

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146568.1 + phase: 0
(610 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
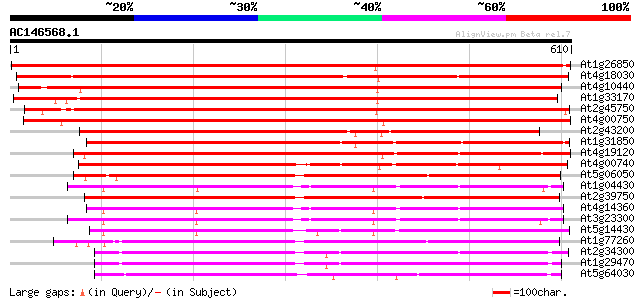
Score E
Sequences producing significant alignments: (bits) Value
At1g26850 unknown protein 1050 0.0
At4g18030 unknown protein 883 0.0
At4g10440 putative protein 735 0.0
At1g33170 unknown protein 704 0.0
At2g45750 hypothetical protein 681 0.0
At4g00750 unknown protein 676 0.0
At2g43200 hypothetical protein 564 e-161
At1g31850 unknown protein 511 e-145
At4g19120 ERD3 protein (ERD3) 507 e-144
At4g00740 unknown protein 483 e-136
At5g06050 ankyrin-like protein 443 e-124
At1g04430 unknown protein 436 e-122
At2g39750 unknown protein 436 e-122
At4g14360 427 e-119
At3g23300 ankyrin repeat protein 425 e-119
At5g14430 unknown protein 421 e-118
At1g77260 unknown protein 415 e-116
At2g34300 unknown protein 401 e-112
At1g29470 unknown protein 400 e-111
At5g64030 ankyrin-like protein 399 e-111
>At1g26850 unknown protein
Length = 616
Score = 1050 bits (2716), Expect = 0.0
Identities = 491/614 (79%), Positives = 548/614 (88%), Gaps = 9/614 (1%)
Query: 3 KPSAADNRTRSSVQIFIVVGLCCFFYILGAWQRSGFGKGDSIALEITKNNAECDVVPNLS 62
K S+AD +TRSSVQIFIV LCCFFYILGAWQRSGFGKGDSIALE+T + A+C++VP+L+
Sbjct: 4 KSSSADGKTRSSVQIFIVFSLCCFFYILGAWQRSGFGKGDSIALEMTNSGADCNIVPSLN 63
Query: 63 FDSHHAGEVSQIDESNS-NTKVFKPCEARYTDYTPCQDQRRAMTFPRENMNYRERHCPPE 121
F++HHAGE S + S + K F+PC+ RYTDYTPCQDQRRAMTFPR++M YRERHC PE
Sbjct: 64 FETHHAGESSLVGASEAAKVKAFEPCDGRYTDYTPCQDQRRAMTFPRDSMIYRERHCAPE 123
Query: 122 EEKLHCMIPAPKGYVTPFPWPKSRDYVPYANAPYKSLTVEKAIQNWIQYEGNVFRFPGGG 181
EKLHC+IPAPKGYVTPF WPKSRDYVPYANAPYK+LTVEKAIQNWIQYEG+VFRFPGGG
Sbjct: 124 NEKLHCLIPAPKGYVTPFSWPKSRDYVPYANAPYKALTVEKAIQNWIQYEGDVFRFPGGG 183
Query: 182 TQFPQGADKYIDQLASVIPINDGTVRTALDTGCGVASWGAYLWSRNVVAMSFAPRDSHEA 241
TQFPQGADKYIDQLASVIP+ +GTVRTALDTGCGVASWGAYLWSRNV AMSFAPRDSHEA
Sbjct: 184 TQFPQGADKYIDQLASVIPMENGTVRTALDTGCGVASWGAYLWSRNVRAMSFAPRDSHEA 243
Query: 242 QVQFALERGVPAVIGVFGTIKLPYPSRAFDMAHCSRCLIPWGANDGMYMMEVDRVLRPGG 301
QVQFALERGVPAVIGV GTIKLPYP+RAFDMAHCSRCLIPWGANDGMY+MEVDRVLRPGG
Sbjct: 244 QVQFALERGVPAVIGVLGTIKLPYPTRAFDMAHCSRCLIPWGANDGMYLMEVDRVLRPGG 303
Query: 302 YWVLSGPPINWKVNYKPWQRPKEELEEEQRKIEEVAKKLCWEKKSEKAEIAIWQKMTDTE 361
YW+LSGPPINWKVNYK WQRPKE+L+EEQRKIEE AK LCWEKK E EIAIWQK + E
Sbjct: 304 YWILSGPPINWKVNYKAWQRPKEDLQEEQRKIEEAAKLLCWEKKYEHGEIAIWQKRVNDE 363
Query: 362 SCRSRQDDSSVEFCESSDPDDVWYKKLKACVTPTP------KVSGGDLKPFPDRLYAIPP 415
+CRSRQDD FC++ D DDVWYKK++AC+TP P +V+GG+L+ FPDRL A+PP
Sbjct: 364 ACRSRQDDPRANFCKTDDTDDVWYKKMEACITPYPETSSSDEVAGGELQAFPDRLNAVPP 423
Query: 416 RVSSGSIPGVSSETYQNDNKMWKKHVNAYKKINSLLDSGRYRNIMDMNAGLGSFAAAIHS 475
R+SSGSI GV+ + Y++DN+ WKKHV AYK+INSLLD+GRYRNIMDMNAG G FAAA+ S
Sbjct: 424 RISSGSISGVTVDAYEDDNRQWKKHVKAYKRINSLLDTGRYRNIMDMNAGFGGFAAALES 483
Query: 476 SKSWVMNVVPTIAEKSTLGVIYERGLIGIYHDWCEGFSTYPRTYDLIHANGLFSLYQDKC 535
K WVMNVVPTIAEK+ LGV+YERGLIGIYHDWCE FSTYPRTYDLIHAN LFSLY++KC
Sbjct: 484 QKLWVMNVVPTIAEKNRLGVVYERGLIGIYHDWCEAFSTYPRTYDLIHANHLFSLYKNKC 543
Query: 536 NTEDILLEMDRILRPEGAVIIRDEVDVLIKVKKLIGGMRWNMKLVDHEDGPLVPEKVLIA 595
N +DILLEMDRILRPEGAVIIRD+VD LIKVK++I GMRW+ KLVDHEDGPLVPEKVLIA
Sbjct: 544 NADDILLEMDRILRPEGAVIIRDDVDTLIKVKRIIAGMRWDAKLVDHEDGPLVPEKVLIA 603
Query: 596 VKQYWVTDGNSTST 609
VKQYWVT NSTST
Sbjct: 604 VKQYWVT--NSTST 615
>At4g18030 unknown protein
Length = 621
Score = 883 bits (2282), Expect = 0.0
Identities = 413/608 (67%), Positives = 490/608 (79%), Gaps = 13/608 (2%)
Query: 8 DNRTRSSVQIFIVVGLCCFFYILGAWQRSGFGKGDSIALEITKNNAECDVVPNLSFDSHH 67
+NR+RS++ + +VVGLCCFFY+LGAWQ+SGFGKGDSIA+EITK D+V +L F+ HH
Sbjct: 10 NNRSRSTLSLLVVVGLCCFFYLLGAWQKSGFGKGDSIAMEITKQAQCTDIVTDLDFEPHH 69
Query: 68 AGEVSQIDESNSNTKVFKPCEARYTDYTPCQDQRRAMTFPRENMNYRERHCPPEEEKLHC 127
V +++ FKPC+ + DYTPCQ+Q RAM FPRENM YRERHCPP+ EKL C
Sbjct: 70 -NTVKIPHKADPKPVSFKPCDVKLKDYTPCQEQDRAMKFPRENMIYRERHCPPDNEKLRC 128
Query: 128 MIPAPKGYVTPFPWPKSRDYVPYANAPYKSLTVEKAIQNWIQYEGNVFRFPGGGTQFPQG 187
++PAPKGY+TPFPWPKSRDYV YANAP+KSLTVEKA QNW+Q++GNVF+FPGGGT FPQG
Sbjct: 129 LVPAPKGYMTPFPWPKSRDYVHYANAPFKSLTVEKAGQNWVQFQGNVFKFPGGGTMFPQG 188
Query: 188 ADKYIDQLASVIPINDGTVRTALDTGCGVASWGAYLWSRNVVAMSFAPRDSHEAQVQFAL 247
AD YI++LASVIPI DG+VRTALDTGCGVASWGAY+ RNV+ MSFAPRD+HEAQVQFAL
Sbjct: 189 ADAYIEELASVIPIKDGSVRTALDTGCGVASWGAYMLKRNVLTMSFAPRDNHEAQVQFAL 248
Query: 248 ERGVPAVIGVFGTIKLPYPSRAFDMAHCSRCLIPWGANDGMYMMEVDRVLRPGGYWVLSG 307
ERGVPA+I V G+I LPYP+RAFDMA CSRCLIPW AN+G Y+MEVDRVLRPGGYWVLSG
Sbjct: 249 ERGVPAIIAVLGSILLPYPARAFDMAQCSRCLIPWTANEGTYLMEVDRVLRPGGYWVLSG 308
Query: 308 PPINWKVNYKPWQRPKEELEEEQRKIEEVAKKLCWEKKSEKAEIAIWQKMTDTESCRSRQ 367
PPINWK +K W R K EL EQ++IE +A+ LCWEKK EK +IAI++K + SC
Sbjct: 309 PPINWKTWHKTWNRTKAELNAEQKRIEGIAESLCWEKKYEKGDIAIFRKKINDRSC---D 365
Query: 368 DDSSVEFCESSDPDDVWYKKLKACVTPTPKVS------GGDLKPFPDRLYAIPPRVSSGS 421
+ V+ C+ D DDVWYK+++ CVTP PKVS GG LK FP+RL+A+PP +S G
Sbjct: 366 RSTPVDTCKRKDTDDVWYKEIETCVTPFPKVSNEEEVAGGKLKKFPERLFAVPPSISKGL 425
Query: 422 IPGVSSETYQNDNKMWKKHVNAYKKINSLLDSGRYRNIMDMNAGLGSFAAAIHSSKSWVM 481
I GV E+YQ D +WKK V YK+IN L+ S RYRN+MDMNAGLG FAAA+ S KSWVM
Sbjct: 426 INGVDEESYQEDINLWKKRVTGYKRINRLIGSTRYRNVMDMNAGLGGFAAALESPKSWVM 485
Query: 482 NVVPTIAEKSTLGVIYERGLIGIYHDWCEGFSTYPRTYDLIHANGLFSLYQDKCNTEDIL 541
NV+PTI K+TL V+YERGLIGIYHDWCEGFSTYPRTYD IHA+G+FSLYQ C EDIL
Sbjct: 486 NVIPTI-NKNTLSVVYERGLIGIYHDWCEGFSTYPRTYDFIHASGVFSLYQHSCKLEDIL 544
Query: 542 LEMDRILRPEGAVIIRDEVDVLIKVKKLIGGMRWNMKLVDHEDGPLVPEKVLIAVKQYWV 601
LE DRILRPEG VI RDEVDVL V+K++ GMRW+ KL+DHEDGPLVPEK+L+A KQYWV
Sbjct: 545 LETDRILRPEGIVIFRDEVDVLNDVRKIVDGMRWDTKLMDHEDGPLVPEKILVATKQYWV 604
Query: 602 T--DGNST 607
DGN++
Sbjct: 605 AGDDGNNS 612
>At4g10440 putative protein
Length = 633
Score = 735 bits (1898), Expect = 0.0
Identities = 341/608 (56%), Positives = 445/608 (73%), Gaps = 23/608 (3%)
Query: 10 RTRSSVQIFIVVGLCCFFYILGAWQRSGFGKGDSIALEITKNNAECDVVPNLSFDSHHAG 69
R + I V GLC FY+LGAWQ +++ I+K E P+ S S +
Sbjct: 15 RRKKLTLILGVSGLCILFYVLGAWQ------ANTVPSSISKLGCETQSNPSSSSSSSSSS 68
Query: 70 EVSQID----------ESNSNTKVFKPCEARYTDYTPCQDQRRAMTFPRENMNYRERHCP 119
E +++D E+N K F+PCE ++YTPC+D++R F R M YRERHCP
Sbjct: 69 ESAELDFKSHNQIELKETNQTIKYFEPCELSLSEYTPCEDRQRGRRFDRNMMKYRERHCP 128
Query: 120 PEEEKLHCMIPAPKGYVTPFPWPKSRDYVPYANAPYKSLTVEKAIQNWIQYEGNVFRFPG 179
++E L+C+IP P Y PF WP+SRDY Y N P+K L+VEKA+QNWIQ EG+ FRFPG
Sbjct: 129 VKDELLYCLIPPPPNYKIPFKWPQSRDYAWYDNIPHKELSVEKAVQNWIQVEGDRFRFPG 188
Query: 180 GGTQFPQGADKYIDQLASVIPINDGTVRTALDTGCGVASWGAYLWSRNVVAMSFAPRDSH 239
GGT FP+GAD YID +A +IP+ DG +RTA+DTGCGVAS+GAYL R+++A+SFAPRD+H
Sbjct: 189 GGTMFPRGADAYIDDIARLIPLTDGGIRTAIDTGCGVASFGAYLLKRDIMAVSFAPRDTH 248
Query: 240 EAQVQFALERGVPAVIGVFGTIKLPYPSRAFDMAHCSRCLIPWGANDGMYMMEVDRVLRP 299
EAQVQFALERGVPA+IG+ G+ +LPYP+RAFD+AHCSRCLIPW NDG+Y+MEVDRVLRP
Sbjct: 249 EAQVQFALERGVPAIIGIMGSRRLPYPARAFDLAHCSRCLIPWFKNDGLYLMEVDRVLRP 308
Query: 300 GGYWVLSGPPINWKVNYKPWQRPKEELEEEQRKIEEVAKKLCWEKKSEKAEIAIWQKMTD 359
GGYW+LSGPPINWK ++ W+R +E+L++EQ IE+VAK LCW+K +EK +++IWQK +
Sbjct: 309 GGYWILSGPPINWKQYWRGWERTEEDLKKEQDSIEDVAKSLCWKKVTEKGDLSIWQKPLN 368
Query: 360 TESCRS-RQDDSSVEFCESSDPDDVWYKKLKACVTPTPKV------SGGDLKPFPDRLYA 412
C+ +Q++ S C S + D WYK L+ C+TP P+ +GG L+ +PDR +A
Sbjct: 369 HIECKKLKQNNKSPPICSSDNADSAWYKDLETCITPLPETNNPDDSAGGALEDWPDRAFA 428
Query: 413 IPPRVSSGSIPGVSSETYQNDNKMWKKHVNAYKKINSLLDSGRYRNIMDMNAGLGSFAAA 472
+PPR+ G+IP +++E ++ DN++WK+ + YKKI L GR+RNIMDMNA LG FAA+
Sbjct: 429 VPPRIIRGTIPEMNAEKFREDNEVWKERIAHYKKIVPELSHGRFRNIMDMNAFLGGFAAS 488
Query: 473 IHSSKSWVMNVVPTIAEKSTLGVIYERGLIGIYHDWCEGFSTYPRTYDLIHANGLFSLYQ 532
+ SWVMNVVP AEK TLGVIYERGLIG Y DWCEGFSTYPRTYD+IHA GLFSLY+
Sbjct: 489 MLKYPSWVMNVVPVDAEKQTLGVIYERGLIGTYQDWCEGFSTYPRTYDMIHAGGLFSLYE 548
Query: 533 DKCNTEDILLEMDRILRPEGAVIIRDEVDVLIKVKKLIGGMRWNMKLVDHEDGPLVPEKV 592
+C+ ILLEMDRILRPEG V++RD V+ L KV+K++ GM+W ++VDHE GP PEK+
Sbjct: 549 HRCDLTLILLEMDRILRPEGTVVLRDNVETLNKVEKIVKGMKWKSQIVDHEKGPFNPEKI 608
Query: 593 LIAVKQYW 600
L+AVK YW
Sbjct: 609 LVAVKTYW 616
>At1g33170 unknown protein
Length = 709
Score = 704 bits (1816), Expect = 0.0
Identities = 333/624 (53%), Positives = 436/624 (69%), Gaps = 36/624 (5%)
Query: 5 SAADNRTRSSVQIFIVVGLCCFFYILGAWQRSGFGKGDSIALEI---------------- 48
S A+ + + I V GLC Y+LG+WQ + S A
Sbjct: 9 SLAEAKRKRLTWILCVSGLCILSYVLGSWQTNTVPTSSSEAYSRMGCDETSTTTRAQTTQ 68
Query: 49 TKNNAECDVVPN---------LSFDSHHAGEVSQIDESNSNTKVFKPCEARYTDYTPCQD 99
T+ N D + L F+SHH E+ +N K F+PC+ ++YTPC+D
Sbjct: 69 TQTNPSSDDTSSSLSSSEPVELDFESHHKLELKI---TNQTVKYFEPCDMSLSEYTPCED 125
Query: 100 QRRAMTFPRENMNYRERHCPPEEEKLHCMIPAPKGYVTPFPWPKSRDYVPYANAPYKSLT 159
+ R F R M YRERHCP ++E L+C+IP P Y PF WP+SRDY Y N P+K L+
Sbjct: 126 RERGRRFDRNMMKYRERHCPSKDELLYCLIPPPPNYKIPFKWPQSRDYAWYDNIPHKELS 185
Query: 160 VEKAIQNWIQYEGNVFRFPGGGTQFPQGADKYIDQLASVIPINDGTVRTALDTGCGVASW 219
+EKAIQNWIQ EG FRFPGGGT FP+GAD YID +A +IP+ DG +RTA+DTGCGVAS+
Sbjct: 186 IEKAIQNWIQVEGERFRFPGGGTMFPRGADAYIDDIARLIPLTDGAIRTAIDTGCGVASF 245
Query: 220 GAYLWSRNVVAMSFAPRDSHEAQVQFALERGVPAVIGVFGTIKLPYPSRAFDMAHCSRCL 279
GAYL R++VAMSFAPRD+HEAQVQFALERGVPA+IG+ G+ +LPYP+RAFD+AHCSRCL
Sbjct: 246 GAYLLKRDIVAMSFAPRDTHEAQVQFALERGVPAIIGIMGSRRLPYPARAFDLAHCSRCL 305
Query: 280 IPWGANDGMYMMEVDRVLRPGGYWVLSGPPINWKVNYKPWQRPKEELEEEQRKIEEVAKK 339
IPW NDG+Y+ EVDRVLRPGGYW+LSGPPINWK +K W+R +E+L++EQ IE+ A+
Sbjct: 306 IPWFQNDGLYLTEVDRVLRPGGYWILSGPPINWKKYWKGWERSQEDLKQEQDSIEDAARS 365
Query: 340 LCWEKKSEKAEIAIWQKMTDTESC-RSRQDDSSVEFCESSD-PDDVWYKKLKACVTPTPK 397
LCW+K +EK +++IWQK + C + ++ + C SD PD WYK L++CVTP P+
Sbjct: 366 LCWKKVTEKGDLSIWQKPINHVECNKLKRVHKTPPLCSKSDLPDFAWYKDLESCVTPLPE 425
Query: 398 V------SGGDLKPFPDRLYAIPPRVSSGSIPGVSSETYQNDNKMWKKHVNAYKKINSLL 451
+GG L+ +P+R +A+PPR+ G+IP +++E ++ DN++WK+ ++ YK+I L
Sbjct: 426 ANSSDEFAGGALEDWPNRAFAVPPRIIGGTIPDINAEKFREDNEVWKERISYYKQIMPEL 485
Query: 452 DSGRYRNIMDMNAGLGSFAAAIHSSKSWVMNVVPTIAEKSTLGVIYERGLIGIYHDWCEG 511
GR+RNIMDMNA LG FAAA+ SWVMNVVP AEK TLGVI+ERG IG Y DWCEG
Sbjct: 486 SRGRFRNIMDMNAYLGGFAAAMMKYPSWVMNVVPVDAEKQTLGVIFERGFIGTYQDWCEG 545
Query: 512 FSTYPRTYDLIHANGLFSLYQDKCNTEDILLEMDRILRPEGAVIIRDEVDVLIKVKKLIG 571
FSTYPRTYDLIHA GLFS+Y+++C+ ILLEMDRILRPEG V+ RD V++L K++ +
Sbjct: 546 FSTYPRTYDLIHAGGLFSIYENRCDVTLILLEMDRILRPEGTVVFRDTVEMLTKIQSITN 605
Query: 572 GMRWNMKLVDHEDGPLVPEKVLIA 595
GMRW +++DHE GP PEK+L+A
Sbjct: 606 GMRWKSRILDHERGPFNPEKILLA 629
>At2g45750 hypothetical protein
Length = 631
Score = 681 bits (1758), Expect = 0.0
Identities = 332/616 (53%), Positives = 429/616 (68%), Gaps = 30/616 (4%)
Query: 17 IFIVVGLCCFFYILGAWQ------RSGFGKGDSIALE-ITKNNAECDVVPNLSFDSHHAG 69
+ +V LC Y+LG WQ R+ F D E T+ N+ D L FD+HH
Sbjct: 19 VTLVALLCIASYLLGIWQNTAVNPRAAFDDSDGTPCEGFTRPNSTKD----LDFDAHH-- 72
Query: 70 EVSQIDESNSNTKVFKPCEARYTDYTPCQDQRRAMTFPRENMNYRERHCPPEEEKLHCMI 129
+ F C A +++TPC+D +R++ F RE + YR+RHCP EE L C I
Sbjct: 73 NIQDPPPVTETAVSFPSCAAALSEHTPCEDAKRSLKFSRERLEYRQRHCPEREEILKCRI 132
Query: 130 PAPKGYVTPFPWPKSRDYVPYANAPYKSLTVEKAIQNWIQYEGNVFRFPGGGTQFPQGAD 189
PAP GY TPF WP SRD +AN P+ LTVEK QNW++YE + F FPGGGT FP+GAD
Sbjct: 133 PAPYGYKTPFRWPASRDVAWFANVPHTELTVEKKNQNWVRYENDRFWFPGGGTMFPRGAD 192
Query: 190 KYIDQLASVIPINDGTVRTALDTGCGVASWGAYLWSRNVVAMSFAPRDSHEAQVQFALER 249
YID + +I ++DG++RTA+DTGCGVAS+GAYL SRN+ MSFAPRD+HEAQVQFALER
Sbjct: 193 AYIDDIGRLIDLSDGSIRTAIDTGCGVASFGAYLLSRNITTMSFAPRDTHEAQVQFALER 252
Query: 250 GVPAVIGVFGTIKLPYPSRAFDMAHCSRCLIPWGANDGMYMMEVDRVLRPGGYWVLSGPP 309
GVPA+IG+ TI+LPYPSRAFD+AHCSRCLIPWG NDG Y+MEVDRVLRPGGYW+LSGPP
Sbjct: 253 GVPAMIGIMATIRLPYPSRAFDLAHCSRCLIPWGQNDGAYLMEVDRVLRPGGYWILSGPP 312
Query: 310 INWKVNYKPWQRPKEELEEEQRKIEEVAKKLCWEKKSEKAEIAIWQKMTDTESC-RSRQD 368
INW+ +K W+R ++L EQ +IE+VA+ LCW+K ++ ++AIWQK + C ++R+
Sbjct: 313 INWQKRWKGWERTMDDLNAEQTQIEQVARSLCWKKVVQRDDLAIWQKPFNHIDCKKTREV 372
Query: 369 DSSVEFC-ESSDPDDVWYKKLKACVTPTPK---------VSGGDLKPFPDRLYAIPPRVS 418
+ EFC DPD WY K+ +C+TP P+ V+GG ++ +P RL AIPPRV+
Sbjct: 373 LKNPEFCRHDQDPDMAWYTKMDSCLTPLPEVDDAEDLKTVAGGKVEKWPARLNAIPPRVN 432
Query: 419 SGSIPGVSSETYQNDNKMWKKHVNAYKKIN-SLLDSGRYRNIMDMNAGLGSFAAAIHSSK 477
G++ ++ E + + K+WK+ V+ YKK++ L ++GRYRN++DMNA LG FAAA+
Sbjct: 433 KGALEEITPEAFLENTKLWKQRVSYYKKLDYQLGETGRYRNLVDMNAYLGGFAAALADDP 492
Query: 478 SWVMNVVPTIAEKSTLGVIYERGLIGIYHDWCEGFSTYPRTYDLIHANGLFSLYQDKCNT 537
WVMNVVP A+ +TLGVIYERGLIG Y +WCE STYPRTYD IHA+ +F+LYQ +C
Sbjct: 493 VWVMNVVPVEAKLNTLGVIYERGLIGTYQNWCEAMSTYPRTYDFIHADSVFTLYQGQCEP 552
Query: 538 EDILLEMDRILRPEGAVIIRDEVDVLIKVKKLIGGMRWNMKLVDHEDGPLVPEKVLIAVK 597
E+ILLEMDRILRP G VIIRD+VDVLIKVK+L G+ W ++ DHE GP EK+ AVK
Sbjct: 553 EEILLEMDRILRPGGGVIIRDDVDVLIKVKELTKGLEWEGRIADHEKGPHEREKIYYAVK 612
Query: 598 QYWVT-----DGNSTS 608
QYW D N+TS
Sbjct: 613 QYWTVPAPDEDKNNTS 628
>At4g00750 unknown protein
Length = 633
Score = 676 bits (1745), Expect = 0.0
Identities = 315/608 (51%), Positives = 425/608 (69%), Gaps = 14/608 (2%)
Query: 16 QIFIVVGLCCFFYILGAWQRSGFGKGDSIALEITKNNAEC----DVVPNLSFDSHHAGEV 71
++ ++ LC FY +G WQ SG G S + C P L+F S H
Sbjct: 24 RVILIAILCVTFYFVGVWQHSGRGISRSSISNHELTSVPCTFPHQTTPILNFASRHTAPD 83
Query: 72 SQIDESNSNTKVFKPCEARYTDYTPCQDQRRAMTFPRENMNYRERHCPPEEEKLHCMIPA 131
+++ C +++YTPC+ R++ FPRE + YRERHCP + E + C IPA
Sbjct: 84 LPPTITDARVVQIPSCGVEFSEYTPCEFVNRSLNFPRERLIYRERHCPEKHEIVRCRIPA 143
Query: 132 PKGYVTPFPWPKSRDYVPYANAPYKSLTVEKAIQNWIQYEGNVFRFPGGGTQFPQGADKY 191
P GY PF WP+SRD +AN P+ LTVEK QNW++YE + F FPGGGT FP+GAD Y
Sbjct: 144 PYGYSLPFRWPESRDVAWFANVPHTELTVEKKNQNWVRYEKDRFLFPGGGTMFPRGADAY 203
Query: 192 IDQLASVIPINDGTVRTALDTGCGVASWGAYLWSRNVVAMSFAPRDSHEAQVQFALERGV 251
ID++ +I + DG++RTA+DTGCGVAS+GAYL SRN+V MSFAPRD+HEAQVQFALERGV
Sbjct: 204 IDEIGRLINLKDGSIRTAIDTGCGVASFGAYLMSRNIVTMSFAPRDTHEAQVQFALERGV 263
Query: 252 PAVIGVFGTIKLPYPSRAFDMAHCSRCLIPWGANDGMYMMEVDRVLRPGGYWVLSGPPIN 311
PA+IGV +I+LP+P+RAFD+AHCSRCLIPWG +G Y++EVDRVLRPGGYW+LSGPPIN
Sbjct: 264 PAIIGVLASIRLPFPARAFDIAHCSRCLIPWGQYNGTYLIEVDRVLRPGGYWILSGPPIN 323
Query: 312 WKVNYKPWQRPKEELEEEQRKIEEVAKKLCWEKKSEKAEIAIWQKMTDTESC-RSRQDDS 370
W+ ++K W+R +++L EQ +IE VA+ LCW K ++ ++A+WQK T+ C R+R
Sbjct: 324 WQRHWKGWERTRDDLNSEQSQIERVARSLCWRKLVQREDLAVWQKPTNHVHCKRNRIALG 383
Query: 371 SVEFCESSDPDDVWYKKLKACVTPTPKVSGGDLKP--------FPDRLYAIPPRVSSGSI 422
FC + P+ WY KL+ C+TP P+V+G ++K +P+RL A+PPR+ SGS+
Sbjct: 384 RPPFCHRTLPNQGWYTKLETCLTPLPEVTGSEIKEVAGGQLARWPERLNALPPRIKSGSL 443
Query: 423 PGVSSETYQNDNKMWKKHVNAYKKIN-SLLDSGRYRNIMDMNAGLGSFAAAIHSSKSWVM 481
G++ + + ++ + W++ V+ YKK + L ++GRYRN +DMNA LG FA+A+ WVM
Sbjct: 444 EGITEDEFVSNTEKWQRRVSYYKKYDQQLAETGRYRNFLDMNAHLGGFASALVDDPVWVM 503
Query: 482 NVVPTIAEKSTLGVIYERGLIGIYHDWCEGFSTYPRTYDLIHANGLFSLYQDKCNTEDIL 541
NVVP A +TLGVIYERGLIG Y +WCE STYPRTYD IHA+ +FSLY+D+C+ EDIL
Sbjct: 504 NVVPVEASVNTLGVIYERGLIGTYQNWCEAMSTYPRTYDFIHADSVFSLYKDRCDMEDIL 563
Query: 542 LEMDRILRPEGAVIIRDEVDVLIKVKKLIGGMRWNMKLVDHEDGPLVPEKVLIAVKQYWV 601
LEMDRILRP+G+VIIRD++DVL KVKK+ M+W ++ DHE+GPL EK+L VK+YW
Sbjct: 564 LEMDRILRPKGSVIIRDDIDVLTKVKKITDAMQWEGRIGDHENGPLEREKILFLVKEYWT 623
Query: 602 TDGNSTST 609
S+
Sbjct: 624 APAPDQSS 631
>At2g43200 hypothetical protein
Length = 611
Score = 564 bits (1454), Expect = e-161
Identities = 269/507 (53%), Positives = 352/507 (69%), Gaps = 10/507 (1%)
Query: 77 SNSNTKVFKPCEARYTDYTPCQDQRRAMTFPRENMNYRERHCPP-EEEKLHCMIPAPKGY 135
S+S + F C +T+Y PC D A + E RERHCP +EK C++P P GY
Sbjct: 84 SSSLSSYFPLCPKNFTNYLPCHDPSTARQYSIERHYRRERHCPDIAQEKFRCLVPKPTGY 143
Query: 136 VTPFPWPKSRDYVPYANAPYKSLTVEKAIQNWIQYEGNVFRFPGGGTQFPQGADKYIDQL 195
TPFPWP+SR Y + N P+K L K QNW++ EG+ F FPGGGT FP G Y+D +
Sbjct: 144 KTPFPWPESRKYAWFRNVPFKRLAELKKTQNWVRLEGDRFVFPGGGTSFPGGVKDYVDVI 203
Query: 196 ASVIPINDGTVRTALDTGCGVASWGAYLWSRNVVAMSFAPRDSHEAQVQFALERGVPAVI 255
SV+P+ G++RT LD GCGVAS+GA+L + ++ MS APRD HEAQVQFALERG+PA++
Sbjct: 204 LSVLPLASGSIRTVLDIGCGVASFGAFLLNYKILTMSIAPRDIHEAQVQFALERGLPAML 263
Query: 256 GVFGTIKLPYPSRAFDMAHCSRCLIPWGANDGMYMMEVDRVLRPGGYWVLSGPPINWKVN 315
GV T KLPYPSR+FDM HCSRCL+ W + DG+Y+MEVDRVLRP GYWVLSGPP+ +V
Sbjct: 264 GVLSTYKLPYPSRSFDMVHCSRCLVNWTSYDGLYLMEVDRVLRPEGYWVLSGPPVASRVK 323
Query: 316 YKPWQRPKEELEEEQRKIEEVAKKLCWEKKSEKAEIAIWQKMTDTESCRSRQDDSSVEF- 374
+K +R +EL+ + K+ +V ++LCWEK +E + IW+K ++ CR R +++F
Sbjct: 324 FKNQKRDSKELQNQMEKLNDVFRRLCWEKIAESYPVVIWRKPSNHLQCRKRL--KALKFP 381
Query: 375 --CESSDPDDVWYKKLKACVTPTPKVSGGD---LKPFPDRLYAIPPRVSSGSIPGVSSET 429
C SSDPD WYK+++ C+TP P V+ + LK +P+RL + PR+ +GSI G +
Sbjct: 382 GLCSSSDPDAAWYKEMEPCITPLPDVNDTNKTVLKNWPERLNHV-PRMKTGSIQGTTIAG 440
Query: 430 YQNDNKMWKKHVNAYKKINSLLDSGRYRNIMDMNAGLGSFAAAIHSSKSWVMNVVPTIAE 489
++ D +W++ V Y L +G+YRN++DMNAGLG FAAA+ WVMNVVP +
Sbjct: 441 FKADTNLWQRRVLYYDTKFKFLSNGKYRNVIDMNAGLGGFAAALIKYPMWVMNVVPFDLK 500
Query: 490 KSTLGVIYERGLIGIYHDWCEGFSTYPRTYDLIHANGLFSLYQDKCNTEDILLEMDRILR 549
+TLGV+Y+RGLIG Y +WCE STYPRTYDLIHANG+FSLY DKC+ DILLEM RILR
Sbjct: 501 PNTLGVVYDRGLIGTYMNWCEALSTYPRTYDLIHANGVFSLYLDKCDIVDILLEMQRILR 560
Query: 550 PEGAVIIRDEVDVLIKVKKLIGGMRWN 576
PEGAVIIRD DVL+KVK + MRWN
Sbjct: 561 PEGAVIIRDRFDVLVKVKAITNQMRWN 587
>At1g31850 unknown protein
Length = 603
Score = 511 bits (1317), Expect = e-145
Identities = 247/534 (46%), Positives = 352/534 (65%), Gaps = 15/534 (2%)
Query: 84 FKPCEARYTDYTPCQDQRRAMTFPRENMNYRERHCPPEEEKLHCMIPAPKGYVTPFPWPK 143
F C + + DYTPC D +R + +++ ERHCPP EK C+IP P GY P WPK
Sbjct: 76 FPECGSEFQDYTPCTDPKRWKKYGVHRLSFLERHCPPVYEKNECLIPPPDGYKPPIRWPK 135
Query: 144 SRDYVPYANAPYKSLTVEKAIQNWIQYEGNVFRFPGGGTQFPQGADKYIDQLASVIP-IN 202
SR+ Y N PY + +K+ Q+W++ EG+ F FPGGGT FP+G Y+D + +IP +
Sbjct: 136 SREQCWYRNVPYDWINKQKSNQHWLKKEGDKFHFPGGGTMFPRGVSHYVDLMQDLIPEMK 195
Query: 203 DGTVRTALDTGCGVASWGAYLWSRNVVAMSFAPRDSHEAQVQFALERGVPAVIGVFGTIK 262
DGTVRTA+DTGCGVASWG L R ++++S APRD+HEAQVQFALERG+PA++G+ T +
Sbjct: 196 DGTVRTAIDTGCGVASWGGDLLDRGILSLSLAPRDNHEAQVQFALERGIPAILGIISTQR 255
Query: 263 LPYPSRAFDMAHCSRCLIPWGANDGMYMMEVDRVLRPGGYWVLSGPPINWKVNYKPWQRP 322
LP+PS AFDMAHCSRCLIPW G+Y++E+ R++RPGG+WVLSGPP+N+ ++ W
Sbjct: 256 LPFPSNAFDMAHCSRCLIPWTEFGGIYLLEIHRIVRPGGFWVLSGPPVNYNRRWRGWNTT 315
Query: 323 KEELEEEQRKIEEVAKKLCWEKKSEKAEIAIWQKMTDTESCRSRQDDSSVEF---CESS- 378
E+ + + K++ + +C++K ++K +IA+WQK++D +SC + + + C+ S
Sbjct: 316 MEDQKSDYNKLQSLLTSMCFKKYAQKDDIAVWQKLSD-KSCYDKIAKNMEAYPPKCDDSI 374
Query: 379 DPDDVWYKKLKAC-VTPTPKV--SG-GDLKPFPDRLYAIPPRVSSGSIPGVSSETYQNDN 434
+PD WY L+ C V PTPKV SG G + +P+RL+ P R+ G + G S+ + ++D+
Sbjct: 375 EPDSAWYTPLRPCVVAPTPKVKKSGLGSIPKWPERLHVAPERI--GDVHGGSANSLKHDD 432
Query: 435 KMWKKHVNAYKKINSLLDSGRYRNIMDMNAGLGSFAAAIHSSKSWVMNVVPTIAEKSTLG 494
WK V YKK+ L + + RN+MDMN G F+AA+ WVMNVV + + S L
Sbjct: 433 GKWKNRVKHYKKVLPALGTDKIRNVMDMNTVYGGFSAALIEDPIWVMNVVSSYSANS-LP 491
Query: 495 VIYERGLIGIYHDWCEGFSTYPRTYDLIHANGLFSLYQDKCNTEDILLEMDRILRPEGAV 554
V+++RGLIG YHDWCE FSTYPRTYDL+H + LF+L +C + ILLEMDRILRP G V
Sbjct: 492 VVFDRGLIGTYHDWCEAFSTYPRTYDLLHLDSLFTLESHRCEMKYILLEMDRILRPSGYV 551
Query: 555 IIRDEVDVLIKVKKLIGGMRWNMKLVDHEDGPLVPEKVLIAVKQYWVTDGNSTS 608
IIR+ + + L G+RW+ + + E + EK+L+ K+ W + N TS
Sbjct: 552 IIRESSYFMDAITTLAKGIRWSCRREETEYA-VKSEKILVCQKKLWFS-SNQTS 603
>At4g19120 ERD3 protein (ERD3)
Length = 600
Score = 507 bits (1305), Expect = e-144
Identities = 243/551 (44%), Positives = 346/551 (62%), Gaps = 15/551 (2%)
Query: 70 EVSQIDESNS---NTKVFKPCEARYTDYTPCQDQRRAMTFPRENMNYRERHCPPEEEKLH 126
E S +D +S + F C + Y DYTPC D R+ + + + ERHCPP ++
Sbjct: 53 ESSSLDVDDSLQVKSVSFSECSSDYQDYTPCTDPRKWKKYGTHRLTFMERHCPPVFDRKQ 112
Query: 127 CMIPAPKGYVTPFPWPKSRDYVPYANAPYKSLTVEKAIQNWIQYEGNVFRFPGGGTQFPQ 186
C++P P GY P WPKS+D Y N PY + +K+ QNW++ EG F FPGGGT FP
Sbjct: 113 CLVPPPDGYKPPIRWPKSKDECWYRNVPYDWINKQKSNQNWLRKEGEKFIFPGGGTMFPH 172
Query: 187 GADKYIDQLASVIP-INDGTVRTALDTGCGVASWGAYLWSRNVVAMSFAPRDSHEAQVQF 245
G Y+D + +IP + DGT+RTA+DTGCGVASWG L R ++ +S APRD+HEAQVQF
Sbjct: 173 GVSAYVDLMQDLIPEMKDGTIRTAIDTGCGVASWGGDLLDRGILTVSLAPRDNHEAQVQF 232
Query: 246 ALERGVPAVIGVFGTIKLPYPSRAFDMAHCSRCLIPWGANDGMYMMEVDRVLRPGGYWVL 305
ALERG+PA++G+ T +LP+PS +FDMAHCSRCLIPW G+Y++EV R+LRPGG+WVL
Sbjct: 233 ALERGIPAILGIISTQRLPFPSNSFDMAHCSRCLIPWTEFGGVYLLEVHRILRPGGFWVL 292
Query: 306 SGPPINWKVNYKPWQRPKEELEEEQRKIEEVAKKLCWEKKSEKAEIAIWQKMTDT--ESC 363
SGPP+N++ +K W EE K++E+ +C++ ++K +IA+WQK D +
Sbjct: 293 SGPPVNYENRWKGWDTTIEEQRSNYEKLQELLSSMCFKMYAKKDDIAVWQKSPDNLCYNK 352
Query: 364 RSRQDDSSVEFCESS-DPDDVWYKKLKAC-VTPTPKVSGGDLK---PFPDRLYAIPPRVS 418
S D+ C+ S +PD WY L+ C V P+PK+ DL+ +P+RL+ P R+S
Sbjct: 353 LSNDPDAYPPKCDDSLEPDSAWYTPLRPCVVVPSPKLKKTDLESTPKWPERLHTTPERIS 412
Query: 419 SGSIPGVSSETYQNDNKMWKKHVNAYKKINSLLDSGRYRNIMDMNAGLGSFAAAIHSSKS 478
+PG + +++D+ WK YKK+ + S + RN+MDMN G AAA+ +
Sbjct: 413 --DVPGGNGNVFKHDDSKWKTRAKHYKKLLPAIGSDKIRNVMDMNTAYGGLAAALVNDPL 470
Query: 479 WVMNVVPTIAEKSTLGVIYERGLIGIYHDWCEGFSTYPRTYDLIHANGLFSLYQDKCNTE 538
WVMNVV + A +TL V+++RGLIG YHDWCE FSTYPRTYDL+H +GLF+ +C+ +
Sbjct: 471 WVMNVVSSYA-ANTLPVVFDRGLIGTYHDWCEAFSTYPRTYDLLHVDGLFTSESQRCDMK 529
Query: 539 DILLEMDRILRPEGAVIIRDEVDVLIKVKKLIGGMRWNMKLVDHEDGPLVPEKVLIAVKQ 598
++LEMDRILRP G IIR+ + + +RW+ + + + EK+LI K+
Sbjct: 530 YVMLEMDRILRPSGYAIIRESSYFADSIASVAKELRWSCR-KEQTESASANEKLLICQKK 588
Query: 599 YWVTDGNSTST 609
W + S+ T
Sbjct: 589 LWYSSNASSET 599
>At4g00740 unknown protein
Length = 600
Score = 483 bits (1242), Expect = e-136
Identities = 249/543 (45%), Positives = 336/543 (61%), Gaps = 28/543 (5%)
Query: 76 ESNSNTKVFKPCEARYTDYTPCQDQRRAMTFPRENMNYRERHCPPEEEKLHCMIPAPKGY 135
E+ + + + C A + PC+D RR RE YRERHCP EE C+IP P GY
Sbjct: 73 EAGQHLQPIEYCPAEAVAHMPCEDPRRNSQLSREMNFYRERHCPLPEETPLCLIPPPSGY 132
Query: 136 VTPFPWPKSRDYVPYANAPYKSLTVEKAIQNWIQYEGNVFRFPGGGTQFPQGADKYIDQL 195
P PWP+S + +AN PY + K Q W++ EG F FPGGGT FP GA +YI++L
Sbjct: 133 KIPVPWPESLHKIWHANMPYNKIADRKGHQGWMKREGEYFTFPGGGTMFPGGAGQYIEKL 192
Query: 196 ASVIPINDGTVRTALDTGCGVASWGAYLWSRNVVAMSFAPRDSHEAQVQFALERGVPAVI 255
A IP+N GT+RTALD GCGVAS+G L S+ ++A+SFAPRDSH++Q+QFALERGVPA +
Sbjct: 193 AQYIPLNGGTLRTALDMGCGVASFGGTLLSQGILALSFAPRDSHKSQIQFALERGVPAFV 252
Query: 256 GVFGTIKLPYPSRAFDMAHCSRCLIPWGANDGMYMMEVDRVLRPGGYWVLSGPPINWKVN 315
+ GT +LP+P+ +FD+ HCSRCLIP+ A + Y +EVDR+LRPGGY V+SGPP+ W
Sbjct: 253 AMLGTRRLPFPAYSFDLMHCSRCLIPFTAYNATYFIEVDRLLRPGGYLVISGPPVQW--- 309
Query: 316 YKPWQRPKEELEEEQRKIEEVAKKLCWEKKSEKAEIAIWQKMTDTESCRSRQDDSSVEFC 375
PK+ ++E ++ VA+ LC+E + IW+K +SC Q++ +E C
Sbjct: 310 ------PKQ--DKEWADLQAVARALCYELIAVDGNTVIWKKPVG-DSCLPSQNEFGLELC 360
Query: 376 -ESSDPDDVWYKKLKACVTPTPKVSG----GDLKPFPDRLYAIPPRVSSGSIPGVSSETY 430
ES P D WY KLK CVT V G G + +P+RL +P R + + +
Sbjct: 361 DESVPPSDAWYFKLKRCVTRPSSVKGEHALGTISKWPERLTKVPSR---AIVMKNGLDVF 417
Query: 431 QNDNKMWKKHVNAYK-KINSLLDSGRYRNIMDMNAGLGSFAAAIHSSKSWVMNVVPTIAE 489
+ D + W + V Y+ +N L S RN+MDMNA G FAA + S WVMNV+P +
Sbjct: 418 EADARRWARRVAYYRDSLNLKLKSPTVRNVMDMNAFFGGFAATLASDPVWVMNVIPA-RK 476
Query: 490 KSTLGVIYERGLIGIYHDWCEGFSTYPRTYDLIHANGLFSLY------QDKCNTEDILLE 543
TL VIY+RGLIG+YHDWCE FSTYPRTYD IH +G+ SL + +C+ D+++E
Sbjct: 477 PLTLDVIYDRGLIGVYHDWCEPFSTYPRTYDFIHVSGIESLIKRQDSSKSRCSLVDLMVE 536
Query: 544 MDRILRPEGAVIIRDEVDVLIKVKKLIGGMRWNMKLVDHEDGPLVPEKVLIAVKQYWVTD 603
MDRILRPEG V+IRD +VL KV ++ +RW+ + + E EK+LIA K W
Sbjct: 537 MDRILRPEGKVVIRDSPEVLDKVARMAHAVRWSSSIHEKEPESHGREKILIATKSLWKLP 596
Query: 604 GNS 606
NS
Sbjct: 597 SNS 599
>At5g06050 ankyrin-like protein
Length = 682
Score = 443 bits (1139), Expect = e-124
Identities = 240/547 (43%), Positives = 336/547 (60%), Gaps = 30/547 (5%)
Query: 70 EVSQIDESNSNT-----KVFKPCEARYTDYTPCQDQRRAMTFPRENMNYR----ERHCPP 120
E S D+ S T + F+ C T+Y PC D A+ R N R ER+CP
Sbjct: 130 ESSDDDDIKSTTARVSVRKFEICSENMTEYIPCLDNVEAIK--RLNSTARGERFERNCPN 187
Query: 121 EEEKLHCMIPAPKGYVTPFPWPKSRDYVPYANAPYKSLTVEKAIQNWIQYEGNVFRFPGG 180
+ L+C +P P+GY +P PWP+SRD V + N P+ L +K QNWI E + F+FPGG
Sbjct: 188 DGMGLNCTVPIPQGYRSPIPWPRSRDEVWFNNVPHTKLVEDKGGQNWIYKENDKFKFPGG 247
Query: 181 GTQFPQGADKYIDQLASVIP-INDGT-VRTALDTGCGVASWGAYLWSRNVVAMSFAPRDS 238
GTQF GAD+Y+DQ++ +IP I+ G R LD GCGVAS+GAYL SRNV+ MS AP+D
Sbjct: 248 GTQFIHGADQYLDQISQMIPDISFGNHTRVVLDIGCGVASFGAYLMSRNVLTMSIAPKDV 307
Query: 239 HEAQVQFALERGVPAVIGVFGTIKLPYPSRAFDMAHCSRCLIPWGANDGMYMMEVDRVLR 298
HE Q+QFALERGVPA++ F T +L YPS+AFD+ HCSRC I W +DG+ ++EV+R+LR
Sbjct: 308 HENQIQFALERGVPAMVAAFTTRRLLYPSQAFDLVHCSRCRINWTRDDGILLLEVNRMLR 367
Query: 299 PGGYWVLSGPPINWKVNYKPWQRPKEELEEEQRKIEEVAKKLCWEKKSEKAEIAIWQKMT 358
GGY+V + P+ + ++ LEE+ ++ + +LCW ++ IAIWQK
Sbjct: 368 AGGYFVWAAQPV---------YKHEKALEEQWEEMLNLTTRLCWVLVKKEGYIAIWQKPV 418
Query: 359 DTESCRSRQDDSSVEFCES-SDPDDVWYKKLKACVTPTPKVS-GGDLKPFPDRLYAIPPR 416
+ SR S C S DPD+VWY LKAC+T + G +L P+P RL P R
Sbjct: 419 NNTCYLSRGAGVSPPLCNSEDDPDNVWYVDLKACITRIEENGYGANLAPWPARLLTPPDR 478
Query: 417 VSSGSIPG--VSSETYQNDNKMWKKHVNAYKKINSLLDSGRYRNIMDMNAGLGSFAAAIH 474
+ + I E + ++K WK+ ++ Y G RN++DM AG G FAAA+
Sbjct: 479 LQTIQIDSYIARKELFVAESKYWKEIISNYVNALHWKQIG-LRNVLDMRAGFGGFAAALA 537
Query: 475 SSK--SWVMNVVPTIAEKSTLGVIYERGLIGIYHDWCEGFSTYPRTYDLIHANGLFSLYQ 532
K WV+NV+P ++ +TL VIY+RGL+G+ HDWCE F TYPRTYDL+HA GLFS+ +
Sbjct: 538 ELKVDCWVLNVIP-VSGPNTLPVIYDRGLLGVMHDWCEPFDTYPRTYDLLHAAGLFSIER 596
Query: 533 DKCNTEDILLEMDRILRPEGAVIIRDEVDVLIKVKKLIGGMRWNMKLVDHEDGPLVPEKV 592
+CN ++LEMDRILRP G V IRD ++V +++++ MRW+ L + +GP +V
Sbjct: 597 KRCNMTTMMLEMDRILRPGGRVYIRDTINVTSELQEIGNAMRWHTSLRETAEGPHSSYRV 656
Query: 593 LIAVKQY 599
L+ K++
Sbjct: 657 LLCEKRF 663
>At1g04430 unknown protein
Length = 623
Score = 436 bits (1122), Expect = e-122
Identities = 235/567 (41%), Positives = 336/567 (58%), Gaps = 43/567 (7%)
Query: 64 DSHHAGEVSQIDESNSNTKVFKPCEARYTDYTPCQDQ----RRAMTFPRENMNYRERHCP 119
D+ V+ ++S K F C+ R+++ PC D+ + + M + ERHCP
Sbjct: 64 DTKQDDSVANAEDSLVVAKSFPVCDDRHSEIIPCLDRNFIYQMRLKLDLSLMEHYERHCP 123
Query: 120 PEEEKLHCMIPAPKGYVTPFPWPKSRDYVPYANAPYKSLTVEKAIQNWIQYEGNVFRFPG 179
P E + +C+IP P GY P WPKSRD V AN P+ L EK+ QNW+ +G FPG
Sbjct: 124 PPERRFNCLIPPPSGYKVPIKWPKSRDEVWKANIPHTHLAKEKSDQNWMVEKGEKISFPG 183
Query: 180 GGTQFPQGADKYIDQLASVIPIN------DGTVRTALDTGCGVASWGAYLWSRNVVAMSF 233
GGT F GADKYI +A+++ + +G +RT LD GCGVAS+GAYL + +++ MS
Sbjct: 184 GGTHFHYGADKYIASIANMLNFSNDVLNDEGRLRTVLDVGCGVASFGAYLLASDIMTMSL 243
Query: 234 APRDSHEAQVQFALERGVPAVIGVFGTIKLPYPSRAFDMAHCSRCLIPWGANDGMYMMEV 293
AP D H+ Q+QFALERG+PA +GV GT +LPYPSR+F+ AHCSRC I W DG+ ++E+
Sbjct: 244 APNDVHQNQIQFALERGIPAYLGVLGTKRLPYPSRSFEFAHCSRCRIDWLQRDGLLLLEL 303
Query: 294 DRVLRPGGYWVLSGPPINWKVNYKPWQRPKEELEEEQRKIEEVAKKLCWEKKSEKAEIAI 353
DRVLRPGGY+ S P + + + +E L + +++ + +++CW ++ + +
Sbjct: 304 DRVLRPGGYFAYSSP--------EAYAQDEENL-KIWKEMSALVERMCWRIAVKRNQTVV 354
Query: 354 WQKMTDTESCRSRQDDSSVEFCES-SDPDDVWYKKLKACVTP----TPKVSGGDLKPFPD 408
WQK + R+ + C S +DPD V ++AC+TP K G L P+P
Sbjct: 355 WQKPLSNDCYLEREPGTQPPLCRSDADPDAVAGVSMEACITPYSKHDHKTKGSGLAPWPA 414
Query: 409 RLYAIPPRVSSGSIPGVSSETYQNDNKMWKKHVNAY-KKINSLLDSGRYRNIMDMNAGLG 467
RL + PPR++ G S++ ++ D ++WK+ V++Y ++S + S RNIMDM A +G
Sbjct: 415 RLTSSPPRLADF---GYSTDMFEKDTELWKQQVDSYWNLMSSKVKSNTVRNIMDMKAHMG 471
Query: 468 SFAAAIHSSKSWVMNVVPTIAEKSTLGVIYERGLIGIYHDWCEGFSTYPRTYDLIHANGL 527
SFAAA+ WVMNVV +TL +IY+RGLIG H+WCE FSTYPRTYDL+HA +
Sbjct: 472 SFAAALKDKDVWVMNVVSPDG-PNTLKLIYDRGLIGTNHNWCEAFSTYPRTYDLLHAWSI 530
Query: 528 FSLYQDK-CNTEDILLEMDRILRPEGAVIIRDEVDVLIKVKKLIGGMRWNMKL------- 579
FS + K C+ ED+L+EMDRILRP G VIIRD+ V+ +KK + + W
Sbjct: 531 FSDIKSKGCSAEDLLIEMDRILRPTGFVIIRDKQSVVESIKKYLQALHWETVASEKVNTS 590
Query: 580 ----VDHEDGPLVPEKVLIAVKQYWVT 602
D EDG V I K+ W+T
Sbjct: 591 SELDQDSEDGE--NNVVFIVQKKLWLT 615
>At2g39750 unknown protein
Length = 694
Score = 436 bits (1120), Expect = e-122
Identities = 224/527 (42%), Positives = 324/527 (60%), Gaps = 21/527 (3%)
Query: 82 KVFKPCEARYTDYTPCQDQRRAMTFPR--ENMNYRERHCPPEEEKLHCMIPAPKGYVTPF 139
K F C +Y PC D + + E ERHCP + + L+C++P PKGY P
Sbjct: 175 KKFGMCPESMREYIPCLDNTDVIKKLKSTERGERFERHCPEKGKGLNCLVPPPKGYRQPI 234
Query: 140 PWPKSRDYVPYANAPYKSLTVEKAIQNWIQYEGNVFRFPGGGTQFPQGADKYIDQLASVI 199
PWPKSRD V ++N P+ L +K QNWI + N F+FPGGGTQF GAD+Y+DQ++ ++
Sbjct: 235 PWPKSRDEVWFSNVPHTRLVEDKGGQNWISRDKNKFKFPGGGTQFIHGADQYLDQMSKMV 294
Query: 200 P-INDGT-VRTALDTGCGVASWGAYLWSRNVVAMSFAPRDSHEAQVQFALERGVPAVIGV 257
I G +R A+D GCGVAS+GAYL SR+V+ MS AP+D HE Q+QFALERGVPA+
Sbjct: 295 SDITFGKHIRVAMDVGCGVASFGAYLLSRDVMTMSVAPKDVHENQIQFALERGVPAMAAA 354
Query: 258 FGTIKLPYPSRAFDMAHCSRCLIPWGANDGMYMMEVDRVLRPGGYWVLSGPPINWKVNYK 317
F T +L YPS+AFD+ HCSRC I W +DG+ ++E++R+LR GGY+ + P+
Sbjct: 355 FATRRLLYPSQAFDLIHCSRCRINWTRDDGILLLEINRMLRAGGYFAWAAQPV------- 407
Query: 318 PWQRPKEELEEEQRKIEEVAKKLCWEKKSEKAEIAIWQKMTDTESCRSRQDDSSVEFC-E 376
+ + LEE+ ++ + LCW+ ++ +AIWQK + + SR+ + C E
Sbjct: 408 --YKHEPALEEQWTEMLNLTISLCWKLVKKEGYVAIWQKPFNNDCYLSREAGTKPPLCDE 465
Query: 377 SSDPDDVWYKKLKACVTPTP-KVSGGDLKPFPDRLYAIPPRVSSGSIPG--VSSETYQND 433
S DPD+VWY LK C++ P K GG++ +P RL+ P R+ + E ++ +
Sbjct: 466 SDDPDNVWYTNLKPCISRIPEKGYGGNVPLWPARLHTPPDRLQTIKFDSYIARKELFKAE 525
Query: 434 NKMWKKHVNAYKKINSLLDSGRYRNIMDMNAGLGSFAAAI--HSSKSWVMNVVPTIAEKS 491
+K W + + Y + + RN++DM AG G FAAA+ H WV++VVP ++ +
Sbjct: 526 SKYWNEIIGGYVRALK-WKKMKLRNVLDMRAGFGGFAAALNDHKLDCWVLSVVP-VSGPN 583
Query: 492 TLGVIYERGLIGIYHDWCEGFSTYPRTYDLIHANGLFSLYQDKCNTEDILLEMDRILRPE 551
TL VIY+RGL+G+ HDWCE F TYPRTYD +HA+GLFS+ + +C ILLEMDRILRP
Sbjct: 584 TLPVIYDRGLLGVMHDWCEPFDTYPRTYDFLHASGLFSIERKRCEMSTILLEMDRILRPG 643
Query: 552 GAVIIRDEVDVLIKVKKLIGGMRWNMKLVDHEDGPLVPEKVLIAVKQ 598
G IRD +DV+ +++++ M W+ L D +GP ++L K+
Sbjct: 644 GRAYIRDSIDVMDEIQEITKAMGWHTSLRDTSEGPHASYRILTCEKR 690
>At4g14360
Length = 608
Score = 427 bits (1097), Expect = e-119
Identities = 225/538 (41%), Positives = 322/538 (59%), Gaps = 32/538 (5%)
Query: 84 FKPCEARYTDYTPCQDQ----RRAMTFPRENMNYRERHCPPEEEKLHCMIPAPKGYVTPF 139
F C+ R+++ PC D+ + + M + ERHCPP E + +C+IP P GY P
Sbjct: 76 FPVCDDRHSELIPCLDRNLIYQMRLKLDLSLMEHYERHCPPPERRFNCLIPPPNGYKVPI 135
Query: 140 PWPKSRDYVPYANAPYKSLTVEKAIQNWIQYEGNVFRFPGGGTQFPQGADKYIDQLASVI 199
WPKSRD V N P+ L EK+ QNW+ +G+ FPGGGT F GADKYI +A+++
Sbjct: 136 KWPKSRDEVWKVNIPHTHLAHEKSDQNWMVVKGDKINFPGGGTHFHYGADKYIASMANML 195
Query: 200 PI------NDGTVRTALDTGCGVASWGAYLWSRNVVAMSFAPRDSHEAQVQFALERGVPA 253
N G +RT D GCGVAS+G YL S +++ MS AP D H+ Q+QFALERG+PA
Sbjct: 196 NYPNNVLNNGGRLRTVFDVGCGVASFGGYLLSSDILTMSLAPNDVHQNQIQFALERGIPA 255
Query: 254 VIGVFGTIKLPYPSRAFDMAHCSRCLIPWGANDGMYMMEVDRVLRPGGYWVLSGPPINWK 313
+GV GT +LPYPSR+F+++HCSRC I W DG+ ++E+DRVLRPGGY+ S P
Sbjct: 256 SLGVLGTKRLPYPSRSFELSHCSRCRIDWLQRDGILLLELDRVLRPGGYFAYSSP----- 310
Query: 314 VNYKPWQRPKEELEEEQRKIEEVAKKLCWEKKSEKAEIAIWQKMTDTESCRSRQDDSSVE 373
+ + + +E+L R++ + +++CW+ +++ + IWQK + R+ +
Sbjct: 311 ---EAYAQDEEDL-RIWREMSALVERMCWKIAAKRNQTVIWQKPLTNDCYLEREPGTQPP 366
Query: 374 FCES-SDPDDVWYKKLKACVTP----TPKVSGGDLKPFPDRLYAIPPRVSSGSIPGVSSE 428
C S +DPD VW ++AC+T K G L P+P RL + PPR++ G S+
Sbjct: 367 LCRSDNDPDAVWGVNMEACITSYSDHDHKTKGSGLAPWPARLTSPPPRLADF---GYSTG 423
Query: 429 TYQNDNKMWKKHVNAY-KKINSLLDSGRYRNIMDMNAGLGSFAAAIHSSKSWVMNVVPTI 487
++ D ++W++ V+ Y ++ ++S RNIMDM A +GSFAAA+ WVMNVVP
Sbjct: 424 MFEKDTELWRQRVDTYWDLLSPRIESDTVRNIMDMKASMGSFAAALKEKDVWVMNVVPED 483
Query: 488 AEKSTLGVIYERGLIGIYHDWCEGFSTYPRTYDLIHANGLFSLYQDK-CNTEDILLEMDR 546
+TL +IY+RGL+G H WCE FSTYPRTYDL+HA + S + K C+ D+LLEMDR
Sbjct: 484 G-PNTLKLIYDRGLMGAVHSWCEAFSTYPRTYDLLHAWDIISDIKKKGCSEVDLLLEMDR 542
Query: 547 ILRPEGAVIIRDEVDVLIKVKKLIGGMRWNM--KLVDHEDGPLVPEKVLIAVKQYWVT 602
ILRP G +IIRD+ V+ VKK + + W D + V I K+ W+T
Sbjct: 543 ILRPSGFIIIRDKQRVVDFVKKYLKALHWEEVGTKTDSDSDQDSDNVVFIVQKKLWLT 600
>At3g23300 ankyrin repeat protein
Length = 611
Score = 425 bits (1092), Expect = e-119
Identities = 228/559 (40%), Positives = 327/559 (57%), Gaps = 34/559 (6%)
Query: 64 DSHHAGEVSQIDESNSNTKVFKPCEARYTDYTPCQDQ----RRAMTFPRENMNYRERHCP 119
D+ + V + + + F C+ R+++ PC D+ + + M + ERHCP
Sbjct: 59 DTSSSFYVEDVVGNGFTPRSFPVCDDRHSELIPCLDRNLIYQMRLKLDLSLMEHYERHCP 118
Query: 120 PEEEKLHCMIPAPKGYVTPFPWPKSRDYVPYANAPYKSLTVEKAIQNWIQYEGNVFRFPG 179
P E + +C+IP P GY P WPKSRD V N P+ L EK+ QNW+ +G FPG
Sbjct: 119 PPERRFNCLIPPPPGYKIPIKWPKSRDEVWKVNIPHTHLAHEKSDQNWMVVKGEKINFPG 178
Query: 180 GGTQFPQGADKYIDQLASVIPI------NDGTVRTALDTGCGVASWGAYLWSRNVVAMSF 233
GGT F GADKYI +A+++ N G +RT LD GCGVAS+G YL + ++ MS
Sbjct: 179 GGTHFHYGADKYIASMANMLNFPNNVLNNGGRLRTFLDVGCGVASFGGYLLASEIMTMSL 238
Query: 234 APRDSHEAQVQFALERGVPAVIGVFGTIKLPYPSRAFDMAHCSRCLIPWGANDGMYMMEV 293
AP D H+ Q+QFALERG+PA +GV GT +LPYPSR+F++AHCSRC I W DG+ ++E+
Sbjct: 239 APNDVHQNQIQFALERGIPAYLGVLGTKRLPYPSRSFELAHCSRCRIDWLQRDGILLLEL 298
Query: 294 DRVLRPGGYWVLSGPPINWKVNYKPWQRPKEELEEEQRKIEEVAKKLCWEKKSEKAEIAI 353
DRVLRPGGY+ S P + + + +E+L R++ + ++CW +++ + I
Sbjct: 299 DRVLRPGGYFAYSSP--------EAYAQDEEDL-RIWREMSALVGRMCWTIAAKRNQTVI 349
Query: 354 WQKMTDTESCRSRQDDSSVEFCES-SDPDDVWYKKLKACVTP----TPKVSGGDLKPFPD 408
WQK + R+ + C S SDPD V+ ++AC+T K G L P+P
Sbjct: 350 WQKPLTNDCYLGREPGTQPPLCNSDSDPDAVYGVNMEACITQYSDHDHKTKGSGLAPWPA 409
Query: 409 RLYAIPPRVSSGSIPGVSSETYQNDNKMWKKHVNAY-KKINSLLDSGRYRNIMDMNAGLG 467
RL + PPR++ G S++ ++ D + W++ V+ Y ++ + S RNIMDM A +G
Sbjct: 410 RLTSPPPRLADF---GYSTDIFEKDTETWRQRVDTYWDLLSPKIQSDTVRNIMDMKASMG 466
Query: 468 SFAAAIHSSKSWVMNVVPTIAEKSTLGVIYERGLIGIYHDWCEGFSTYPRTYDLIHANGL 527
SFAAA+ WVMNVVP +TL +IY+RGL+G H WCE FSTYPRTYDL+HA +
Sbjct: 467 SFAAALKEKDVWVMNVVPEDG-PNTLKLIYDRGLMGAVHSWCEAFSTYPRTYDLLHAWDI 525
Query: 528 FS-LYQDKCNTEDILLEMDRILRPEGAVIIRDEVDVLIKVKKLIGGMRW---NMKLVDHE 583
S + + C+ ED+LLEMDRILRP G ++IRD+ V+ VKK + + W K
Sbjct: 526 ISDIKKRGCSAEDLLLEMDRILRPSGFILIRDKQSVVDLVKKYLKALHWEAVETKTASES 585
Query: 584 DGPLVPEKVLIAVKQYWVT 602
D +LI K+ W+T
Sbjct: 586 DQD-SDNVILIVQKKLWLT 603
>At5g14430 unknown protein
Length = 612
Score = 421 bits (1082), Expect = e-118
Identities = 231/548 (42%), Positives = 323/548 (58%), Gaps = 44/548 (8%)
Query: 87 CEARYTDYTPCQDQ----RRAMTFPRENMNYRERHCPPEEEKLHCMIPAPKGYVTPFPWP 142
C++R+++ PC D+ + + M + E HCPP E + +C++P P GY P WP
Sbjct: 83 CDSRHSELIPCLDRNLHYQLKLKLNLSLMEHYEHHCPPSERRFNCLVPPPVGYKIPLRWP 142
Query: 143 KSRDYVPYANAPYKSLTVEKAIQNWIQYEGNVFRFPGGGTQFPQGADKYIDQLASVIPI- 201
SRD V AN P+ L EK+ QNW+ G+ FPGGGT F GADKYI LA ++
Sbjct: 143 VSRDEVWKANIPHTHLAQEKSDQNWMVVNGDKINFPGGGTHFHNGADKYIVSLAQMLKFP 202
Query: 202 -----NDGTVRTALDTGCGVASWGAYLWSRNVVAMSFAPRDSHEAQVQFALERGVPAVIG 256
N G++R LD GCGVAS+GAYL S +++AMS AP D H+ Q+QFALERG+P+ +G
Sbjct: 203 GDKLNNGGSIRNVLDVGCGVASFGAYLLSHDIIAMSLAPNDVHQNQIQFALERGIPSTLG 262
Query: 257 VFGTIKLPYPSRAFDMAHCSRCLIPWGANDGMYMMEVDRVLRPGGYWVLSGPPINWKVNY 316
V GT +LPYPSR+F++AHCSRC I W DG+ ++E+DR+LRPGGY+V S P
Sbjct: 263 VLGTKRLPYPSRSFELAHCSRCRIDWLQRDGILLLELDRLLRPGGYFVYSSP-------- 314
Query: 317 KPWQRPKEELEEEQRKI----EEVAKKLCWEKKSEKAEIAIWQKMTDTESCRSRQDDSSV 372
+ E RKI ++ K++CW+ +++ + IW K + SC ++D +
Sbjct: 315 -----EAYAHDPENRKIGNAMHDLFKRMCWKVVAKRDQSVIWGKPI-SNSCYLKRDPGVL 368
Query: 373 -EFCES-SDPDDVWYKKLKACVTP----TPKVSGGDLKPFPDRLYAIPPRVSSGSIPGVS 426
C S DPD W +KAC++P K L P+P RL A PPR+ GV+
Sbjct: 369 PPLCPSGDDPDATWNVSMKACISPYSVRMHKERWSGLVPWPRRLTAPPPRLEE---IGVT 425
Query: 427 SETYQNDNKMWKKHVNAY-KKINSLLDSGRYRNIMDMNAGLGSFAAAIHSSKSWVMNVVP 485
E ++ D + W+ V Y K + ++ RN+MDM++ LG FAAA++ WVMNV+P
Sbjct: 426 PEQFREDTETWRLRVIEYWKLLKPMVQKNSIRNVMDMSSNLGGFAAALNDKDVWVMNVMP 485
Query: 486 TIAEKSTLGVIYERGLIGIYHDWCEGFSTYPRTYDLIHANGLFSLYQDK-CNTEDILLEM 544
+ + +IY+RGLIG HDWCE F TYPRT+DLIHA F+ Q + C+ ED+L+EM
Sbjct: 486 -VQSSPRMKIIYDRGLIGATHDWCEAFDTYPRTFDLIHAWNTFTETQARGCSFEDLLIEM 544
Query: 545 DRILRPEGAVIIRDEVDVLIKVKKLIGGMRWNMKLVD--HEDGPL--VPEKVLIAVKQYW 600
DRILRPEG VIIRD D + +KK + ++W+ + + PL E VLIA K+ W
Sbjct: 545 DRILRPEGFVIIRDTTDNISYIKKYLTLLKWDKWSTETTPKGDPLSTKDEIVLIARKKLW 604
Query: 601 VTDGNSTS 608
S S
Sbjct: 605 SLPAISVS 612
>At1g77260 unknown protein
Length = 655
Score = 415 bits (1067), Expect = e-116
Identities = 230/573 (40%), Positives = 339/573 (59%), Gaps = 36/573 (6%)
Query: 48 ITKNNAECDVVPNLSFDSHHAGEV------SQIDESNSNTKVF-----KPCEARYTDYTP 96
I +N A D FD E+ S ++E S F K C+ DY P
Sbjct: 96 INENGAMSDSFEIGGFDPDSIDELKSATGNSSVEEKESPEVGFQIEKLKLCDKTKIDYIP 155
Query: 97 CQDQR---RAMTFPRENMNYRERHCPPEEEKLHCMIPAPKGYVTPFPWPKSRDYVPYANA 153
C D + + NY ERHCP ++ L C+IP P GY P WP+SRD + + N
Sbjct: 156 CLDNEEEIKRLNNTDRGENY-ERHCP--KQSLDCLIPPPDGYKKPIQWPQSRDKIWFNNV 212
Query: 154 PYKSLTVEKAIQNWIQYEGNVFRFPGGGTQFPQGADKYIDQLASVIP-INDGT-VRTALD 211
P+ L +K QNWI+ E + F FPGGGTQF GAD+Y+DQ++ +IP I G+ R ALD
Sbjct: 213 PHTRLVEDKGGQNWIRREKDKFVFPGGGTQFIHGADQYLDQISQMIPDITFGSRTRVALD 272
Query: 212 TGCGVASWGAYLWSRNVVAMSFAPRDSHEAQVQFALERGVPAVIGVFGTIKLPYPSRAFD 271
GCGVAS+GA+L RN +S AP+D HE Q+QFALERGVPA++ VF T +L YPS++F+
Sbjct: 273 IGCGVASFGAFLMQRNTTTLSVAPKDVHENQIQFALERGVPAMVAVFATRRLLYPSQSFE 332
Query: 272 MAHCSRCLIPWGANDGMYMMEVDRVLRPGGYWVLSGPPINWKVNYKPWQRPKEELEEEQR 331
M HCSRC I W +DG+ ++EV+R+LR GGY+V + P+ + ++ L+E+ +
Sbjct: 333 MIHCSRCRINWTRDDGILLLEVNRMLRAGGYFVWAAQPV---------YKHEDNLQEQWK 383
Query: 332 KIEEVAKKLCWEKKSEKAEIAIWQKMTDTESCRSRQDDSSVEFCE-SSDPDDVWYKKLKA 390
++ ++ ++CWE ++ IA+W+K + SR+ + C DPDDVWY +K
Sbjct: 384 EMLDLTNRICWELIKKEGYIAVWRKPLNNSCYVSREAGTKPPLCRPDDDPDDVWYVDMKP 443
Query: 391 CVTPTPKVS-GGDLKPFPDRLYAIPPRVSSGSIPGVSS--ETYQNDNKMWKKHVNAYKKI 447
C+T P G ++ +P RL+ P R+ S + S E + +++ W + V +Y ++
Sbjct: 444 CITRLPDNGYGANVSTWPARLHDPPERLQSIQMDAYISRKEIMKAESRFWLEVVESYVRV 503
Query: 448 NSLLDSGRYRNIMDMNAGLGSFAAAIH--SSKSWVMNVVPTIAEKSTLGVIYERGLIGIY 505
+ + RN++DM AG G FAAA++ WVMN+VP ++ +TL VIY+RGL G
Sbjct: 504 FRWKEF-KLRNVLDMRAGFGGFAAALNDLGLDCWVMNIVP-VSGFNTLPVIYDRGLQGAM 561
Query: 506 HDWCEGFSTYPRTYDLIHANGLFSLYQDKCNTEDILLEMDRILRPEGAVIIRDEVDVLIK 565
HDWCE F TYPRTYDLIHA LFS+ + +CN +I+LEMDR+LRP G V IRD + ++ +
Sbjct: 562 HDWCEPFDTYPRTYDLIHAAFLFSVEKKRCNITNIMLEMDRMLRPGGHVYIRDSLSLMDQ 621
Query: 566 VKKLIGGMRWNMKLVDHEDGPLVPEKVLIAVKQ 598
++++ + W + D +GP ++LI K+
Sbjct: 622 LQQVAKAIGWTAGVHDTGEGPHASVRILICDKR 654
>At2g34300 unknown protein
Length = 770
Score = 401 bits (1031), Expect = e-112
Identities = 221/532 (41%), Positives = 308/532 (57%), Gaps = 34/532 (6%)
Query: 93 DYTPCQDQRRAMTFPRENMNY--RERHCPPEEEKLHCMIPAPKGYVTPFPWPKSRDYVPY 150
DY PC D +A+ M+Y RERHCP EE HC++ P GY WPKSR+ + Y
Sbjct: 250 DYIPCLDNWQAIKKLHTTMHYEHRERHCP--EESPHCLVSLPDGYKRSIKWPKSREKIWY 307
Query: 151 ANAPYKSLTVEKAIQNWIQYEGNVFRFPGGGTQFPQGADKYIDQLASVIPIN--DGTVRT 208
N P+ L K QNW++ G FPGGGTQF GA YID + P R
Sbjct: 308 NNVPHTKLAEIKGHQNWVKMSGEHLTFPGGGTQFKNGALHYIDFIQQSHPAIAWGNRTRV 367
Query: 209 ALDTGCGVASWGAYLWSRNVVAMSFAPRDSHEAQVQFALERGVPAVIGVFGTIKLPYPSR 268
LD GCGVAS+G YL+ R+V+A+SFAP+D HEAQVQFALERG+PA++ V GT +LP+P
Sbjct: 368 ILDVGCGVASFGGYLFERDVLALSFAPKDEHEAQVQFALERGIPAMLNVMGTKRLPFPGS 427
Query: 269 AFDMAHCSRCLIPWGANDGMYMMEVDRVLRPGGYWVLSGPPINWKVNYKPWQRPKEELEE 328
FD+ HC+RC +PW G ++E++R LRPGG++V S P+ R EE
Sbjct: 428 VFDLIHCARCRVPWHIEGGKLLLELNRALRPGGFFVWSATPV---------YRKNEEDSG 478
Query: 329 EQRKIEEVAKKLCWE----KKSEKAEI--AIWQKMTDTESCRSRQDDSSVEFCESSDPDD 382
+ + E+ K +CW+ KK + E+ AI+QK T + C +++ + C+ SD +
Sbjct: 479 IWKAMSELTKAMCWKLVTIKKDKLNEVGAAIYQKPT-SNKCYNKRPQNEPPLCKDSDDQN 537
Query: 383 -VWYKKLKACVTPTPKVSG--GDLKP--FPDRLYAIPPRVSS--GSIPGVSSETYQNDNK 435
W L+AC+ + S G + P +P+R+ P + S G + E + D +
Sbjct: 538 AAWNVPLEACMHKVTEDSSKRGAVWPNMWPERVETAPEWLDSQEGVYGKPAPEDFTADQE 597
Query: 436 MWKKHVNAYKKINSLLDSGRYRNIMDMNAGLGSFAAAIHSSKSWVMNVVPTIAEKSTLGV 495
WK V+ + +D RN+MDM A G FAAA+ K WVMNVVP A TL +
Sbjct: 598 KWKTIVSKAYLNDMGIDWSNVRNVMDMRAVYGGFAAALKDLKLWVMNVVPVDA-PDTLPI 656
Query: 496 IYERGLIGIYHDWCEGFSTYPRTYDLIHANGLFSLYQDKCNTEDILLEMDRILRPEGAVI 555
IYERGL GIYHDWCE F+TYPRTYDL+HA+ LFS + +CN ++ E+DRILRP+G I
Sbjct: 657 IYERGLFGIYHDWCESFNTYPRTYDLLHADHLFSTLRKRCNLVSVMAEIDRILRPQGTFI 716
Query: 556 IRDEVDVLIKVKKLIGGMRWNMKLVDHEDGPLVPEKVLIAVKQYWVTDGNST 607
IRD+++ L +V+K++ M+W +K+ +D E +L K +W + T
Sbjct: 717 IRDDMETLGEVEKMVKSMKWKVKMTQSKDN----EGLLSIEKSWWRPEETET 764
>At1g29470 unknown protein
Length = 768
Score = 400 bits (1028), Expect = e-111
Identities = 218/525 (41%), Positives = 305/525 (57%), Gaps = 34/525 (6%)
Query: 93 DYTPCQDQRRAMTFPRENMNY--RERHCPPEEEKLHCMIPAPKGYVTPFPWPKSRDYVPY 150
DY PC D +A+ +Y RERHCP EE C++ P+GY WPKSR+ + Y
Sbjct: 248 DYIPCLDNWQAIRKLHSTKHYEHRERHCP--EESPRCLVSLPEGYKRSIKWPKSREKIWY 305
Query: 151 ANAPYKSLTVEKAIQNWIQYEGNVFRFPGGGTQFPQGADKYIDQLASVIP--INDGTVRT 208
N P+ L K QNW++ G FPGGGTQF GA YID L P R
Sbjct: 306 TNIPHTKLAEVKGHQNWVKMSGEYLTFPGGGTQFKNGALHYIDFLQESYPDIAWGNRTRV 365
Query: 209 ALDTGCGVASWGAYLWSRNVVAMSFAPRDSHEAQVQFALERGVPAVIGVFGTIKLPYPSR 268
LD GCGVAS+G YL+ R+V+A+SFAP+D HEAQVQFALERG+PA+ V GT +LP+P
Sbjct: 366 ILDVGCGVASFGGYLFDRDVLALSFAPKDEHEAQVQFALERGIPAMSNVMGTKRLPFPGS 425
Query: 269 AFDMAHCSRCLIPWGANDGMYMMEVDRVLRPGGYWVLSGPPINWKVNYKPWQRPKEELEE 328
FD+ HC+RC +PW G ++E++R LRPGG++V S P+ R EE
Sbjct: 426 VFDLIHCARCRVPWHIEGGKLLLELNRALRPGGFFVWSATPV---------YRKTEEDVG 476
Query: 329 EQRKIEEVAKKLCWE----KKSEKAEI--AIWQKMTDTESCRSRQDDSSVEFCESSDPDD 382
+ + ++ K +CWE KK E E+ AI+QK + C + + + C+ SD +
Sbjct: 477 IWKAMSKLTKAMCWELMTIKKDELNEVGAAIYQKPM-SNKCYNERSQNEPPLCKDSDDQN 535
Query: 383 -VWYKKLKACVTPTPKVSG--GDLKP--FPDRLYAIPPRVSS--GSIPGVSSETYQNDNK 435
W L+AC+ + S G + P +P+R+ +P + S G + E + D++
Sbjct: 536 AAWNVPLEACIHKVTEDSSKRGAVWPESWPERVETVPQWLDSQEGVYGKPAQEDFTADHE 595
Query: 436 MWKKHVNAYKKINSLLDSGRYRNIMDMNAGLGSFAAAIHSSKSWVMNVVPTIAEKSTLGV 495
WK V+ +D RN+MDM A G FAAA+ K WVMNVVP I TL +
Sbjct: 596 RWKTIVSKSYLNGMGIDWSYVRNVMDMRAVYGGFAAALKDLKLWVMNVVP-IDSPDTLPI 654
Query: 496 IYERGLIGIYHDWCEGFSTYPRTYDLIHANGLFSLYQDKCNTEDILLEMDRILRPEGAVI 555
IYERGL GIYHDWCE FSTYPRTYDL+HA+ LFS + +CN ++ E+DRILRP+G I
Sbjct: 655 IYERGLFGIYHDWCESFSTYPRTYDLLHADHLFSSLKKRCNLVGVMAEVDRILRPQGTFI 714
Query: 556 IRDEVDVLIKVKKLIGGMRWNMKLVDHEDGPLVPEKVLIAVKQYW 600
+RD+++ + +++K++ M+WN+++ +DG E +L K +W
Sbjct: 715 VRDDMETIGEIEKMVKSMKWNVRMTHSKDG----EGLLSVQKSWW 755
>At5g64030 ankyrin-like protein
Length = 829
Score = 399 bits (1024), Expect = e-111
Identities = 217/526 (41%), Positives = 301/526 (56%), Gaps = 35/526 (6%)
Query: 93 DYTPCQDQRRAMTFPRENMNY--RERHCPPEEEKLHCMIPAPKGYVTPFPWPKSRDYVPY 150
DY PC D +A+ +Y RERHCP C++P P GY P WPKSR+ + Y
Sbjct: 308 DYIPCLDNVQAIRSLPSTKHYEHRERHCPDSPPT--CLVPLPDGYKRPIEWPKSREKIWY 365
Query: 151 ANAPYKSLTVEKAIQNWIQYEGNVFRFPGGGTQFPQGADKYIDQLASVIPIN--DGTVRT 208
N P+ L K QNW++ G FPGGGTQF GA YID + +P R
Sbjct: 366 TNVPHTKLAEYKGHQNWVKVTGEYLTFPGGGTQFKHGALHYIDFIQESVPAIAWGKRSRV 425
Query: 209 ALDTGCGVASWGAYLWSRNVVAMSFAPRDSHEAQVQFALERGVPAVIGVFGTIKLPYPSR 268
LD GCGVAS+G +L+ R+V+ MS AP+D HEAQVQFALERG+PA+ V GT +LP+P R
Sbjct: 426 VLDVGCGVASFGGFLFDRDVITMSLAPKDEHEAQVQFALERGIPAISAVMGTTRLPFPGR 485
Query: 269 AFDMAHCSRCLIPWGANDGMYMMEVDRVLRPGGYWVLSGPPINWKVNYKPWQRPKEELEE 328
FD+ HC+RC +PW G ++E++RVLRPGG++V S P+ K K E E
Sbjct: 486 VFDIVHCARCRVPWHIEGGKLLLELNRVLRPGGFFVWSATPVYQK---------KTEDVE 536
Query: 329 EQRKIEEVAKKLCWEKKSEKAE------IAIWQKMTDTESCRSRQDDSSVEFCESSDPDD 382
+ + E+ KK+CWE S + +A ++K T E ++R + +S DP+
Sbjct: 537 IWKAMSELIKKMCWELVSINKDTINGVGVATYRKPTSNECYKNRSEPVPPICADSDDPNA 596
Query: 383 VWYKKLKACV--TPTPKVSGGDLKP--FPDRLYAIPPRVSS---GSIPGVSSETYQNDNK 435
W L+AC+ P K G P +P RL P +SS G + E + D +
Sbjct: 597 SWKVPLQACMHTAPEDKTQRGSQWPEQWPARLEKAPFWLSSSQTGVYGKAAPEDFSADYE 656
Query: 436 MWKKHVNAYKKINSL-LDSGRYRNIMDMNAGLGSFAAAIHSSKSWVMNVVPTIAEKSTLG 494
WK+ V +N L ++ RN+MDM A G FAAA+ K WVMNVVP I TL
Sbjct: 657 HWKRVVTK-SYLNGLGINWASVRNVMDMRAVYGGFAAALRDLKVWVMNVVP-IDSPDTLA 714
Query: 495 VIYERGLIGIYHDWCEGFSTYPRTYDLIHANGLFSLYQDKCNTEDILLEMDRILRPEGAV 554
+IYERGL GIYHDWCE FSTYPR+YDL+HA+ LFS + +CN ++ E+DR+LRPEG +
Sbjct: 715 IIYERGLFGIYHDWCESFSTYPRSYDLLHADHLFSKLKQRCNLTAVIAEVDRVLRPEGKL 774
Query: 555 IIRDEVDVLIKVKKLIGGMRWNMKLVDHEDGPLVPEKVLIAVKQYW 600
I+RD+ + + +V+ ++ M+W +++ ++ E +L K W
Sbjct: 775 IVRDDAETIQQVEGMVKAMKWEVRMTYSKE----KEGLLSVQKSIW 816
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.136 0.429
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,513,478
Number of Sequences: 26719
Number of extensions: 739139
Number of successful extensions: 2509
Number of sequences better than 10.0: 51
Number of HSP's better than 10.0 without gapping: 38
Number of HSP's successfully gapped in prelim test: 13
Number of HSP's that attempted gapping in prelim test: 2224
Number of HSP's gapped (non-prelim): 64
length of query: 610
length of database: 11,318,596
effective HSP length: 105
effective length of query: 505
effective length of database: 8,513,101
effective search space: 4299116005
effective search space used: 4299116005
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 63 (28.9 bits)
Medicago: description of AC146568.1


