

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146569.5 + phase: 0
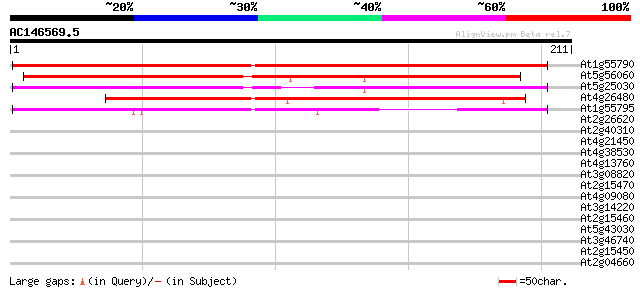
(211 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g55790 hypothetical protein 201 3e-52
At5g56060 putative protein 179 7e-46
At5g25030 putative protein 155 2e-38
At4g26480 putative protein 142 1e-34
At1g55795 unknown protein 132 1e-31
At2g26620 putative polygalacturonase 34 0.064
At2g40310 putative polygalacturonase 32 0.32
At4g21450 unknown protein 30 0.71
At4g38530 phosphoinositide-specific phospholipase C 30 0.93
At4g13760 putative polygalacturonase 30 0.93
At3g08820 unknown protein 30 1.2
At2g15470 putative polygalacturonase 29 1.6
At4g09080 outer envelope membrane protein OEP75 precursor homolog 28 4.6
At3g14220 myrosinase-associated protein like 28 4.6
At2g15460 putative polygalacturonase 28 4.6
At5g43030 CHP-rich zinc finger protein-like 27 6.0
At3g46740 chloroplast import-associated channel protein homolog 27 6.0
At2g15450 putative polygalacturonase 27 6.0
At2g04660 unknown protein 27 7.9
>At1g55790 hypothetical protein
Length = 383
Score = 201 bits (510), Expect = 3e-52
Identities = 100/201 (49%), Positives = 134/201 (65%), Gaps = 1/201 (0%)
Query: 2 EEKIVKHYSSFHNILLVGEGDFSFALCLAKAFGSAVNMVATSLDDRGSLAMKYRGAIRNL 61
EE VKHYSS H ILLVGEGDFSF+ LA FGSA N+ A+SLD + KY+ A NL
Sbjct: 17 EEVWVKHYSSNHQILLVGEGDFSFSHSLATLFGSASNICASSLDSYDVVVRKYKKARSNL 76
Query: 62 IELEGLGCTIMHEVDVHNMNQHHQLKHHNFFDRIIFNFPHSGFFQNESDAWVIGEHKKLV 121
L+ LG ++H VD ++ H L++ F DR+IFNFPH+GF ESD+ +I +H++LV
Sbjct: 77 KTLKRLGALLLHGVDATTLHFHPDLRYRRF-DRVIFNFPHAGFHGRESDSSLIRKHRELV 135
Query: 122 SGFLGSAKYMLNVGGEIHITHKTAHPFSNWNIKNLAENEKLLFIEEVTFYQHFYPGYGNK 181
GF A +L GE+H++HK PFS WN++ LA L+ I+ V F ++ YPGY NK
Sbjct: 136 FGFFNGASRLLRANGEVHVSHKNKAPFSEWNLEELASRCFLVLIQRVAFEKNNYPGYENK 195
Query: 182 KGAGFKCDKSFPIGKCSTFKF 202
+G G +CD+ F +G+CSTFKF
Sbjct: 196 RGDGRRCDQPFLLGECSTFKF 216
>At5g56060 putative protein
Length = 220
Score = 179 bits (455), Expect = 7e-46
Identities = 95/189 (50%), Positives = 126/189 (66%), Gaps = 5/189 (2%)
Query: 6 VKHYSSFHNILLVGEGDFSFALCLAKAFGSAVNMVATSLDDRGSLAMKYRGAIRNLIELE 65
++ YS+ ILLVGEGDFSF+L LA+ FGSA N+ ATSLD R L +KY N+ LE
Sbjct: 9 LQQYSNKQKILLVGEGDFSFSLSLARVFGSATNITATSLDTREELGIKYTDGKANVEGLE 68
Query: 66 GLGCTIMHEVDVHNMNQHHQLKHHNFFDRIIFNFPHSGF-FQNESDAWVIGEHKKLVSGF 124
GCT++H V+VH+M+ ++L +DRIIFNFPHSG F +E D + I H+ LV GF
Sbjct: 69 LFGCTVVHGVNVHSMSSDYRLGR---YDRIIFNFPHSGLGFGSEHDIFFIMLHQGLVRGF 125
Query: 125 LGSAKYML-NVGGEIHITHKTAHPFSNWNIKNLAENEKLLFIEEVTFYQHFYPGYGNKKG 183
L SA+ ML + GEIH+THKT PF+ W I+ LA + L I E+ F++ +PGY NKKG
Sbjct: 126 LESARKMLKDEDGEIHVTHKTTDPFNRWGIETLAGEKGLRLIGEIEFHKWAFPGYSNKKG 185
Query: 184 AGFKCDKSF 192
G C+ +F
Sbjct: 186 GGSNCNSTF 194
>At5g25030 putative protein
Length = 193
Score = 155 bits (391), Expect = 2e-38
Identities = 86/202 (42%), Positives = 118/202 (57%), Gaps = 16/202 (7%)
Query: 2 EEKIVKHYSSFHNILLVGEGDFSFALCLAKAFGSAVNMVATSLDDRGSLAMKYRGAIRNL 61
E K + YS+ IL+VGEG+FSF+L LAKA GSA N+ A SLD R L Y N+
Sbjct: 5 ESKRLSRYSNEQKILVVGEGEFSFSLSLAKALGSATNITAISLDIREDLGRNYNNGKGNV 64
Query: 62 IELEGLGCTIMHEVDVHNMNQHHQLKHHNFFDRIIFNFPHSGFFQNESDAWVIGEHKKLV 121
ELE LGCT++ V+VH+M +L H +D IIFNFPH+ G+ K+
Sbjct: 65 EELERLGCTVVRGVNVHSMKSDDRLAH---YDIIIFNFPHA------------GKRNKVF 109
Query: 122 SGFLGSAKYML-NVGGEIHITHKTAHPFSNWNIKNLAENEKLLFIEEVTFYQHFYPGYGN 180
GF+ SA+ M+ + GEIHIT T +PF+ W++K LAE L I+ + F + +P N
Sbjct: 110 GGFMESAREMMKDEDGEIHITLNTLNPFNKWDLKALAEESGLRLIQRMQFIKWAFPSSSN 169
Query: 181 KKGAGFKCDKSFPIGKCSTFKF 202
K+ +G CD +PIG T+ F
Sbjct: 170 KRESGSNCDFIYPIGSAITYMF 191
>At4g26480 putative protein
Length = 555
Score = 142 bits (359), Expect = 1e-34
Identities = 76/161 (47%), Positives = 101/161 (62%), Gaps = 4/161 (2%)
Query: 37 VNMVATSLDDRGSLAMKYRGAIRNLIELEGLGCTIMHEVDVHNMNQHHQLKHHNFFDRII 96
+N+ ATSLD L++KY A+ N+ L+ GC I HEVDVH M+ + L + DRI+
Sbjct: 1 MNITATSLDSEDELSIKYMDAVDNINILKRYGCDIQHEVDVHTMSFDNSLSLQRY-DRIV 59
Query: 97 FNFPHSG--FFQNESDAWVIGEHKKLVSGFLGSAKYMLNVGGEIHITHKTAHPFSNWNIK 154
FNFPH+G FF E + I HK+LV GFL +AK ML GEIHITHKT +PFS+W IK
Sbjct: 60 FNFPHAGSRFFGRELSSRAIESHKELVRGFLENAKEMLEEDGEIHITHKTTYPFSDWGIK 119
Query: 155 NLAENEKLLFIEEVTFYQHFYPGYGNKKGA-GFKCDKSFPI 194
L + E L +++ F YPGY K+G+ G + D FP+
Sbjct: 120 KLGKGEGLKLLKKSKFELSHYPGYITKRGSGGRRSDDYFPV 160
>At1g55795 unknown protein
Length = 314
Score = 132 bits (333), Expect = 1e-31
Identities = 87/232 (37%), Positives = 113/232 (48%), Gaps = 61/232 (26%)
Query: 2 EEKIVKHYSSFHNILLVGEGDFSFALCLAKAFGSAVNMVATSLD-------DRG------ 48
EE VKHYSS H ILLVGEGDFSF+ LA FGSA N+ A+SLD ++G
Sbjct: 15 EEVWVKHYSSKHQILLVGEGDFSFSCSLATCFGSASNIYASSLDSYDYKPVNKGCSFKLD 74
Query: 49 --------------SLAMKYRGAIRNLIELEGLGCTIMHEVDVHNMNQHHQLKHHNFFDR 94
+ KY+ A NL L+ LG ++H VD ++ H L++ F DR
Sbjct: 75 FLSCCMSFMVIKPDDVVRKYKNARSNLETLKRLGAFLLHGVDATTLHFHPDLRYRRF-DR 133
Query: 95 IIFNFPHSGFFQNESDAWVI----GEHKKLVSGFLGSAKYMLNVGGEIHITHKTAHPFSN 150
+IFNFPH+GF + ESD I + L FL A +ML GE+
Sbjct: 134 VIFNFPHTGFHRKESDPCQIQPAAATLRNLFKDFLHGASHMLRADGELE----------- 182
Query: 151 WNIKNLAENEKLLFIEEVTFYQHFYPGYGNKKGAGFKCDKSFPIGKCSTFKF 202
F + YPGY NK+G G +CD+ F +G+CSTFKF
Sbjct: 183 ------------------AFEKRNYPGYENKRGDGSRCDQPFLLGECSTFKF 216
>At2g26620 putative polygalacturonase
Length = 402
Score = 33.9 bits (76), Expect = 0.064
Identities = 33/135 (24%), Positives = 52/135 (38%), Gaps = 13/135 (9%)
Query: 62 IELEGLGCTIMHEVDVHNMNQHHQLKHHN--FFDRIIFNFPHSGFFQNESDAWVIGEHKK 119
+ + + C H + V ++ ++ K F +IFN G W K
Sbjct: 235 LNISNINCGPGHGISVGSLGKNKDEKDVKDLFVRDVIFNGTSDGI---RIKTWESSASKI 291
Query: 120 LVSGFLGSAKYMLNVGGEIHITHK-TAHPFSNWNIKNLAENEKLLFIEEVTFYQHFYPGY 178
LVS F+ M++VG I+I K HP K+ + + L ++ Y
Sbjct: 292 LVSNFVYENIQMIDVGKPINIDQKYCPHPPCEHERKSHVQIQDLKL-------KNIYGTS 344
Query: 179 GNKKGAGFKCDKSFP 193
NK +C KSFP
Sbjct: 345 KNKVAVNLQCSKSFP 359
>At2g40310 putative polygalacturonase
Length = 404
Score = 31.6 bits (70), Expect = 0.32
Identities = 34/135 (25%), Positives = 58/135 (42%), Gaps = 11/135 (8%)
Query: 62 IELEGLGCTIMHEVDVHNMNQHHQLKH-HNFFDR-IIFNFPHSGFFQNESDAWVIGEHKK 119
+++ + C H + V ++ ++ K N R +IFN G W K
Sbjct: 235 LDISNVKCGPGHGISVGSLGKNKDEKDVKNLTVRDVIFNGTSDGI---RIKTWESSASKI 291
Query: 120 LVSGFLGSAKYMLNVGGEIHITHK-TAHPFSNWNIKNLAENEKLLFIEEVTFYQHFYPGY 178
LVS F+ M++VG I+I K HP ++ + E + I+++ ++ Y
Sbjct: 292 LVSNFVYENIQMIDVGKPINIDQKYCPHP----PCEHEQKGESHVQIQDLKL-KNIYGTS 346
Query: 179 GNKKGAGFKCDKSFP 193
NK +C KSFP
Sbjct: 347 KNKVAMNLQCSKSFP 361
>At4g21450 unknown protein
Length = 295
Score = 30.4 bits (67), Expect = 0.71
Identities = 13/33 (39%), Positives = 19/33 (57%), Gaps = 1/33 (3%)
Query: 77 VHNMNQHHQLKHHNFFDRIIFNFPH-SGFFQNE 108
+HN +QHH HH ++ +N PH G QN+
Sbjct: 54 LHNHHQHHHQHHHQHHHQLGYNGPHGDGSGQNQ 86
>At4g38530 phosphoinositide-specific phospholipase C
Length = 526
Score = 30.0 bits (66), Expect = 0.93
Identities = 14/32 (43%), Positives = 18/32 (55%)
Query: 71 IMHEVDVHNMNQHHQLKHHNFFDRIIFNFPHS 102
I H V HN+ HH L H N F R +F+ +S
Sbjct: 27 IFHSVKHHNVFHHHGLVHLNAFYRYLFSDTNS 58
>At4g13760 putative polygalacturonase
Length = 375
Score = 30.0 bits (66), Expect = 0.93
Identities = 32/135 (23%), Positives = 54/135 (39%), Gaps = 11/135 (8%)
Query: 62 IELEGLGCTIMHEVDVHNMNQHHQLKHHNFFD--RIIFNFPHSGFFQNESDAWVIGEHKK 119
+++ + C H + V ++ ++ K +IFN G W K
Sbjct: 206 LDISNVNCGPGHGISVGSLGKNKDEKDVKDLTIRDVIFNGTSDGI---RIKTWESSASKI 262
Query: 120 LVSGFLGSAKYMLNVGGEIHITHK-TAHPFSNWNIKNLAENEKLLFIEEVTFYQHFYPGY 178
LVS FL M++VG I+I K HP ++ + E + I+ + ++ Y
Sbjct: 263 LVSNFLYENIQMIDVGKPINIDQKYCPHP----PCEHEQKGESHVQIQNLKL-KNIYGTS 317
Query: 179 GNKKGAGFKCDKSFP 193
NK +C K FP
Sbjct: 318 KNKVAVNLQCSKRFP 332
>At3g08820 unknown protein
Length = 685
Score = 29.6 bits (65), Expect = 1.2
Identities = 19/91 (20%), Positives = 42/91 (45%), Gaps = 4/91 (4%)
Query: 58 IRNLIELEGLGCTIMHEVDVHNMNQHHQLKHHNFFDRIIFNFPHSGFFQNESDAWVIGEH 117
++ + L + CT+ H +H +H L H F ++ + FF+ ++++ H
Sbjct: 13 VQQIKTLISVACTVNHLKQIHVSLINHHLHHDTFLVNLLLK--RTLFFRQTKYSYLLFSH 70
Query: 118 KKLVSGFLGSAKYMLNVGGEIHITHKTAHPF 148
+ + FL ++ ++N H+ H+T F
Sbjct: 71 TQFPNIFLYNS--LINGFVNNHLFHETLDLF 99
>At2g15470 putative polygalacturonase
Length = 402
Score = 29.3 bits (64), Expect = 1.6
Identities = 31/135 (22%), Positives = 50/135 (36%), Gaps = 13/135 (9%)
Query: 62 IELEGLGCTIMHEVDVHNMNQHHQLKHHN--FFDRIIFNFPHSGFFQNESDAWVIGEHKK 119
+ + + C H + V ++ ++ K +IFN G W K
Sbjct: 235 LNISNVNCGPGHGISVGSLGKNKDEKDAKDLIVRDVIFNGTSDGI---RIKTWESSASKI 291
Query: 120 LVSGFLGSAKYMLNVGGEIHITHK-TAHPFSNWNIKNLAENEKLLFIEEVTFYQHFYPGY 178
LVS F+ M++VG I+I K HP K+ + + L ++ Y
Sbjct: 292 LVSNFVYENIQMIDVGKPINIDQKYCPHPPCEHERKSHVQIQNLKL-------KNIYGTS 344
Query: 179 GNKKGAGFKCDKSFP 193
NK +C K FP
Sbjct: 345 KNKVAMNLQCSKIFP 359
>At4g09080 outer envelope membrane protein OEP75 precursor homolog
Length = 407
Score = 27.7 bits (60), Expect = 4.6
Identities = 20/65 (30%), Positives = 30/65 (45%), Gaps = 2/65 (3%)
Query: 123 GFLGSAKYMLNVGGEIHITHKTAHPFSNWNIKNLAENEKLLFIEEVTFYQHFYPGYGNKK 182
G LG+AK +L +G EI I K H ++ N + K + Y+ G+G+
Sbjct: 318 GELGAAKNILELGAEIRIPVKNTHVYAFAEHGNDLGSSKDVKGNPTGLYRKM--GHGSSY 375
Query: 183 GAGFK 187
G G K
Sbjct: 376 GLGVK 380
>At3g14220 myrosinase-associated protein like
Length = 363
Score = 27.7 bits (60), Expect = 4.6
Identities = 12/34 (35%), Positives = 18/34 (52%)
Query: 117 HKKLVSGFLGSAKYMLNVGGEIHITHKTAHPFSN 150
HK+ A YM+N+G E ++ AHP +N
Sbjct: 138 HKQWTDKERAEAIYMVNIGAEDYLNFAKAHPNAN 171
>At2g15460 putative polygalacturonase
Length = 402
Score = 27.7 bits (60), Expect = 4.6
Identities = 27/100 (27%), Positives = 41/100 (41%), Gaps = 9/100 (9%)
Query: 95 IIFNFPHSGFFQNESDAWVIGEHKKLVSGFLGSAKYMLNVGGEIHITHK-TAHPFSNWNI 153
+IFN G W K LVS F+ M++VG I+I K HP
Sbjct: 270 VIFNGTSDGI---RIKTWESSASKILVSNFVYENIQMIDVGKPINIDQKYCPHP----PC 322
Query: 154 KNLAENEKLLFIEEVTFYQHFYPGYGNKKGAGFKCDKSFP 193
++ + E + I+ + ++ Y NK +C K FP
Sbjct: 323 EHERKGESHVQIQNLKL-KNIYGTSKNKVAVNLQCSKIFP 361
>At5g43030 CHP-rich zinc finger protein-like
Length = 564
Score = 27.3 bits (59), Expect = 6.0
Identities = 10/22 (45%), Positives = 16/22 (72%)
Query: 1 MEEKIVKHYSSFHNILLVGEGD 22
+EE ++KH++ HN+ LV E D
Sbjct: 244 VEEGLIKHFTHEHNLRLVNEDD 265
>At3g46740 chloroplast import-associated channel protein homolog
Length = 818
Score = 27.3 bits (59), Expect = 6.0
Identities = 24/81 (29%), Positives = 36/81 (43%), Gaps = 5/81 (6%)
Query: 110 DAWVIGEHKKLVS---GFLGSAKYMLNVGGEIHITHKTAHPFSNWNIKNLAENEKLLFIE 166
DA+V+G + G LG+A+ + VG EI I K H ++ N + K +
Sbjct: 713 DAFVLGGPYSVRGYNMGELGAARNIAEVGAEIRIPVKNTHVYAFVEHGNDLGSSKDVKGN 772
Query: 167 EVTFYQHFYPGYGNKKGAGFK 187
Y+ G G+ GAG K
Sbjct: 773 PTAVYRR--TGQGSSYGAGVK 791
>At2g15450 putative polygalacturonase
Length = 404
Score = 27.3 bits (59), Expect = 6.0
Identities = 31/135 (22%), Positives = 53/135 (38%), Gaps = 11/135 (8%)
Query: 62 IELEGLGCTIMHEVDVHNMNQHHQLKHHN--FFDRIIFNFPHSGFFQNESDAWVIGEHKK 119
+ + + C H + V ++ ++ K +IFN G W K
Sbjct: 235 LNISNVNCGPGHGISVGSLGKNKDEKDVKDLIVRDVIFNGTSDGI---RIKNWESSASKI 291
Query: 120 LVSGFLGSAKYMLNVGGEIHITHK-TAHPFSNWNIKNLAENEKLLFIEEVTFYQHFYPGY 178
LVS F+ M++VG I+I K HP ++ + E + I+ + ++ Y
Sbjct: 292 LVSNFVYENIQMIDVGKPINIDQKYCPHP----PCEHERKGESHVQIQNLKL-KNIYGTS 346
Query: 179 GNKKGAGFKCDKSFP 193
NK +C K FP
Sbjct: 347 KNKVAVNLQCSKIFP 361
>At2g04660 unknown protein
Length = 865
Score = 26.9 bits (58), Expect = 7.9
Identities = 11/22 (50%), Positives = 13/22 (59%)
Query: 115 GEHKKLVSGFLGSAKYMLNVGG 136
G+H KLV F+ S KY L G
Sbjct: 393 GQHSKLVESFISSLKYRLLTAG 414
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.324 0.141 0.437
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,196,353
Number of Sequences: 26719
Number of extensions: 221666
Number of successful extensions: 610
Number of sequences better than 10.0: 19
Number of HSP's better than 10.0 without gapping: 10
Number of HSP's successfully gapped in prelim test: 9
Number of HSP's that attempted gapping in prelim test: 586
Number of HSP's gapped (non-prelim): 21
length of query: 211
length of database: 11,318,596
effective HSP length: 95
effective length of query: 116
effective length of database: 8,780,291
effective search space: 1018513756
effective search space used: 1018513756
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.5 bits)
S2: 58 (26.9 bits)
Medicago: description of AC146569.5


