

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146568.7 - phase: 0
(703 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
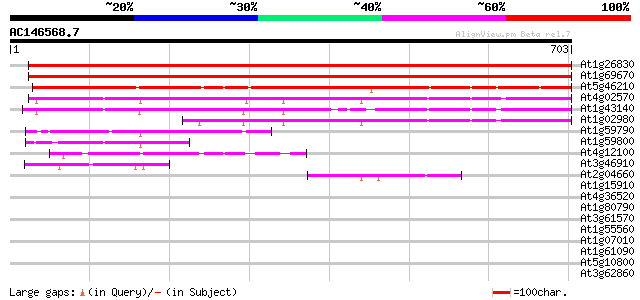
Score E
Sequences producing significant alignments: (bits) Value
At1g26830 cullin 3-like protein 1141 0.0
At1g69670 putative cullin 1132 0.0
At5g46210 cullin 486 e-137
At4g02570 putative cullin-like 1 protein 413 e-115
At1g43140 hypothetical protein 336 3e-92
At1g02980 296 3e-80
At1g59790 hypothetical protein 120 2e-27
At1g59800 hypothetical protein 100 4e-21
At4g12100 unknown protein (At4g12100) 80 5e-15
At3g46910 putative protein 59 9e-09
At2g04660 unknown protein 59 1e-08
At1g15910 39 0.010
At4g36520 trichohyalin like protein 37 0.029
At1g80790 hypothetical protein 35 0.11
At3g61570 unknown protein 33 0.55
At1g55560 unknown protein 33 0.71
At1g07010 unknown protein 32 0.93
At1g61090 hypothetical protein 32 1.2
At5g10800 putative protein 31 2.1
At3g62860 unknown protein 31 2.1
>At1g26830 cullin 3-like protein
Length = 732
Score = 1141 bits (2952), Expect = 0.0
Identities = 553/680 (81%), Positives = 621/680 (91%)
Query: 24 FRNAYNMVLHKFGDRLYSGLVATMTAHLKEIAKSIEAAQGGSFLEELNRKWNDHNKALQM 83
+RNAYNMVLHKFG++LY+G +ATMT+HLKE +K IEAAQGGSFLEELN+KWN+HNKAL+M
Sbjct: 53 YRNAYNMVLHKFGEKLYTGFIATMTSHLKEKSKLIEAAQGGSFLEELNKKWNEHNKALEM 112
Query: 84 IRDILMYMDRTFIPSAKKTPVHELGLNLWRESVIYSNQIRTRLLNTLLELVQSERTGEVI 143
IRDILMYMDRT+I S KKT VH +GLNLWR++V++ +I TRLLNTLL+LVQ ER GEVI
Sbjct: 113 IRDILMYMDRTYIESTKKTHVHPMGLNLWRDNVVHFTKIHTRLLNTLLDLVQKERIGEVI 172
Query: 144 DRGIMRNITKMLMDLGPAVYGQDFEAHFLQVSAEFYQVESQRFIECCDCGDYLKKAERRL 203
DRG+MRN+ KM MDLG +VY +DFE FL S+EFY+VESQ FIE CDCGDYLKK+E+RL
Sbjct: 173 DRGLMRNVIKMFMDLGESVYQEDFEKPFLDASSEFYKVESQEFIESCDCGDYLKKSEKRL 232
Query: 204 NEEMDRVGHYMDPETEKKINKVVETQMIENHMLRLIHMENSGLVNMLCDDKYEDLGRMYN 263
EE++RV HY+D ++E+KI VVE +MI NHM RL+HMENSGLVNML +DKYEDLGRMYN
Sbjct: 233 TEEIERVAHYLDAKSEEKITSVVEKEMIANHMQRLVHMENSGLVNMLLNDKYEDLGRMYN 292
Query: 264 LFRRVADGLLKIREVMTLHIRESGKQLVTDPERLKDPVEFVQRLLDEKDKYDKIINQAFN 323
LFRRV +GL+ +R+VMT H+RE GKQLVTDPE+ KDPVEFVQRLLDE+DKYDKIIN AF
Sbjct: 293 LFRRVTNGLVTVRDVMTSHLREMGKQLVTDPEKSKDPVEFVQRLLDERDKYDKIINTAFG 352
Query: 324 NDKSFQNALNSSFEYFINLNPRSPEFISLFVDDKLRKGLKGVNEDDVEVTLDKVMMLFRY 383
NDK+FQNALNSSFEYFINLN RSPEFISLFVDDKLRKGLKG+ + DVEV LDKVMMLFRY
Sbjct: 353 NDKTFQNALNSSFEYFINLNARSPEFISLFVDDKLRKGLKGITDVDVEVILDKVMMLFRY 412
Query: 384 LQEKDVFEKYYKQHLAKRLLSGKTVSDDAERSLIVKLKTECGYQFTSKLEGMFTDMKTSQ 443
LQEKDVFEKYYKQHLAKRLLSGKTVSDDAERSLIVKLKTECGYQFTSKLEGMFTDMKTS+
Sbjct: 413 LQEKDVFEKYYKQHLAKRLLSGKTVSDDAERSLIVKLKTECGYQFTSKLEGMFTDMKTSE 472
Query: 444 DTMQGFYASHPDLGDGPTLTVQVLTTGSWPTQSSITCNLPVEISALCEKFRSYYLGTHTG 503
DTM+GFY SHP+L +GPTL VQVLTTGSWPTQ ++ CNLP E+S LCEKFRSYYLGTHTG
Sbjct: 473 DTMRGFYGSHPELSEGPTLIVQVLTTGSWPTQPAVPCNLPAEVSVLCEKFRSYYLGTHTG 532
Query: 504 RRLSWQTNMGFADLKATFGKGQKHELNVSTYQMCVLMLFNNADKLSYKEIEQATEIPAPD 563
RRLSWQTNMG AD+KA FGKGQKHELNVST+QMCVLMLFNN+D+LSYKEIEQATEIPA D
Sbjct: 533 RRLSWQTNMGTADIKAIFGKGQKHELNVSTFQMCVLMLFNNSDRLSYKEIEQATEIPAAD 592
Query: 564 LKRCLQSLALVKGRNVLRKEPMSKDVGEDDAFSVNDKFSSKLYKVKIGTVVAQKESEPEK 623
LKRCLQSLA VKG+NV++KEPMSKD+GE+D F VNDKF+SK YKVKIGTVVAQKE+EPEK
Sbjct: 593 LKRCLQSLACVKGKNVIKKEPMSKDIGEEDLFVVNDKFTSKFYKVKIGTVVAQKETEPEK 652
Query: 624 QETRQRVEEDRKPQIEAAIVRIMKSRRLLDHNNLIAEVTKQLQLRFLANPTEVKKRIESL 683
QETRQRVEEDRKPQIEAAIVRIMKSR++LDHNN+IAEVTKQLQ RFLANPTE+KKRIESL
Sbjct: 653 QETRQRVEEDRKPQIEAAIVRIMKSRKILDHNNIIAEVTKQLQPRFLANPTEIKKRIESL 712
Query: 684 IERDFLERDDNDRKMYRYLA 703
IERDFLERD DRK+YRYLA
Sbjct: 713 IERDFLERDSTDRKLYRYLA 732
>At1g69670 putative cullin
Length = 732
Score = 1132 bits (2928), Expect = 0.0
Identities = 550/680 (80%), Positives = 621/680 (90%)
Query: 24 FRNAYNMVLHKFGDRLYSGLVATMTAHLKEIAKSIEAAQGGSFLEELNRKWNDHNKALQM 83
+RNAYNMVLHK+GD+LY+GLV TMT HLKEI KSIE AQGG+FLE LNRKWNDHNKALQM
Sbjct: 53 YRNAYNMVLHKYGDKLYTGLVTTMTFHLKEICKSIEEAQGGAFLELLNRKWNDHNKALQM 112
Query: 84 IRDILMYMDRTFIPSAKKTPVHELGLNLWRESVIYSNQIRTRLLNTLLELVQSERTGEVI 143
IRDILMYMDRT++ + KKT VHELGL+LWR++V+YS++I+TRLLNTLL+LV ERTGEVI
Sbjct: 113 IRDILMYMDRTYVSTTKKTHVHELGLHLWRDNVVYSSKIQTRLLNTLLDLVHKERTGEVI 172
Query: 144 DRGIMRNITKMLMDLGPAVYGQDFEAHFLQVSAEFYQVESQRFIECCDCGDYLKKAERRL 203
DR +MRN+ KM MDLG +VY DFE FL+ SAEFY+VES FIE CDCG+YLKKAE+ L
Sbjct: 173 DRVLMRNVIKMFMDLGESVYQDDFEKPFLEASAEFYKVESMEFIESCDCGEYLKKAEKPL 232
Query: 204 NEEMDRVGHYMDPETEKKINKVVETQMIENHMLRLIHMENSGLVNMLCDDKYEDLGRMYN 263
EE++RV +Y+D ++E KI VVE +MI NH+ RL+HMENSGLVNML +DKYED+GRMY+
Sbjct: 233 VEEVERVVNYLDAKSEAKITSVVEREMIANHVQRLVHMENSGLVNMLLNDKYEDMGRMYS 292
Query: 264 LFRRVADGLLKIREVMTLHIRESGKQLVTDPERLKDPVEFVQRLLDEKDKYDKIINQAFN 323
LFRRVA+GL+ +R+VMTLH+RE GKQLVTDPE+ KDPVEFVQRLLDE+DKYD+IIN AFN
Sbjct: 293 LFRRVANGLVTVRDVMTLHLREMGKQLVTDPEKSKDPVEFVQRLLDERDKYDRIINMAFN 352
Query: 324 NDKSFQNALNSSFEYFINLNPRSPEFISLFVDDKLRKGLKGVNEDDVEVTLDKVMMLFRY 383
NDK+FQNALNSSFEYF+NLN RSPEFISLFVDDKLRKGLKGV E+DV++ LDKVMMLFRY
Sbjct: 353 NDKTFQNALNSSFEYFVNLNTRSPEFISLFVDDKLRKGLKGVGEEDVDLILDKVMMLFRY 412
Query: 384 LQEKDVFEKYYKQHLAKRLLSGKTVSDDAERSLIVKLKTECGYQFTSKLEGMFTDMKTSQ 443
LQEKDVFEKYYKQHLAKRLLSGKTVSDDAER+LIVKLKTECGYQFTSKLEGMFTDMKTS
Sbjct: 413 LQEKDVFEKYYKQHLAKRLLSGKTVSDDAERNLIVKLKTECGYQFTSKLEGMFTDMKTSH 472
Query: 444 DTMQGFYASHPDLGDGPTLTVQVLTTGSWPTQSSITCNLPVEISALCEKFRSYYLGTHTG 503
DT+ GFY SHP+L +GPTL VQVLTTGSWPTQ +I CNLP E+S LCEKFRSYYLGTHTG
Sbjct: 473 DTLLGFYNSHPELSEGPTLVVQVLTTGSWPTQPTIQCNLPAEVSVLCEKFRSYYLGTHTG 532
Query: 504 RRLSWQTNMGFADLKATFGKGQKHELNVSTYQMCVLMLFNNADKLSYKEIEQATEIPAPD 563
RRLSWQTNMG AD+KA FGKGQKHELNVST+QMCVLMLFNN+D+LSYKEIEQATEIP PD
Sbjct: 533 RRLSWQTNMGTADIKAVFGKGQKHELNVSTFQMCVLMLFNNSDRLSYKEIEQATEIPTPD 592
Query: 564 LKRCLQSLALVKGRNVLRKEPMSKDVGEDDAFSVNDKFSSKLYKVKIGTVVAQKESEPEK 623
LKRCLQS+A VKG+NVLRKEPMSK++ E+D F VND+F+SK YKVKIGTVVAQKE+EPEK
Sbjct: 593 LKRCLQSMACVKGKNVLRKEPMSKEIAEEDWFVVNDRFASKFYKVKIGTVVAQKETEPEK 652
Query: 624 QETRQRVEEDRKPQIEAAIVRIMKSRRLLDHNNLIAEVTKQLQLRFLANPTEVKKRIESL 683
QETRQRVEEDRKPQIEAAIVRIMKSRR+LDHNN+IAEVTKQLQ RFLANPTE+KKRIESL
Sbjct: 653 QETRQRVEEDRKPQIEAAIVRIMKSRRVLDHNNIIAEVTKQLQTRFLANPTEIKKRIESL 712
Query: 684 IERDFLERDDNDRKMYRYLA 703
IERDFLERD+ DRK+YRYLA
Sbjct: 713 IERDFLERDNTDRKLYRYLA 732
>At5g46210 cullin
Length = 792
Score = 486 bits (1251), Expect = e-137
Identities = 278/682 (40%), Positives = 426/682 (61%), Gaps = 21/682 (3%)
Query: 29 NMVLHKFGDRLYSGLVATMTAHLKEIAKSIEAAQGGS--FLEELNRKWNDHNKALQMIRD 86
N+ LHK +LY + H+ +S+ FL + + W D + MIR
Sbjct: 125 NLCLHKLDGKLYDQIEKECEEHISAALQSLVGQNTDLTVFLSRVEKCWQDFCDQMLMIRS 184
Query: 87 ILMYMDRTF-IPSAKKTPVHELGLNLWRESVIYSNQIRTRLLNTLLELVQSERTGEVIDR 145
I + +DR + I + + E+GL L+R+ + + ++ R + LL +++ ER E ++R
Sbjct: 185 IALTLDRKYVIQNPNVRSLWEMGLQLFRKHLSLAPEVEQRTVKGLLSMIEKERLAEAVNR 244
Query: 146 GIMRNITKMLMDLGPAVYGQDFEAHFLQVSAEFYQVESQRFIECCDCGDYLKKAERRLNE 205
++ ++ KM LG +Y + FE FL+ ++EFY E ++++ D +YLK E RL+E
Sbjct: 245 TLLSHLLKMFTALG--IYMESFEKPFLEGTSEFYAAEGMKYMQQSDVPEYLKHVEGRLHE 302
Query: 206 EMDRVGHYMDPETEKKINKVVETQMIENHMLRLIHMENSGLVNMLCDDKYEDLGRMYNLF 265
E +R Y+D T K + VE Q++E H+L ++ G ++ + EDL RM LF
Sbjct: 303 ENERCILYIDAVTRKPLITTVERQLLERHILVVLE---KGFTTLMDGRRTEDLQRMQTLF 359
Query: 266 RRVADGLLKIREVMTLHIRESGKQLVTDPERLKDPVEFVQRLLDEKDKYDKIINQAFNND 325
RV + L +R+ ++ ++R++G+++V D E+ KD VQ LLD K D I ++F +
Sbjct: 360 SRV-NALESLRQALSSYVRKTGQKIVMDEEKDKD---MVQSLLDFKASLDIIWEESFYKN 415
Query: 326 KSFQNALNSSFEYFINLNPRSP-EFISLFVDDKLRKGLKGVNEDDVEVTLDKVMMLFRYL 384
+SF N + SFE+ INL P E I+ F+D+KLR G KG +E+++E L+KV++LFR++
Sbjct: 416 ESFGNTIKDSFEHLINLRQNRPAELIAKFLDEKLRAGNKGTSEEELESVLEKVLVLFRFI 475
Query: 385 QEKDVFEKYYKQHLAKRLLSGKTVSDDAERSLIVKLKTECGYQFTSKLEGMFTDMKTSQD 444
Q KDVFE +YK+ LAKRLL GK+ S DAE+S+I KLKTECG QFT+KLEGMF D++ S++
Sbjct: 476 QGKDVFEAFYKKDLAKRLLLGKSASIDAEKSMISKLKTECGSQFTNKLEGMFKDIELSKE 535
Query: 445 TMQGFYAS---HPDLGDGPTLTVQVLTTGSWPTQSSITCNLPVEISALCEKFRSYYLGTH 501
+ F S L G ++V VLTTG WPT + LP E++ + F+ +YL +
Sbjct: 536 INESFKQSSQARTKLPSGIEMSVHVLTTGYWPTYPPMDVKLPHELNVYQDIFKEFYLSKY 595
Query: 502 TGRRLSWQTNMGFADLKATFGKGQKHELNVSTYQMCVLMLFNNADKLSYKEIEQATEIPA 561
+GRRL WQ ++G LKA F KG+K EL VS +Q VLMLFN+A KLS+++I+ +T I
Sbjct: 596 SGRRLMWQNSLGHCVLKADFSKGKK-ELAVSLFQAVVLMLFNDAMKLSFEDIKDSTSIED 654
Query: 562 PDLKRCLQSLALVKGRNVLRKEPMSKDVGEDDAFSVNDKFSSKLYKVKIGTVVAQKESEP 621
+L+R LQSLA K R VL+K P +DV + D F ND+F++ LY++K+ + KE+
Sbjct: 655 KELRRTLQSLACGKVR-VLQKNPKGRDVEDGDEFEFNDEFAAPLYRIKV-NAIQMKETVE 712
Query: 622 EKQETRQRVEEDRKPQIEAAIVRIMKSRRLLDHNNLIAEVTKQLQLRFLANPTEVKKRIE 681
E T +RV +DR+ QI+AAIVRIMK+R++L H LI E+ + QL+F P ++KKRIE
Sbjct: 713 ENTSTTERVFQDRQYQIDAAIVRIMKTRKVLSHTLLITELFQ--QLKFPIKPADLKKRIE 770
Query: 682 SLIERDFLERDDNDRKMYRYLA 703
SLI+R++LER+ ++ ++Y YLA
Sbjct: 771 SLIDREYLEREKSNPQIYNYLA 792
>At4g02570 putative cullin-like 1 protein
Length = 738
Score = 413 bits (1062), Expect = e-115
Identities = 239/706 (33%), Positives = 397/706 (55%), Gaps = 36/706 (5%)
Query: 24 FRNAYNMVL----HKFGDRLYSGLVATMTAHLKE-IAKSIEAAQGGSFLEELNRKWNDHN 78
+ YNM H + +LY ++ + ++ L EL ++W++H
Sbjct: 43 YTTIYNMCTQKPPHDYSQQLYDKYREAFEEYINSTVLPALREKHDEFMLRELFKRWSNHK 102
Query: 79 KALQMIRDILMYMDRTFIPSAKKTPVHELGLNLWRESVIYSNQIRTRLLNTLLELVQSER 138
++ + Y+DR FI P++E+GL +R+ V N++ +++ ++ LV ER
Sbjct: 103 VMVRWLSRFFYYLDRYFIARRSLPPLNEVGLTCFRDLVY--NELHSKVKQAVIALVDKER 160
Query: 139 TGEVIDRGIMRNITKMLMDLGPAV---YGQDFEAHFLQVSAEFYQVESQRFIECCDCGDY 195
GE IDR +++N+ + +++G Y +DFE+ LQ ++ +Y ++ +I+ C DY
Sbjct: 161 EGEQIDRALLKNVLDIYVEIGMGQMERYEEDFESFMLQDTSSYYSRKASSWIQEDSCPDY 220
Query: 196 LKKAERRLNEEMDRVGHYMDPETEKKINKVVETQMIENHMLRLIHMENSGLVNMLCDDKY 255
+ K+E L +E +RV HY+ +E K+ + V+ +++ +L+ E+SG +L DDK
Sbjct: 221 MLKSEECLKKERERVAHYLHSSSEPKLVEKVQHELLVVFASQLLEKEHSGCRALLRDDKV 280
Query: 256 EDLGRMYNLFRRVADGLLKIREVMTLHIRESGKQLVTDPE-----------RLKDPVEFV 304
+DL RMY L+ ++ GL + + H+ G LV E +++ V +
Sbjct: 281 DDLSRMYRLYHKILRGLEPVANIFKQHVTAEGNALVQQAEDTATNQVANTASVQEQV-LI 339
Query: 305 QRLLDEKDKYDKIINQAFNNDKSFQNALNSSFEYFIN---LNPRSPEFISLFVDDKLRK- 360
+++++ DKY + + F N F AL +FE F N S E ++ F D+ L+K
Sbjct: 340 RKVIELHDKYMVYVTECFQNHTLFHKALKEAFEIFCNKTVAGSSSAELLATFCDNILKKG 399
Query: 361 GLKGVNEDDVEVTLDKVMMLFRYLQEKDVFEKYYKQHLAKRLLSGKTVSDDAERSLIVKL 420
G + ++++ +E TL+KV+ L Y+ +KD+F ++Y++ LA+RLL ++ +DD ERS++ KL
Sbjct: 400 GSEKLSDEAIEDTLEKVVKLLAYISDKDLFAEFYRKKLARRLLFDRSANDDHERSILTKL 459
Query: 421 KTECGYQFTSKLEGMFTDM---KTSQDTMQGFYASHPDLGDGPTLTVQVLTTGSWPTQSS 477
K +CG QFTSK+EGM TD+ + +Q++ + + S+P G LTV VLTTG WP+ S
Sbjct: 460 KQQCGGQFTSKMEGMVTDLTLARENQNSFEDYLGSNPAANPGIDLTVTVLTTGFWPSYKS 519
Query: 478 ITCNLPVEISALCEKFRSYYLGTHTGRRLSWQTNMGFADLKATFGKGQKHELNVSTYQMC 537
NLP E+ E F+ +Y R+L+W ++G + F + + EL VSTYQ
Sbjct: 520 FDINLPSEMIKCVEVFKGFYETKTKHRKLTWIYSLGTCHINGKFDQ-KAIELIVSTYQAA 578
Query: 538 VLMLFNNADKLSYKEIEQATEIPAPDLKRCLQSLALVKGRNVLRKEPMSKDVGEDDAFSV 597
VL+LFN DKLSY EI + DL R L SL+ K + +L KEP +K V ++DAF
Sbjct: 579 VLLLFNTTDKLSYTEILAQLNLSHEDLVRLLHSLSCAKYK-ILLKEPNTKTVSQNDAFEF 637
Query: 598 NDKFSSKLYKVKIGTVVAQKESEPEKQETRQRVEEDRKPQIEAAIVRIMKSRRLLDHNNL 657
N KF+ ++ ++KI E+++ + V++DR+ I+AAIVRIMKSR++L H L
Sbjct: 638 NSKFTDRMRRIKIPLPPVD-----ERKKVVEDVDKDRRYAIDAAIVRIMKSRKVLGHQQL 692
Query: 658 IAEVTKQLQLRFLANPTEVKKRIESLIERDFLERDDNDRKMYRYLA 703
++E +QL F + +KKR+E LI RD+LERD + M+RYLA
Sbjct: 693 VSECVEQLSRMFKPDIKAIKKRMEDLITRDYLERDKENPNMFRYLA 738
>At1g43140 hypothetical protein
Length = 711
Score = 336 bits (861), Expect = 3e-92
Identities = 222/711 (31%), Positives = 371/711 (51%), Gaps = 54/711 (7%)
Query: 17 PPFCLCGFRNAYNMVL--------HKFGDRLYSGLVATMTAHLKE-IAKSIEAAQGGSFL 67
PPF + N Y ++ + + LY+ + + KE + S+ G L
Sbjct: 31 PPFDPGQYINLYTIIYDMCLQQPPNDYSQELYNKYRGVVDHYNKETVLPSMRERHGEYML 90
Query: 68 EELNRKWNDHNKALQMIRDILMYMDRTFIPSAKKTPVHELGLNLWRESVIYSNQIRTRLL 127
EL ++W +H ++ + Y+DR ++ ++++G + + V +I++
Sbjct: 91 RELVKRWANHKILVRWLSRFCFYLDRFYVARRGLPTLNDVGFTSFHDLVY--QEIQSEAK 148
Query: 128 NTLLELVQSERTGEVIDRGIMRNITKMLMDLGPA---VYGQDFEAHFLQVSAEFYQVESQ 184
+ LL L+ ER GE IDR +++N+ + G +Y +DFE+ LQ +A +Y ++
Sbjct: 149 DVLLALIHKEREGEQIDRTLVKNVIDVYCGNGVGQMVIYEEDFESFLLQDTASYYSRKAS 208
Query: 185 RFIECCDCGDYLKKAERRLNEEMDRVGHYMDPETEKKINKVVETQMIENHMLRLIHMENS 244
R+ + C DY+ KAE L E +RV +Y+ TE K+ + V+ +++ +LI E+S
Sbjct: 209 RWSQEDSCPDYMLKAEECLKLEKERVTNYLHSTTEPKLVEKVQNELLVVVAKQLIENEHS 268
Query: 245 GLVNMLCDDKYEDLGRMYNLFRRVADGLLKIREVMTLHIRESGKQLV-------TDPERL 297
G + +L DDK DL RMY L+R + GL I ++ H+ G L+ T+ +
Sbjct: 269 GCLALLRDDKMGDLSRMYRLYRLIPQGLEPIADLFKQHVTAEGNALIKQAADAATNQDAS 328
Query: 298 KDPVEFVQRLLDEKDKYDKIINQAFNNDKSFQNALNSSFEYFIN---LNPRSPEFISLFV 354
V V++ ++ DKY +++ F F L +FE F N S E ++ +
Sbjct: 329 ASQV-LVRKEIELHDKYMVYVDECFQKHSLFHKLLKEAFEVFCNKTVAGASSAEILATYC 387
Query: 355 DD--KLRKGLKGVNEDDVEVTLDKVMMLFRYLQEKDVFEKYYKQHLAKRLLSGKTVSDDA 412
D+ K R G + ++++ E+TL+KV+ L Y+ +KD+F ++Y++ A+RLL
Sbjct: 388 DNILKTRGGSEKLSDEATEITLEKVVNLLVYISDKDLFAEFYRKKQARRLL--------F 439
Query: 413 ERSLIVKLKTECGYQFTSKLEGMFTDMKTSQDTMQGFYASHPDLGDGPTLTVQVLTTGSW 472
+RS I+K T+ +L+ F D ++ T + G TV VLTTG W
Sbjct: 440 DRSGIMKEVTD--ITLARELQTNFVDYLSANMTTK----------LGIDFTVTVLTTGFW 487
Query: 473 PTQSSITCNLPVEISALCEKFRSYYLGTHTGRRLSWQTNMGFADLKATFGKGQKHELNVS 532
P+ + NLP E+ E F+ +Y RRLSW ++G + F K + EL VS
Sbjct: 488 PSYKTTDLNLPTEMVNCVEAFKVFYGTKTNSRRLSWIYSLGTCHILGKFEK-KTMELVVS 546
Query: 533 TYQMCVLMLFNNADKLSYKEIEQATEIPAPDLKRCLQSLALVKGRNVLRKEPMSKDVGED 592
TYQ VL+LFNNA++LSY EI + + DL R L SL+ +K + +L KEPMS+ + +
Sbjct: 547 TYQAAVLLLFNNAERLSYTEISEQLNLSHEDLVRLLHSLSCLKYK-ILIKEPMSRTISKT 605
Query: 593 DAFSVNDKFSSKLYKVKIGTVVAQKESEPEKQETRQRVEEDRKPQIEAAIVRIMKSRRLL 652
D F N KF+ K+ K+++ E+++ + V++DR+ I+AA+VRIMKSR++L
Sbjct: 606 DTFEFNSKFTDKMRKIRV-----PLPPMDERKKVVEDVDKDRRYAIDAALVRIMKSRKVL 660
Query: 653 DHNNLIAEVTKQLQLRFLANPTEVKKRIESLIERDFLERDDNDRKMYRYLA 703
H L++E + L F + +KKRIE LI RD+LERD + ++Y+A
Sbjct: 661 AHQQLVSECVEHLSKMFKPDIKMIKKRIEDLINRDYLERDTENANTFKYVA 711
>At1g02980
Length = 648
Score = 296 bits (757), Expect = 3e-80
Identities = 181/513 (35%), Positives = 285/513 (55%), Gaps = 34/513 (6%)
Query: 217 ETEKKINKVVETQMIENHML-----RLIHMENSGLVNMLCDDKYEDLGRMYNLFRRVADG 271
E + K V ++N +L +LI E+SG +L DDK +DL RMY L+ + G
Sbjct: 144 ELQSKAKDAVLALKVQNELLVVVAKQLIENEHSGCRALLRDDKMDDLARMYRLYHPIPQG 203
Query: 272 LLKIREVMTLHIRESGKQLV-------------TDPERLKDPVEFVQRLLDEKDKYDKII 318
L + ++ HI G L+ T +++D V +++L+D DK+ +
Sbjct: 204 LDPVADLFKQHITVEGSALIKQATEAATDKAASTSGLKVQDQV-LIRQLIDLHDKFMVYV 262
Query: 319 NQAFNNDKSFQNALNSSFEYFIN---LNPRSPEFISLFVDDKLRKG--LKGVNEDDVEVT 373
++ F F AL +FE F N S E ++ + D+ L+ G ++ + +D+E+T
Sbjct: 263 DECFQKHSLFHKALKEAFEVFCNKTVAGVSSAEILATYCDNILKTGGGIEKLENEDLELT 322
Query: 374 LDKVMMLFRYLQEKDVFEKYYKQHLAKRLLSGKTVSDDAERSLIVKLKTECGYQFTSKLE 433
L+KV+ L Y+ +KD+F +++++ A+RLL + +D ERSL+ K K G QFTSK+E
Sbjct: 323 LEKVVKLLVYISDKDLFAEFFRKKQARRLLFDRNGNDYHERSLLTKFKELLGAQFTSKME 382
Query: 434 GMFTDM---KTSQDTMQGFYASHPDLGDGPTLTVQVLTTGSWPTQSSITCNLPVEISALC 490
GM TDM K Q F + + G TV VLTTG WP+ + NLP+E+
Sbjct: 383 GMLTDMTLAKEHQTNFVEFLSVNKTKKLGMDFTVTVLTTGFWPSYKTTDLNLPIEMVNCV 442
Query: 491 EKFRSYYLGTHTGRRLSWQTNMGFADLKATFGKGQKHELNVSTYQMCVLMLFNNADKLSY 550
E F++YY RRLSW ++G L F K + E+ V+TYQ VL+LFNN ++LSY
Sbjct: 443 EAFKAYYGTKTNSRRLSWIYSLGTCQLAGKFDK-KTIEIVVTTYQAAVLLLFNNTERLSY 501
Query: 551 KEIEQATEIPAPDLKRCLQSLALVKGRNVLRKEPMSKDVGEDDAFSVNDKFSSKLYKVKI 610
EI + + DL R L SL+ +K + +L KEPMS+++ D F N KF+ K+ ++++
Sbjct: 502 TEILEQLNLGHEDLARLLHSLSCLKYK-ILIKEPMSRNISNTDTFEFNSKFTDKMRRIRV 560
Query: 611 GTVVAQKESEPEKQETRQRVEEDRKPQIEAAIVRIMKSRRLLDHNNLIAEVTKQLQLRFL 670
E+++ + V++DR+ I+AA+VRIMKSR++L H L++E + L F
Sbjct: 561 -----PLPPMDERKKIVEDVDKDRRYAIDAALVRIMKSRKVLGHQQLVSECVEHLSKMFK 615
Query: 671 ANPTEVKKRIESLIERDFLERDDNDRKMYRYLA 703
+ +KKRIE LI RD+LERD ++ ++YLA
Sbjct: 616 PDIKMIKKRIEDLISRDYLERDTDNPNTFKYLA 648
>At1g59790 hypothetical protein
Length = 374
Score = 120 bits (302), Expect = 2e-27
Identities = 81/313 (25%), Positives = 152/313 (47%), Gaps = 17/313 (5%)
Query: 21 LCGFRNAYNMVLHKFGDRLYSGLVATMTAHLKEIAKSIEAAQGGSFLEELNRKWNDHNKA 80
+C R+ Y+ L++ Y ++ T ++ + S+ L EL ++WN+H
Sbjct: 55 MCVQRSDYSQQLYE----KYRKVIEDYT--IQTVLPSLREKHDEDMLRELVKRWNNHKIM 108
Query: 81 LQMIRDILMYMDRTFIPSAKKT--PVHELGLNLWRESVIYSNQIRTRLLNTLLELVQSER 138
++ + +Y+DR + +K + E+GL + + V Q + ++ L+ ER
Sbjct: 109 VKWLSKFFVYIDRHLVRRSKIPIPSLDEVGLTCFLDLVYCEMQSTAK--EVVIALIHKER 166
Query: 139 TGEVIDRGIMRNITKMLMDLGPAV---YGQDFEAHFLQVSAEFYQVESQRFIECCDCGDY 195
GE IDR +++N+ + ++ G Y +DFE+ LQ +A +Y ++ R+ E C DY
Sbjct: 167 EGEQIDRALVKNVLDIYVENGMGTLEKYEEDFESFMLQDTASYYSRKASRWTEEDSCPDY 226
Query: 196 LKKAERRLNEEMDRVGHYMDPETEKKINKVVETQMIENHMLRLIHMENSGLVNMLCDDKY 255
+ K E L E +RV HY+ TE K+ + ++ +++ + E+SG +L DDK
Sbjct: 227 MIKVEECLKMERERVTHYLHSITEPKLVEKIQNELLVMVTKNRLENEHSGFSALLRDDKK 286
Query: 256 EDLGRMYNLFRRVADGLLKIREVMTLHIRESGKQLVTDPERLKDPVEFVQRLLDEKDKYD 315
DL R+Y L+ + L ++ ++ HI E G L+ + D Q L++ +K+
Sbjct: 287 NDLSRIYRLYLPIPKRLGRVADLFKKHITEEGNALI----KQADDKTTNQLLIELHNKFI 342
Query: 316 KIINQAFNNDKSF 328
+ + F N F
Sbjct: 343 VYVIECFQNHTLF 355
>At1g59800 hypothetical protein
Length = 255
Score = 100 bits (248), Expect = 4e-21
Identities = 59/208 (28%), Positives = 108/208 (51%), Gaps = 13/208 (6%)
Query: 21 LCGFRNAYNMVLHKFGDRLYSGLVATMTAHLKEIAKSIEAAQGGSFLEELNRKWNDHNKA 80
+C ++N + K+ + + + + T+ L+E L EL ++WN H
Sbjct: 50 ICAYKNPQQLY-DKYRELIENYAIQTVLPSLRE-------KHDECMLRELAKRWNAHKLL 101
Query: 81 LQMIRDILMYMDRTFIPSAKKTPVHELGLNLWRESVIYSNQIRTRLLNTLLELVQSERTG 140
+++ L+Y+D +F+ + E+GLN +R+ V ++++ +L L+ ER G
Sbjct: 102 VRLFSRRLVYLDDSFLSKKGLPSLREVGLNCFRDQVY--REMQSMAAEAILALIHKEREG 159
Query: 141 EVIDRGIMRNITKMLMDLGPAV---YGQDFEAHFLQVSAEFYQVESQRFIECCDCGDYLK 197
E IDR ++RN+ + ++ G Y +DFE LQ +A +Y ++ R+I+ C DY
Sbjct: 160 EQIDRELVRNVIDVFVENGMGTLKKYEEDFERLMLQDTASYYSSKASRWIQEESCLDYTL 219
Query: 198 KAERRLNEEMDRVGHYMDPETEKKINKV 225
K ++ L E +RV HY+ P TE K+ +V
Sbjct: 220 KPQQCLQRERERVTHYLHPTTEPKLFEV 247
>At4g12100 unknown protein (At4g12100)
Length = 434
Score = 79.7 bits (195), Expect = 5e-15
Identities = 71/331 (21%), Positives = 143/331 (42%), Gaps = 44/331 (13%)
Query: 50 HLKEIAKSIEAAQGGS-----FLEELNRKWNDHNKALQMIRDILMYMDRTFIPSAKKTPV 104
H+ E+ +S+E GS FL + +W D + + ++ D+ MY + +
Sbjct: 138 HIAELIQSLEKNCSGSDDPSVFLPHVYNRWLDFKRKMSLVSDVAMYQ------TLNGLTL 191
Query: 105 HELGLNLWRESVIYSNQIRTRLLNTLLELVQSERTGEVIDR--GIMRNITKMLMDLGPAV 162
++G L+ + + + Q++ +++ +L L+ ER G+ + +++N+ M
Sbjct: 192 WDVGQKLFHKQLSMAPQLQDQVITGILRLITDERLGKAANNTSDLLKNLMDMFRMQWQCT 251
Query: 163 YGQDFEAHFLQVSAEFYQVESQRFIECCDCGDYLKKAERRLNEEMDRVG--HYMDPETEK 220
Y ++ FL +++FY E+++ ++ D YLK ER E ++ ++ +
Sbjct: 252 YV--YKDPFLDSTSKFYAEEAEQVLQRSDISHYLKYVERTFLAEEEKCDKHYFFFSSSRS 309
Query: 221 KINKVVETQMIENHMLRLIHMENSGLVNMLCDDKYEDLGRMYNLFRRVADGLLKIREVMT 280
++ KV+++Q++E H L G + ++ + +DL RMY LF V D I ++
Sbjct: 310 RLMKVLKSQLLEAHSSFL----EEGFMLLMDESLIDDLRRMYRLFSMV-DSEDYIDRILR 364
Query: 281 LHIRESGKQLVTDPERLKDPVEFVQRLLDEKDKYDKIINQAFNNDKSFQNALNSSFEYFI 340
+I G+ + L + DKI +Q F D + FE F
Sbjct: 365 AYILAKGEGARQEGS-----------LQELHTSIDKIWHQCFGQDDLLDKTIRDCFEGF- 412
Query: 341 NLNPRSPEFISLFVDDKLRKGLKGVNEDDVE 371
L V + L+ +++DD E
Sbjct: 413 ----------GLHVPGEFSDQLQWIDDDDDE 433
>At3g46910 putative protein
Length = 247
Score = 58.9 bits (141), Expect = 9e-09
Identities = 51/201 (25%), Positives = 90/201 (44%), Gaps = 23/201 (11%)
Query: 19 FCLCGFRNAYNMVL--HKFGDRLYSGLVATMTAHLKEIAKSIEA---AQGGSFLEELNRK 73
F G NA N G+ LY ++ ++ +S+E+ FL L +
Sbjct: 48 FACSGLFNAVNKSWCEKSSGEALYKLILEECEIYISAAIQSLESQCDTDPSLFLSLLEKC 107
Query: 74 WNDHNKALQMIRDILMYMDRTFIPSAKKTPVHELGLNLWRESVIYSNQIRTRLLNTLLEL 133
W D + LQ + I +T P + V +LG L + + + ++R +LL+ +L+L
Sbjct: 108 WLDFRRKLQFLCSIAGGEGQTVGPHS----VWDLGSELSPKHLFSAQKVRDKLLSIILQL 163
Query: 134 VQSERTGEVIDRGIMRNITKMLM--------DLGPAVYGQD------FEAHFLQVSAEFY 179
++ +R+ +D ++N T+ +M +L YGQ F+ F+ + EFY
Sbjct: 164 IRDQRSFMSVDMTQLKNTTRPVMSVHMTQLNNLRGLFYGQSLYKSPFFKKPFIDCAVEFY 223
Query: 180 QVESQRFIECCDCGDYLKKAE 200
E+ +F E D YLK+ E
Sbjct: 224 SAEAMQFKEQSDIPLYLKRVE 244
>At2g04660 unknown protein
Length = 865
Score = 58.5 bits (140), Expect = 1e-08
Identities = 48/200 (24%), Positives = 82/200 (41%), Gaps = 8/200 (4%)
Query: 374 LDKVMMLFRYLQEKDVFEKYYKQHLAKRLLSGKTVSDDAERSLIVKLKTECGYQFTSKLE 433
+D + ML + K+ Y+ LA++LL+ D E + LK G + E
Sbjct: 537 VDILGMLVDIIGSKEQLVNEYRVMLAEKLLNKTDYDIDTEIRTVELLKIHFGEASMQRCE 596
Query: 434 GMFTDM---KTSQDTMQGFYASHPDLGDGP----TLTVQVLTTGSWPTQSSITCNLPVEI 486
M D+ K ++ + +L + TLT +L+T WP LP +
Sbjct: 597 IMLNDLIDSKRVNTNIKKASQTGAELRENELSVDTLTSTILSTNFWPPIQDEPLELPGPV 656
Query: 487 SALCEKFRSYYLGTHTGRRLSWQTNMGFADLKATFGKGQKHELNVSTYQMCVLMLFNNAD 546
L + + Y T R+L W+ N+G L+ F + + + VS ++M F
Sbjct: 657 DKLLSDYANRYHEIKTPRKLLWKKNLGTVKLELQF-EDRAMQFTVSPTHAAIIMQFQEKK 715
Query: 547 KLSYKEIEQATEIPAPDLKR 566
+YK++ + IP L R
Sbjct: 716 SWTYKDLAEVIGIPIDALNR 735
>At1g15910
Length = 634
Score = 38.9 bits (89), Expect = 0.010
Identities = 35/162 (21%), Positives = 74/162 (45%), Gaps = 13/162 (8%)
Query: 263 NLFRRVADGLLKIREVMTLHIRE---SGKQLVTDPERLKDPVEFVQRLLDEKDKYDKIIN 319
NL + AD K++++ HI++ ++L + +R +E + L++ + ++
Sbjct: 293 NLHQAFADETKKMQQMSLRHIQKILYDKEKLSNELDRKMRDLESRAKQLEKHEALTELDR 352
Query: 320 QAFNNDKSFQNALNSSFEYFINLNPRSPEFISLFVDDKLRKGLKGVNEDDVEVTLDKVMM 379
Q + DK +A+N S + ++ E + V++ R+ E L+K+++
Sbjct: 353 QKLDEDKRKSDAMNKSLQLASREQKKADESVLRLVEEHQRQ---------KEDALNKILL 403
Query: 380 LFRYLQEKDVFEKYYKQHLAKRLLSGKTVSDDAERSLIVKLK 421
L + L K E Q L +L K + DD + ++ K+K
Sbjct: 404 LEKQLDTKQTLEMEI-QELKGKLQVMKHLGDDDDEAVQKKMK 444
>At4g36520 trichohyalin like protein
Length = 1432
Score = 37.4 bits (85), Expect = 0.029
Identities = 28/98 (28%), Positives = 53/98 (53%), Gaps = 10/98 (10%)
Query: 609 KIGTVVAQKESEPEKQETRQRVEEDRKPQIEAAIVRIMKSRRLLDHNNLIAEVTKQLQLR 668
K+ + QKE+E +ETR++ E+ K ++ AI K +RL++ AE+ ++L+
Sbjct: 804 KLKEALEQKENERRLKETREK--EENKKKLREAIELEEKEKRLIEAFER-AEIERRLKED 860
Query: 669 FLANPTEVKKRIESLIERDFLERD-----DNDRKMYRY 701
E++ R++ ER+ L R+ +N+RK + Y
Sbjct: 861 L--EQEEMRMRLQEAKERERLHRENQEHQENERKQHEY 896
>At1g80790 hypothetical protein
Length = 634
Score = 35.4 bits (80), Expect = 0.11
Identities = 45/224 (20%), Positives = 96/224 (42%), Gaps = 23/224 (10%)
Query: 223 NKVVETQMIENHMLRLIHMENSGLVNMLCDDKYEDLGRMYNLFRRVADGLLKIREVMTLH 282
NK+ T N + + + + L +L + ++L R+Y + L RE +
Sbjct: 261 NKIAMTNEDLNKLQYMNNEKTLSLRRVLIEK--DELDRVYKQETKKMQELS--REKINRI 316
Query: 283 IRESGKQLVTDPERLKDPVEFVQRLLDEKDKYDKIINQAFNNDKSFQNALNSSFEYFINL 342
RE ++L + E + ++ + LD+K ++ Q + DK + +NSS +
Sbjct: 317 FREK-ERLTNELEAKMNNLKIWSKQLDKKQALTELERQKLDEDKKKSDVMNSSLQLASLE 375
Query: 343 NPRSPEFISLFVDDKLRKGLKGVNEDDVEVTLDKVMMLFRYLQEKDVFEKYYKQHLAK-R 401
++ + + VD+ RK E TL+K++ L + L K + ++ K +
Sbjct: 376 QKKTDDRVLRLVDEHKRK---------KEETLNKILQLEKELDSKQKLQMEIQELKGKLK 426
Query: 402 LLSGKTVSDDAERSLIVKLKTECGYQFTSKLEGMFTDMKTSQDT 445
++ + D+ + + K+K E LE ++++ +DT
Sbjct: 427 VMKHEDEDDEGIKKKMKKMKEE--------LEEKCSELQDLEDT 462
>At3g61570 unknown protein
Length = 712
Score = 33.1 bits (74), Expect = 0.55
Identities = 37/179 (20%), Positives = 77/179 (42%), Gaps = 17/179 (9%)
Query: 522 GKGQKHELNVSTYQMCVLMLFNNADKLSYKEIEQATEIPAPDLKRCLQSLALVKGRNVLR 581
G G +H V+ L NN KL E+E A E + ++ S L
Sbjct: 273 GPGMEHLKEVNK----ALEKENNELKLKRSELEAALE----ESRKLTNSKVFPDATESLT 324
Query: 582 KEPMSKDVGEDDAFSVNDKFSSKLYKVKIGTVVAQKESEPEKQETRQRVEEDRKPQIEAA 641
+ P + D + ++F ++ L ++++ Q+E + +QE ++ + + + E
Sbjct: 325 RHPSTLDKEKPESFPGKEEMEQSLQRLEMDLKETQRERDKARQELKRLKQHLLEKETE-- 382
Query: 642 IVRIMKSRRLLDHNNLIAEV--TKQLQLRFLANPTEVKKRIESLIERDFLERDDNDRKM 698
+S ++ + + LI E+ T + Q +++ + K+ S E + L D+ RK+
Sbjct: 383 -----ESEKMDEDSRLIEELRQTNEYQRSQISHLEKSLKQAISNQEDNRLSNDNQIRKL 436
>At1g55560 unknown protein
Length = 549
Score = 32.7 bits (73), Expect = 0.71
Identities = 61/267 (22%), Positives = 108/267 (39%), Gaps = 46/267 (17%)
Query: 231 IENHMLRLIHMENSGLVNMLCDDKYEDLGRMYNL----------FRRVADGLLKIREVMT 280
I+NH ++L+ ME S ++ D +G+ +++ + VA +E+ T
Sbjct: 232 IQNHKMKLVEMEGSHVIQNDYDSLDVHVGQCFSVLVTANQAAKDYYMVASTRFLKKELST 291
Query: 281 LH-IRESGKQLVTDPERLKDPVEFVQRLLDEKD-KYDKIINQAFNNDKSFQNALNSSFEY 338
+ IR G + E K PV + L + +++ N A N + S+ Y
Sbjct: 292 VGVIRYEGSNVQASTELPKAPVGWAWSLNQFRSFRWNLTSNAARPNPQG-------SYHY 344
Query: 339 F-INLNPRSPEFISL--FVDDKLRKGLKGVNEDDVEVTLDKVMMLFRYLQEKDVFEKYYK 395
IN+ RS + ++ VD K+R G GV+ D E L L Y Q + EK +K
Sbjct: 345 GKINIT-RSIKLVNSKSVVDGKVRFGFNGVSHVDTETPL----KLAEYFQ---MSEKVFK 396
Query: 396 QHLAKRLLSGKTVSDDAERSLIVKLKTECGYQFTSKLEGMFTDMKTSQDTMQGFYASHPD 455
++ K + K + + +++ F + +E +F + + TMQ F+
Sbjct: 397 YNVIKDEPAAKITALTVQPNVL-------NITFRTFVEIIF---ENHEKTMQSFHL---- 442
Query: 456 LGDGPTLTVQVLTTGSWPTQSSITCNL 482
DG + G W + NL
Sbjct: 443 --DGYSFFAVASEPGRWTPEKRENYNL 467
>At1g07010 unknown protein
Length = 389
Score = 32.3 bits (72), Expect = 0.93
Identities = 18/68 (26%), Positives = 34/68 (49%), Gaps = 8/68 (11%)
Query: 138 RTGEVIDRG--------IMRNITKMLMDLGPAVYGQDFEAHFLQVSAEFYQVESQRFIEC 189
+ G+++DRG ++R++ G AV+ + + V +F V+++ F EC
Sbjct: 99 QVGDILDRGDDEIAILSLLRSLDDQAKANGGAVFQVNGNHETMNVEGDFRYVDARAFDEC 158
Query: 190 CDCGDYLK 197
D DYL+
Sbjct: 159 TDFLDYLE 166
>At1g61090 hypothetical protein
Length = 238
Score = 32.0 bits (71), Expect = 1.2
Identities = 21/100 (21%), Positives = 42/100 (42%), Gaps = 8/100 (8%)
Query: 357 KLRKGLKGVNEDDVEVTLDKVMMLFRYLQEKD--------VFEKYYKQHLAKRLLSGKTV 408
K++ + + +++V ++D++M F +D +F Y + + K V
Sbjct: 15 KIKAAQRNMTDNEVLRSMDEIMFCFENCYTEDETFTHIAQMFPSYVLKIVKSFFQISKAV 74
Query: 409 SDDAERSLIVKLKTECGYQFTSKLEGMFTDMKTSQDTMQG 448
+D+ R + EC S+ + K SQDT+ G
Sbjct: 75 TDEESREAYLTDAEECASVRRSQARDTSEEAKRSQDTLMG 114
>At5g10800 putative protein
Length = 957
Score = 31.2 bits (69), Expect = 2.1
Identities = 37/155 (23%), Positives = 62/155 (39%), Gaps = 25/155 (16%)
Query: 547 KLSYKEIE--------QATEIPAPDLKRCLQSLALVKGRNVLRKEPMSKDVGEDDAFSVN 598
K SY ++E IP P+LK A VK + P S+ EDD
Sbjct: 717 KTSYDKVETEEPVDLASTIPIPQPELK------AFVKKEKIDLILPTSRWAREDDETDDE 770
Query: 599 DKFSSKLYKVKIGTVVAQKESEPEKQETRQRVEEDRKPQIEAAIVRIMKSRRLLDHNNL- 657
K S G + + + E K + RV+ + + +E R+ L H +
Sbjct: 771 QKKSYSSGSDNAGGITFKTDEEDLKADPSVRVQPENEIDVE--------QRQKLRHIEIA 822
Query: 658 IAEVTKQLQLRFLANPTEVKKRIESLIERDFLERD 692
+ E + L+ + + N E+++++ I R LE D
Sbjct: 823 LIEYRESLEEQGMKNSEEIERKV--AIHRKRLEAD 855
>At3g62860 unknown protein
Length = 348
Score = 31.2 bits (69), Expect = 2.1
Identities = 43/191 (22%), Positives = 72/191 (37%), Gaps = 37/191 (19%)
Query: 148 MRNITKMLMDLGPAVYGQDFEAHFLQVSAEFYQVESQRFIECCDCGDYLKKAERRLNEEM 207
MR L G AV+G D+E H A Y + + DC DY + EE
Sbjct: 50 MRECGIRLASAGYAVFGMDYEGHGRSKGARCYIKKFSNIVN--DCFDYYTSISAQ--EEY 105
Query: 208 DRVGHYMDPETEKKINKVVETQMIENHMLRLIHMENSG------LVNMLC--DDKYEDLG 259
G ++ E+ + + L+H ++ LV +C +K +
Sbjct: 106 KEKGRFLYGES------------MGGAVALLLHKKDPSFWNGALLVAPMCKISEKVKPHP 153
Query: 260 RMYNLFRRVADGLLKIREVMTLHIRE-------------SGKQLVTDPERLKDPVEFVQR 306
+ NL RV D + K + V T + + + K + D RLK +E ++
Sbjct: 154 VVINLLTRVEDIIPKWKIVPTKDVIDAAFKDPVKREEIRNNKLIYQDKPRLKTALEMLRT 213
Query: 307 LLDEKDKYDKI 317
+D +D +I
Sbjct: 214 SMDLEDTLHEI 224
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.136 0.389
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,331,479
Number of Sequences: 26719
Number of extensions: 655647
Number of successful extensions: 2214
Number of sequences better than 10.0: 36
Number of HSP's better than 10.0 without gapping: 11
Number of HSP's successfully gapped in prelim test: 25
Number of HSP's that attempted gapping in prelim test: 2152
Number of HSP's gapped (non-prelim): 45
length of query: 703
length of database: 11,318,596
effective HSP length: 106
effective length of query: 597
effective length of database: 8,486,382
effective search space: 5066370054
effective search space used: 5066370054
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 64 (29.3 bits)
Medicago: description of AC146568.7


