

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146568.5 - phase: 0
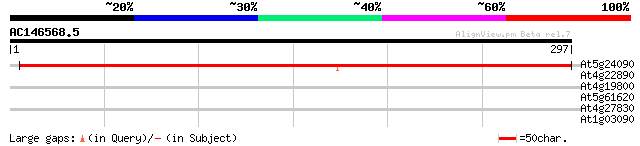
(297 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g24090 acidic endochitinase (dbj|BAA21861.1) 378 e-105
At4g22890 unknown protein 29 3.5
At4g19800 chitinase - like protein 29 3.5
At5g61620 transcriptional activator - like protein 28 5.9
At4g27830 beta-glucosidase like protein 28 5.9
At1g03090 putative 3-methylcrotonyl-CoA carboxylase 28 7.8
>At5g24090 acidic endochitinase (dbj|BAA21861.1)
Length = 302
Score = 378 bits (971), Expect = e-105
Identities = 180/295 (61%), Positives = 219/295 (74%), Gaps = 3/295 (1%)
Query: 6 KSTISFTFFSLLVLALANGSNAGKIAIYWGQNGNEGTLAEACATGNYEYVIIAFLPTFGD 65
K I F FF L+ + ++ G IAIYWGQNGNEG L+ CATG Y YV +AFL FG+
Sbjct: 8 KHVIYFLFFISCSLSKPSDASRGGIAIYWGQNGNEGNLSATCATGRYAYVNVAFLVKFGN 67
Query: 66 GQTPMINLAGHCDPYSNGCTGLSSDIKSCQAKGIKVLLSIGGGAGSYSIASTKDANSVAT 125
GQTP +NLAGHC+P +N CT S +K CQ++GIKV+LS+GGG G+YSI S +DA +A
Sbjct: 68 GQTPELNLAGHCNPAANTCTHFGSQVKDCQSRGIKVMLSLGGGIGNYSIGSREDAKVIAD 127
Query: 126 YLWNNFLGGKSSSRPLGPAVLDGIDFDIEGGSSQHWGDLARYLKGFN---KKVYITAAPQ 182
YLWNNFLGGKSSSRPLG AVLDGIDF+IE GS QHW DLAR L F+ +K+Y+T APQ
Sbjct: 128 YLWNNFLGGKSSSRPLGDAVLDGIDFNIELGSPQHWDDLARTLSKFSHRGRKIYLTGAPQ 187
Query: 183 CPFPDAWIGNALTTGLFDYVWVQFYNNPPCQYNPDAFMNFEDAWKQWTSGIPANKIFLGL 242
CPFPD +G+AL T FDYVW+QFYNNPPC Y+ N D+W +WT+ I A K FLGL
Sbjct: 188 CPFPDRLMGSALNTKRFDYVWIQFYNNPPCSYSSGNTQNLFDSWNKWTTSIAAQKFFLGL 247
Query: 243 PASPTAAGSGFISADDLTSTVLPVIKGSSKYGGVMLWSRYDDVQSGYSSSIKSHV 297
PA+P AAGSG+I D LTS +LP +K S KYGGVMLWS++ D ++GYSSSI + V
Sbjct: 248 PAAPEAAGSGYIPPDVLTSQILPTLKKSRKYGGVMLWSKFWDDKNGYSSSILASV 302
>At4g22890 unknown protein
Length = 324
Score = 28.9 bits (63), Expect = 3.5
Identities = 24/70 (34%), Positives = 29/70 (41%), Gaps = 8/70 (11%)
Query: 228 QWTSGIPANKIFLG-----LPASPTAAGSGFISADDLTSTVLP---VIKGSSKYGGVMLW 279
QWT P I L LPA T SG + D++ S VLP + K K G M
Sbjct: 38 QWTQLSPGKSISLRRRVFLLPAKATTEQSGPVGGDNVDSNVLPYCSINKAEKKTIGEMEQ 97
Query: 280 SRYDDVQSGY 289
+QS Y
Sbjct: 98 EFLQALQSFY 107
>At4g19800 chitinase - like protein
Length = 398
Score = 28.9 bits (63), Expect = 3.5
Identities = 43/174 (24%), Positives = 68/174 (38%), Gaps = 36/174 (20%)
Query: 99 IKVLLSIGGG-AGSYSIAS-TKDANSVATYLWNNFLGGKSSSRPLGPAVLDGIDFDIE-G 155
+K LLSIGGG A + AS + +S A+++ + +S G+D D E
Sbjct: 69 VKTLLSIGGGNADKDAFASMASNPDSRASFIQSTITVARSYG-------FHGLDLDWEYP 121
Query: 156 GSSQHWGDLARYLKGFNKKV------------YITAA-------PQCPFPDAWIGNAL-- 194
+ + D + L+ + V +TAA P+P I N+L
Sbjct: 122 RNEEEMYDFGKLLEEWRSAVEAESNSSGTTALILTAAVYYSSNYQGVPYPVLAISNSLDW 181
Query: 195 -TTGLFDYV---WVQFYNNPPCQYNPDAFMNFEDAWKQWT-SGIPANKIFLGLP 243
+D+ W P Y P + + + WT +G+PA K LG P
Sbjct: 182 INLMAYDFYGPGWSTVTGPPASLYLPTDGRSGDSGVRDWTEAGLPAKKAVLGFP 235
>At5g61620 transcriptional activator - like protein
Length = 317
Score = 28.1 bits (61), Expect = 5.9
Identities = 24/77 (31%), Positives = 35/77 (45%), Gaps = 7/77 (9%)
Query: 206 FYNNPPCQYNPDAFMNFEDAWKQ----WTSGIPANK-IFLGLPASPTAAGSGFISAD--D 258
+YN PP Y+P+ M + + SGIP + I +GLP S + S + D D
Sbjct: 234 YYNFPPIMYHPNYPMYYANPQVPVRFVHPSGIPVPRHIPIGLPLSQPSEASNMTNKDGLD 293
Query: 259 LTSTVLPVIKGSSKYGG 275
L + P G+S G
Sbjct: 294 LHIGLPPQATGASDLTG 310
>At4g27830 beta-glucosidase like protein
Length = 508
Score = 28.1 bits (61), Expect = 5.9
Identities = 25/98 (25%), Positives = 39/98 (39%), Gaps = 23/98 (23%)
Query: 197 GLFDYVWVQFYNNPPCQYNPDAFMNFEDAWKQWTSGIPANKIFLGLPASPTAAGSGFISA 256
G+ +Y+ Q YNNPP + D+ Q T I + ++G
Sbjct: 380 GILEYI-KQSYNNPPIYILENGMPMGRDSTLQDTQRIEFIQAYIG--------------- 423
Query: 257 DDLTSTVLPVIKGSSKYGGVMLWSRYD--DVQSGYSSS 292
+L IK S G +WS D ++ SGY++S
Sbjct: 424 -----AMLNAIKNGSDTRGYFVWSMIDLYELLSGYTTS 456
>At1g03090 putative 3-methylcrotonyl-CoA carboxylase
Length = 734
Score = 27.7 bits (60), Expect = 7.8
Identities = 13/41 (31%), Positives = 20/41 (48%)
Query: 206 FYNNPPCQYNPDAFMNFEDAWKQWTSGIPANKIFLGLPASP 246
+Y+NPP + + +A E W G +N I LG+ P
Sbjct: 534 WYSNPPFRVHHEAKQTIELEWNNECEGTGSNLISLGVRYQP 574
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.317 0.136 0.426
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,477,662
Number of Sequences: 26719
Number of extensions: 341891
Number of successful extensions: 680
Number of sequences better than 10.0: 6
Number of HSP's better than 10.0 without gapping: 2
Number of HSP's successfully gapped in prelim test: 4
Number of HSP's that attempted gapping in prelim test: 677
Number of HSP's gapped (non-prelim): 6
length of query: 297
length of database: 11,318,596
effective HSP length: 99
effective length of query: 198
effective length of database: 8,673,415
effective search space: 1717336170
effective search space used: 1717336170
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 60 (27.7 bits)
Medicago: description of AC146568.5


