

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146567.12 + phase: 0 /pseudo
(307 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
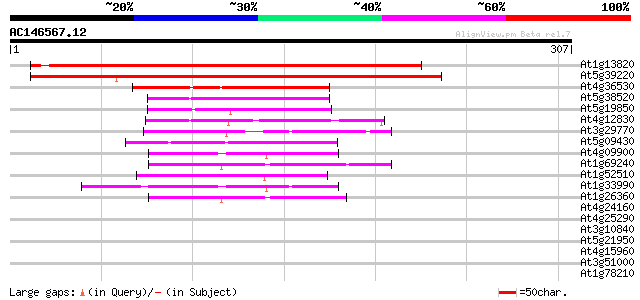
At1g13820 hypothetical protein 335 2e-92
At5g39220 unknown protein 285 2e-77
At4g36530 unknown protein 75 6e-14
At5g38520 unknown protein 49 3e-06
At5g19850 unknown protein 46 3e-05
At4g12830 hydrolase like protein 46 3e-05
At3g29770 alpha/beta hydrolase, putative 45 6e-05
At5g09430 putative hydrolase 44 1e-04
At4g09900 putative host response protein 44 1e-04
At1g69240 alpha/beta hydrolase like protein 44 1e-04
At1g52510 unknown protein (At1g52510) 42 5e-04
At1g33990 unknown protein 42 5e-04
At1g26360 hypothetical protein 41 7e-04
At4g24160 unknown protein 40 0.002
At4g25290 hypothetical protein 39 0.005
At3g10840 alpha/beta hydrolase like protein 39 0.005
At5g21950 unknown protein 37 0.010
At4g15960 unknown protein 37 0.013
At3g51000 epoxide hydrolase-like protein 37 0.018
At1g78210 unknown protein 36 0.030
>At1g13820 hypothetical protein
Length = 339
Score = 335 bits (859), Expect = 2e-92
Identities = 159/214 (74%), Positives = 183/214 (85%), Gaps = 4/214 (1%)
Query: 12 FKVVAETDADSFPSFLPKDVNRIKDPFARTLATRIQRLPVSVKFSENPIMSSCVKPLVQN 71
FKV AE FP+FLP DV +IKD FA LA RI+RLPVSV F E+ IMSSCV PL++N
Sbjct: 25 FKVTAE----KFPTFLPTDVEKIKDTFALKLAARIERLPVSVSFREDSIMSSCVTPLMRN 80
Query: 72 KENPIVLLHGFDSSCLEWRYTYPLLEEAGIETWAIDILGWGFSDLEKLPSCDVVSKREHF 131
+ P+VLLHGFDSSCLEWRYTYPLLEEAG+ETWA DILGWGFSDL+KLP CDV SKREHF
Sbjct: 81 ETTPVVLLHGFDSSCLEWRYTYPLLEEAGLETWAFDILGWGFSDLDKLPPCDVASKREHF 140
Query: 132 YQFWKSYIKKPMILVGPSLGSAVAIDFAINYPEAVEKLVLIDASVYTEGTGNLATLPRAA 191
Y+FWKS+IK+P++LVGPSLG+AVAID A+N+PEAVE LVL+DASVY EGTGNLATLP+AA
Sbjct: 141 YKFWKSHIKRPVVLVGPSLGAAVAIDIAVNHPEAVESLVLMDASVYAEGTGNLATLPKAA 200
Query: 192 AYAGVYLLKSVPLRVYANYLSFTNISFSTSLDWT 225
AYAGVYLLKS+PLR+Y N++ F IS TS DWT
Sbjct: 201 AYAGVYLLKSIPLRLYVNFICFNGISLETSWDWT 234
>At5g39220 unknown protein
Length = 330
Score = 285 bits (729), Expect = 2e-77
Identities = 137/228 (60%), Positives = 172/228 (75%), Gaps = 3/228 (1%)
Query: 12 FKVVAETDADSFPSFLPKDVNRIKDPFARTLATRIQRLPVSVKFSE--NPIMSSCVKPLV 69
F+ + D FP+FLPK++ IKDPFAR LA RI R+PV ++ +MSSC+KPLV
Sbjct: 17 FRQRVTANGDGFPTFLPKEIQNIKDPFARALAQRIVRIPVPLQMGNFRGSVMSSCIKPLV 76
Query: 70 Q-NKENPIVLLHGFDSSCLEWRYTYPLLEEAGIETWAIDILGWGFSDLEKLPSCDVVSKR 128
Q + ++P+VLLH FDSSCLEWR TYPLLE+A +ETWAID+LGWGFSDLEKLP CD SKR
Sbjct: 77 QLHDKSPVVLLHCFDSSCLEWRRTYPLLEQACLETWAIDVLGWGFSDLEKLPPCDAASKR 136
Query: 129 EHFYQFWKSYIKKPMILVGPSLGSAVAIDFAINYPEAVEKLVLIDASVYTEGTGNLATLP 188
H ++ WK+YIK+PMILVGPSLG+ VA+DF YPEAV+KLVLI+A+ Y+EGTG L LP
Sbjct: 137 HHLFELWKTYIKRPMILVGPSLGATVAVDFTATYPEAVDKLVLINANAYSEGTGRLKELP 196
Query: 189 RAAAYAGVYLLKSVPLRVYANYLSFTNISFSTSLDWTNATMLLRR*KW 236
++ AYAGV LLKS PLR+ AN L+F + S ++DWTN L + W
Sbjct: 197 KSIAYAGVKLLKSFPLRLLANVLAFCSSPLSENIDWTNIGRLHCQMPW 244
>At4g36530 unknown protein
Length = 378
Score = 74.7 bits (182), Expect = 6e-14
Identities = 38/108 (35%), Positives = 65/108 (60%), Gaps = 2/108 (1%)
Query: 68 LVQNKENPIVLLHGFDSSCLEWRYTYPLLEEAGIETWAIDILGWGFSDLEKLPSCDVVSK 127
+VQ + +P+VL+HGF +S WRY P L + + +A+D+LG+G+SD + L D +
Sbjct: 94 VVQGEGSPLVLIHGFGASVFHWRYNIPELAKK-YKVYALDLLGFGWSD-KALIEYDAMVW 151
Query: 128 REHFYQFWKSYIKKPMILVGPSLGSAVAIDFAINYPEAVEKLVLIDAS 175
+ F K +K+P ++VG SLG A+ A+ PE V + L++++
Sbjct: 152 TDQVIDFMKEVVKEPAVVVGNSLGGFTALSVAVGLPEQVTGVALLNSA 199
>At5g38520 unknown protein
Length = 362
Score = 48.9 bits (115), Expect = 3e-06
Identities = 34/101 (33%), Positives = 55/101 (53%), Gaps = 2/101 (1%)
Query: 76 IVLLHGFDSSCLEWRYTYPLLEEAGIETWAIDILGWGFSDLEKLPSCDVVSKREHFYQFW 135
++L+HGF +S WR L + +AID+LG+G SD S + S E F
Sbjct: 93 VLLVHGFGASIPHWRRNINALSK-NHTVYAIDLLGFGASDKPPGFSYTMESWAELILNFL 151
Query: 136 KSYIKKPMILVGPSLGS-AVAIDFAINYPEAVEKLVLIDAS 175
+ ++KP IL+G S+GS A I + + + V+ LVL++ +
Sbjct: 152 EEVVQKPTILIGNSVGSLACVIAASESRGDLVKGLVLLNCA 192
>At5g19850 unknown protein
Length = 359
Score = 45.8 bits (107), Expect = 3e-05
Identities = 29/107 (27%), Positives = 51/107 (47%), Gaps = 7/107 (6%)
Query: 76 IVLLHGFDSSCLEWRYTYPLLEEAGIETWAIDILGWGFSDLEKL------PSCDVVSKRE 129
+VL+HGF ++ WR P+L + ++ID++G+G+SD P + E
Sbjct: 97 LVLVHGFGANSDHWRKNTPILGKTH-RVYSIDLIGYGYSDKPNPREFGGEPFYTFETWGE 155
Query: 130 HFYQFWKSYIKKPMILVGPSLGSAVAIDFAINYPEAVEKLVLIDASV 176
F +K + S+G V + A++ PE L+LI+ S+
Sbjct: 156 QLNDFCLDVVKDEAFFICNSIGGLVGLQAAVSKPEICRGLMLINISL 202
>At4g12830 hydrolase like protein
Length = 393
Score = 45.8 bits (107), Expect = 3e-05
Identities = 40/141 (28%), Positives = 62/141 (43%), Gaps = 18/141 (12%)
Query: 75 PIVLLHGFDSSCLEWRYTYPLLEEAGIETWAIDILGWGFSDLEK------LPSCDVVSKR 128
P++L+HGF S +R T P+L + A D LG+GFSD + + VS
Sbjct: 135 PVILIHGFPSQAYSYRKTIPVLSK-NYRAIAFDWLGFGFSDKPQAGYGFNYTMDEFVSSL 193
Query: 129 EHFYQFWKSYIKKPMILVGPSLGSAVAIDFAINYPEAVEKLVLIDASVYTEGTGNLATLP 188
E F + LV SA + +A N P+ ++ L+L++ + T A LP
Sbjct: 194 ESFID---EVTTSKVSLVVQGYFSAAVVKYARNRPDKIKNLILLNPPL----TPEHAKLP 246
Query: 189 RAAAYAGVYLLKSV----PLR 205
+ +LL + PLR
Sbjct: 247 STLSVFSNFLLGEIFSQDPLR 267
>At3g29770 alpha/beta hydrolase, putative
Length = 390
Score = 44.7 bits (104), Expect = 6e-05
Identities = 40/147 (27%), Positives = 60/147 (40%), Gaps = 23/147 (15%)
Query: 74 NPIVLLHGFDSSCLEWRYTYPLLEEAGIETWAIDILGWGFSDLE----------KLPSCD 123
N VL+HG W T LLEE G + AID+ G G + + P D
Sbjct: 137 NHFVLVHGGSFGAWCWYKTIALLEEDGFKVTAIDLAGCGINSININGIASLSQYVKPLTD 196
Query: 124 VVSKREHFYQFWKSYIKKPMILVGPSLGSAVAIDFAIN-YPEAVEKLVLIDASVYTEGTG 182
++ K I + +ILVG G A I +A+ +P + K V + A++ T G
Sbjct: 197 ILEKLP---------IGEKVILVGHDFGGA-CISYAMELFPSKISKAVFLAAAMLTNGQS 246
Query: 183 NLATLPRAAAYAGVYLLKSVPLRVYAN 209
L A L++ + +Y N
Sbjct: 247 TLDMFSLKAGQND--LMRKAQIFIYTN 271
>At5g09430 putative hydrolase
Length = 303
Score = 43.9 bits (102), Expect = 1e-04
Identities = 36/118 (30%), Positives = 56/118 (46%), Gaps = 4/118 (3%)
Query: 64 CVKPLVQNKENP-IVLLHGFDSSCLEWRYTYPLLEEAG-IETWAIDILGWGFSDLEKLPS 121
C P N+ P ++LLHGF ++ + W+Y L G + D+L +G S + P+
Sbjct: 50 CWIPKSPNRSKPNLLLLHGFGANAM-WQYGEHLRAFTGRFNVYVPDLLFFGLSSTSE-PN 107
Query: 122 CDVVSKREHFYQFWKSYIKKPMILVGPSLGSAVAIDFAINYPEAVEKLVLIDASVYTE 179
+ + +++ + M +VG S G V A +PE VEKLVL A V E
Sbjct: 108 RTESFQARCLMRLMEAHGVQRMNIVGISYGGFVGYSLAAQFPENVEKLVLCCAGVCLE 165
>At4g09900 putative host response protein
Length = 256
Score = 43.5 bits (101), Expect = 1e-04
Identities = 32/109 (29%), Positives = 51/109 (46%), Gaps = 9/109 (8%)
Query: 77 VLLHGFDSSCLEWRYTYPLLEEAGIETWAIDILGWGFSDLEKLPSCDVVSKREHFYQFWK 136
VL+HG W T LEE+G+ +D+ G GF+ + + VS E + +
Sbjct: 7 VLVHGEGFGAWCWYKTIASLEESGLSPVTVDLAGSGFN----MTDANSVSTLEEYSKPLI 62
Query: 137 SYI-----KKPMILVGPSLGSAVAIDFAINYPEAVEKLVLIDASVYTEG 180
I ++ +ILVG S G A +PE + K + I A++ T+G
Sbjct: 63 ELIQNLPAEEKVILVGHSTGGACVSYALERFPEKISKAIFICATMVTDG 111
>At1g69240 alpha/beta hydrolase like protein
Length = 444
Score = 43.5 bits (101), Expect = 1e-04
Identities = 41/136 (30%), Positives = 57/136 (41%), Gaps = 6/136 (4%)
Query: 77 VLLHGFDSSCLEWRYTYPLLEEAGIETWAIDILGWGFS--DLEKLPS-CDVVSKREHFYQ 133
VL+HG W T LLE+ G + A+D+ G G S D + S V HF+
Sbjct: 189 VLVHGGGFGAWCWYKTITLLEKHGFQVDAVDLTGSGVSSFDTNNITSLAQYVKPLLHFFD 248
Query: 134 FWKSYIKKPMILVGPSLGSAVAIDFAINYPEAVEKLVLIDASVYTEGTGNLATLPRAAAY 193
K K +ILVG G A YP + K + I A++ L L
Sbjct: 249 TLKPTEK--VILVGHDFGGACMSYAMEMYPSKIAKAIFISAAMLANAQSTL-DLFNQQPD 305
Query: 194 AGVYLLKSVPLRVYAN 209
+ L++ V L +YAN
Sbjct: 306 SNYDLMEQVHLFLYAN 321
>At1g52510 unknown protein (At1g52510)
Length = 380
Score = 41.6 bits (96), Expect = 5e-04
Identities = 29/110 (26%), Positives = 53/110 (47%), Gaps = 5/110 (4%)
Query: 70 QNKENPIVLLHGFDSSCLEWRYTYPLLEEAGIETWAIDILGWGFSDLEKLPSCDVVSKRE 129
+++ IV +HG + +R L +AG +A D +G+GFSD + +++E
Sbjct: 122 ESRRGTIVFVHGAPTQSFSYRTVMSELSDAGFHCFAPDWIGFGFSDKPQPGYGFNYTEKE 181
Query: 130 HFYQFWKSY----IKKPMILVGPS-LGSAVAIDFAINYPEAVEKLVLIDA 174
+ F K +K P LV L + + +A+ P VEKL ++++
Sbjct: 182 YHEAFDKLLEVLEVKSPFFLVVQGFLVGSYGLTWALKNPSKVEKLAILNS 231
>At1g33990 unknown protein
Length = 348
Score = 41.6 bits (96), Expect = 5e-04
Identities = 37/147 (25%), Positives = 69/147 (46%), Gaps = 14/147 (9%)
Query: 40 RTLATRIQRLPVSVKFSENPIMSSCVKPLVQNKENPIVLLHGFDSSCLEWRYTYPLLEEA 99
R +T ++ +S FS + + L+ K VL+HG W LEE+
Sbjct: 65 RVGSTSTRKRTLSDPFSNGKQVPDFSESLIVKK---FVLVHGEGFGAWCWYKMVASLEES 121
Query: 100 GIETWAIDILGWGFSDLEKLPSCDVVSKREHFYQFWKSYI-----KKPMILVGPSLGSAV 154
G+ +D+ G GF+ + + VS E + + + ++ +ILVG S G A
Sbjct: 122 GLSPVTVDLTGCGFN----MTDTNTVSTLEEYSKPLIDLLENLPEEEKVILVGHSTGGA- 176
Query: 155 AIDFAI-NYPEAVEKLVLIDASVYTEG 180
+I +A+ +PE + K + + A++ ++G
Sbjct: 177 SISYALERFPEKISKAIFVCATMVSDG 203
>At1g26360 hypothetical protein
Length = 554
Score = 41.2 bits (95), Expect = 7e-04
Identities = 35/111 (31%), Positives = 49/111 (43%), Gaps = 5/111 (4%)
Query: 77 VLLHGFDSSCLEWRYTYPLLEEAGIETWAIDILGWGFS--DLEKLPSCDVVSK-REHFYQ 133
VL+HG W T LLE+ G + A+++ G G S D + S SK HF++
Sbjct: 188 VLVHGGGFGAWCWYKTITLLEKHGFQVDAVELTGSGVSSIDTNNITSLAHYSKPLLHFFE 247
Query: 134 FWKSYIKKPMILVGPSLGSAVAIDFAINYPEAVEKLVLIDASVYTEGTGNL 184
K K +ILVG G A +P + K V I A++ G L
Sbjct: 248 SLKPTEK--VILVGHDFGGACMSYAMEMFPTKIAKAVFISAAMLANGQSTL 296
>At4g24160 unknown protein
Length = 418
Score = 40.0 bits (92), Expect = 0.002
Identities = 30/108 (27%), Positives = 53/108 (48%), Gaps = 7/108 (6%)
Query: 76 IVLLHGFDSSCLEWRYTYPLLEEAGIETWAIDILGWGFSDLEKLPSCDVVSKRE-----H 130
+V++HG+ +S + + L AID LGWG S +C + E
Sbjct: 123 LVMVHGYGASQGFFFRNFDALASR-FRVIAIDQLGWGGSSRPDF-TCRSTEETEAWFIDS 180
Query: 131 FYQFWKSYIKKPMILVGPSLGSAVAIDFAINYPEAVEKLVLIDASVYT 178
F ++ K+ IL+G S G VA +A+ +PE V+ L+L+ ++ ++
Sbjct: 181 FEEWRKAQNLSNFILLGHSFGGYVAAKYALKHPEHVQHLILVGSAGFS 228
>At4g25290 hypothetical protein
Length = 581
Score = 38.5 bits (88), Expect = 0.005
Identities = 25/108 (23%), Positives = 51/108 (47%), Gaps = 1/108 (0%)
Query: 68 LVQNKENPIVLLHGFDSSCLEWRYTYPLLEEAGIETWAIDILGWGFSDLEKLPSCDVVSK 127
+V N+ ++L+HGF + +R + + W I +LG+G S+ + +++
Sbjct: 338 VVGNEGPAVLLVHGFGAFLEHYRDNVDNIVNSKNRVWTITVLGFGKSEKPNIIYTELL-W 396
Query: 128 REHFYQFWKSYIKKPMILVGPSLGSAVAIDFAINYPEAVEKLVLIDAS 175
E F + +P VG S+G A +P V+ +VL++++
Sbjct: 397 AELLRDFMAEVVGEPAHCVGNSIGGYFVALMAFLWPALVKSVVLVNSA 444
>At3g10840 alpha/beta hydrolase like protein
Length = 429
Score = 38.5 bits (88), Expect = 0.005
Identities = 32/116 (27%), Positives = 46/116 (39%), Gaps = 13/116 (11%)
Query: 75 PIVLLHGFDSSCLEW-RYTYPLLEEAGIETWAIDILGWGFS------------DLEKLPS 121
P++LLHGF +S W R PL + A D +G + D + L
Sbjct: 95 PMILLHGFGASVFSWNRVMKPLARLVSSKVLAFDRPAFGLTSRIFHPFSGATNDAKPLNP 154
Query: 122 CDVVSKREHFYQFWKSYIKKPMILVGPSLGSAVAIDFAINYPEAVEKLVLIDASVY 177
+V F ILVG S G VA+D PE V L+L+ +++
Sbjct: 155 YSMVYSVLTTLYFIDVLAADKAILVGHSAGCPVALDAYFEAPERVAALILVAPAIF 210
>At5g21950 unknown protein
Length = 220
Score = 37.4 bits (85), Expect = 0.010
Identities = 43/163 (26%), Positives = 75/163 (45%), Gaps = 19/163 (11%)
Query: 70 QNKENP-IVLLHGFDSSCLEWRYTYPLLEEAGI-ETWAIDILGWGFSDLEKLPSCDVVSK 127
+N + P ++LLHGF S + W++++ + + + D++ +G S S ++
Sbjct: 50 ENTQKPSLLLLHGFGPSAV-WQWSHQVKPLSHFFRLYVPDLVFFGGS------SSSGENR 102
Query: 128 REHFYQFWKSYIKKPM-----ILVGPSLGSAVAIDFAINYPEAVEKLVLIDASVYTEGTG 182
E F + + + +VG S G VA + A +PE VEK+VL + V +
Sbjct: 103 SEMFQALCMGKLMEKLEVERFSVVGTSYGGFVAYNMAKMFPEKVEKVVLASSGVNLRRSD 162
Query: 183 NLATLPRAAAYAGVYLLKSVPLRVYANYL-SFTNISFSTSLDW 224
N A + RA + +K V L A L F+ + S LD+
Sbjct: 163 NEAFIARAKCHR----IKEVMLPASATDLRRFSGMVSSKRLDY 201
>At4g15960 unknown protein
Length = 375
Score = 37.0 bits (84), Expect = 0.013
Identities = 31/109 (28%), Positives = 49/109 (44%), Gaps = 4/109 (3%)
Query: 66 KPLVQNKENPIVL-LHGFDSSCLEWRYTYPLLEEAGIETWAIDILGWGFSDL-EKLPSCD 123
KP + E+PI+L LHGF WR+ L G T A D+ G+G ++ EK+
Sbjct: 71 KPGSGSGEDPIILFLHGFPELWYTWRHQMVALSSLGYRTIAPDLRGYGDTEAPEKVEDYT 130
Query: 124 VVSKREHFYQFWKSYI--KKPMILVGPSLGSAVAIDFAINYPEAVEKLV 170
++ + + K + +VG G+ +A PE V+ LV
Sbjct: 131 LLKRGRSVVALIVAVTGGDKAVSVVGHDWGAMIAWQLCQYRPEKVKALV 179
>At3g51000 epoxide hydrolase-like protein
Length = 323
Score = 36.6 bits (83), Expect = 0.018
Identities = 24/72 (33%), Positives = 34/72 (46%), Gaps = 13/72 (18%)
Query: 71 NKENPIVLL-HGFDSSCLEWRYTYPLLEEAGIETWAIDILGWGFSDLEKLPS-------- 121
++E P+VLL HGF + WR+ L G A D+ G+G SD LPS
Sbjct: 24 DEEGPLVLLLHGFPETWYSWRHQIDFLSSHGYHVVAPDLRGYGDSD--SLPSHESYTVSH 81
Query: 122 --CDVVSKREHF 131
DV+ +H+
Sbjct: 82 LVADVIGLLDHY 93
>At1g78210 unknown protein
Length = 314
Score = 35.8 bits (81), Expect = 0.030
Identities = 33/133 (24%), Positives = 58/133 (42%), Gaps = 5/133 (3%)
Query: 50 PVSVKFSENPIMS---SCVKPLVQNKENPIVLLHGFDSSCLEWRYTYPLLEEAGIETWAI 106
PV++ + +++ S KP + K N ++L+HG ++ + Y +
Sbjct: 26 PVTIDLKDGTVVNFWVSKTKPESKPKPN-LLLIHGLGATAIWQWYDVARRLSRYFNLYIP 84
Query: 107 DILGWGFSDLEKLPSCDVVSKREHFYQFWKSYIKKPMILVGPSLGSAVAIDFAINYPEAV 166
D++ +G S + D+ + +KK LVG S G V A Y +AV
Sbjct: 85 DLVFFGGSSTTRPERSDIFQAQTLMRALEAQSVKK-FSLVGLSYGGFVGYRMASMYADAV 143
Query: 167 EKLVLIDASVYTE 179
EK+V+ A+V E
Sbjct: 144 EKVVICCAAVCVE 156
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.335 0.145 0.460
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,375,805
Number of Sequences: 26719
Number of extensions: 263764
Number of successful extensions: 1107
Number of sequences better than 10.0: 59
Number of HSP's better than 10.0 without gapping: 28
Number of HSP's successfully gapped in prelim test: 31
Number of HSP's that attempted gapping in prelim test: 1061
Number of HSP's gapped (non-prelim): 60
length of query: 307
length of database: 11,318,596
effective HSP length: 99
effective length of query: 208
effective length of database: 8,673,415
effective search space: 1804070320
effective search space used: 1804070320
T: 11
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.6 bits)
S2: 60 (27.7 bits)
Medicago: description of AC146567.12


