 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 406
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 401 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 428
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 406 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 433
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 407 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 434
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 408 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 435
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 366 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 393
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 374 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 401
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 387 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 414
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 414 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 441
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 386 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 413
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 384 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 411
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 381 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 408
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 372 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 399
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 422 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 449
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 415 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 442
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 390 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 417
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 421 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 448
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 420 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 447
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 367 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 394
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 425 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 452
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 426 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 453
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 427 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 454
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 428 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 455
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 429 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 456
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 430 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 457
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 431 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 458
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 432 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 459
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 402 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 429
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 434 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 461
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 435 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 462
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 436 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 463
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 409 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 436
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 451 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 478
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 385 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 412
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 453 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 480
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 441 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 468
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 442 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 469
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 443 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 470
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 444 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 471
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 445 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 472
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 446 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 473
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 447 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 474
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 400 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 427
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 419 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 446
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 454 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 481
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 411 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 438
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 449 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 476
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 388 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 415
Score = 29.6 bits (65), Expect = 0.78
Identities = 12/28 (42%), Positives = 24/28 (84%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKKMME 42
EE +++ EEE+++EEE+++EEE+++ E
Sbjct: 452 EEEEEEEEEEEEEEEEEEEEEEEEEEEE 479
Score = 27.3 bits (59), Expect = 3.9
Identities = 13/25 (52%), Positives = 19/25 (76%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKKK 39
EE +++ EEE+++EEEKK KKK
Sbjct: 466 EEEEEEEEEEEEEEEEKKYMIVKKK 490
Score = 26.6 bits (57), Expect = 6.6
Identities = 13/29 (44%), Positives = 23/29 (78%), Gaps = 4/29 (13%)
Query: 15 EEVKKKTEEEKKKEEEKKK----EEEKKK 39
EE +++ EEE+++EEE+KK +++KKK
Sbjct: 465 EEEEEEEEEEEEEEEEEKKYMIVKKKKKK 493
>At1g02580 MEDEA (MEA)
Length = 689
Score = 33.9 bits (76), Expect = 0.041
Identities = 29/105 (27%), Positives = 40/105 (37%), Gaps = 13/105 (12%)
Query: 17 VKKKTEEEKKKEEEKKKEEEKKKMMEACRTVLFGSC--NSC-CKTNCNGKCEKCTCGSAK 73
+KK T E K + K K + C + +C C C +CN + C C +
Sbjct: 433 LKKTTSGEAKFYKHYTPCTCKSKCGQQCPCLTHENCCEKYCGCSKDCNNRFGGCNCAIGQ 492
Query: 74 CDGIQCVTICFKCENPKSCCECKP-IIKSCCNNKKCDDGCTPKKP 117
C QC CF EC P + +SC C DG + P
Sbjct: 493 CTNRQCP--CFAANR-----ECDPDLCRSC--PLSCGDGTLGETP 528
>At5g41320 unknown protein
Length = 515
Score = 33.5 bits (75), Expect = 0.054
Identities = 21/56 (37%), Positives = 28/56 (49%), Gaps = 2/56 (3%)
Query: 18 KKKTEEEK--KKEEEKKKEEEKKKMMEACRTVLFGSCNSCCKTNCNGKCEKCTCGS 71
KKK EEEK KKEEE+ KE EK+ ++ + NG+ +C GS
Sbjct: 168 KKKEEEEKMKKKEEEETKESEKQSKPGESGLEMYIPEEKSSEIAHNGRDRECNAGS 223
Score = 30.4 bits (67), Expect = 0.46
Identities = 17/40 (42%), Positives = 24/40 (59%), Gaps = 2/40 (5%)
Query: 5 KFNCVTILSVEEVKKKTEEEKKKEEE--KKKEEEKKKMME 42
K + + VKK+ ++KK+EEE KKKEEE+ K E
Sbjct: 149 KRGVAAVAAKRPVKKRMTKKKKEEEEKMKKKEEEETKESE 188
>At3g58050 putative protein
Length = 1209
Score = 33.5 bits (75), Expect = 0.054
Identities = 14/24 (58%), Positives = 20/24 (83%)
Query: 15 EEVKKKTEEEKKKEEEKKKEEEKK 38
EE K+K EEE++KE+++ KE EKK
Sbjct: 534 EEEKEKREEEERKEKKRSKEREKK 557
Score = 31.6 bits (70), Expect = 0.21
Identities = 13/35 (37%), Positives = 25/35 (71%)
Query: 8 CVTILSVEEVKKKTEEEKKKEEEKKKEEEKKKMME 42
C I+++E+ K EEE+K++ E+++ +EKK+ E
Sbjct: 519 CKEIITLEKQVKLLEEEEKEKREEEERKEKKRSKE 553
Score = 28.9 bits (63), Expect = 1.3
Identities = 14/32 (43%), Positives = 23/32 (71%), Gaps = 2/32 (6%)
Query: 9 VTILSVEEVKKKTEEEKKKEEEKKKEEEKKKM 40
V +L EE +K+ EEE+K E+K+ +E +KK+
Sbjct: 529 VKLLEEEEKEKREEEERK--EKKRSKEREKKL 558
>At3g11450 putative cell division related protein
Length = 663
Score = 33.5 bits (75), Expect = 0.054
Identities = 31/122 (25%), Positives = 58/122 (47%), Gaps = 11/122 (9%)
Query: 3 KKKFNCVTILSVEEVKKKTEEEKKK-----EEEKKKEEEKKKMMEACRT---VLFGSCNS 54
KKK ++ EE K++ EEE+K+ +++KK +E +KK++ R L +
Sbjct: 336 KKKQEEDAAIAAEEEKRRKEEEEKRAAESAQQQKKTKEREKKLLRKERNRLRTLSAPLVA 395
Query: 55 CCKTNCNGKCEKCTCGSAKCDGIQCVTICFKCENPKSCCECKPIIKSCCNNKKCDDGCTP 114
+ + + + C S + +Q +C K N K E +IK CN+ + D+ +
Sbjct: 396 QRLLDISEEDIENLCMSLNTEQLQ--NLCDKMGN-KEGLELAKVIKDGCNSSRNDEAESK 452
Query: 115 KK 116
+K
Sbjct: 453 EK 454
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.135 0.478
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,195,907
Number of Sequences: 26719
Number of extensions: 280214
Number of successful extensions: 7874
Number of sequences better than 10.0: 359
Number of HSP's better than 10.0 without gapping: 251
Number of HSP's successfully gapped in prelim test: 115
Number of HSP's that attempted gapping in prelim test: 4090
Number of HSP's gapped (non-prelim): 2374
length of query: 166
length of database: 11,318,596
effective HSP length: 92
effective length of query: 74
effective length of database: 8,860,448
effective search space: 655673152
effective search space used: 655673152
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 56 (26.2 bits)
Medicago: description of AC146569.7


