

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146567.1 - phase: 0
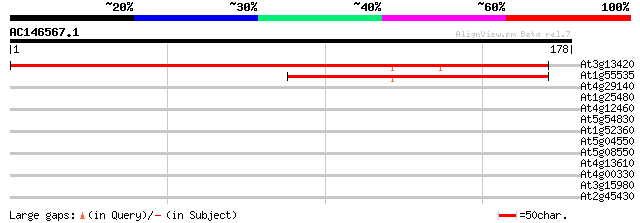
(178 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g13420 unknown protein 215 1e-56
At1g55535 putative protein 120 5e-28
At4g29140 unknown protein 29 1.2
At1g25480 hypothetical protein 28 2.0
At4g12460 putative SWH1 protein 28 3.4
At5g54830 unknown protein 27 4.4
At1g52360 coatomer complex subunit, putative 27 4.4
At5g04550 unknown protein 27 5.8
At5g08550 putative protein 27 7.6
At4g13610 DNA (cytosine-5-)-methyltransferase - like protein 26 9.9
At4g00330 unknown protein 26 9.9
At3g15980 putative coatomer complex subunit 26 9.9
At2g45430 putative AT-hook DNA-binding protein 26 9.9
>At3g13420 unknown protein
Length = 242
Score = 215 bits (547), Expect = 1e-56
Identities = 118/190 (62%), Positives = 139/190 (73%), Gaps = 19/190 (10%)
Query: 1 MISMKIGCALLGSLGASYNGVSLINLAISLFALVAIESSSQSLGRTYAFLLFCSILLDIS 60
+I +IGCAL+GSLGA YNGV LINLAI+LF LVAIES+SQSLGRTYA LLFC+ILLD+S
Sbjct: 27 LIYAQIGCALIGSLGALYNGVVLINLAIALFGLVAIESNSQSLGRTYAVLLFCAILLDVS 86
Query: 61 WFILFTHEIWNISSDDYAKFFIFSVKLTLAMQIIGFIVRLSSSLLWIQIYRLGASYVDTA 120
WFILF+ EIWNISSD Y F+IFSVKLTLAM+I GF+VRLSSSLLW QIYRLGAS +D+
Sbjct: 87 WFILFSKEIWNISSDMYQVFYIFSVKLTLAMEIAGFVVRLSSSLLWFQIYRLGASIIDSP 146
Query: 121 -SRAADFDLRSSFLSP------------------VTPAVARQISNSNEILGGSIYDPAYY 161
R +D DLR+SFL P PA+A+Q S S+EIL SI +PA Y
Sbjct: 147 FPRQSDSDLRNSFLEPPLLARQRSRDPELRNSFLQAPAIAKQRSRSDEILEDSIDEPASY 206
Query: 162 SSLFEDSQEN 171
+ L + N
Sbjct: 207 TPLLDGGLSN 216
>At1g55535 putative protein
Length = 116
Score = 120 bits (300), Expect = 5e-28
Identities = 60/84 (71%), Positives = 71/84 (84%), Gaps = 1/84 (1%)
Query: 89 LAMQIIGFIVRLSSSLLWIQIYRLGASYVDTA-SRAADFDLRSSFLSPVTPAVARQISNS 147
+AM++IGF VRLSSSLLW QIYRLGA+ VDT+ R D DLR+SFL+P TPA+ARQ S +
Sbjct: 1 MAMEMIGFFVRLSSSLLWFQIYRLGAAIVDTSLPRETDSDLRNSFLNPPTPAIARQCSGA 60
Query: 148 NEILGGSIYDPAYYSSLFEDSQEN 171
EILGGSIYDPAYY+SLFE+SQ N
Sbjct: 61 EEILGGSIYDPAYYTSLFEESQTN 84
>At4g29140 unknown protein
Length = 532
Score = 29.3 bits (64), Expect = 1.2
Identities = 28/109 (25%), Positives = 44/109 (39%), Gaps = 28/109 (25%)
Query: 40 SQSLGRTYAFLLFCSILLDISWFILFTHEIWNISSDDYAK----FFIFSV---------- 85
S +L RT FLL C + + + WF + ++ D AK + IFS+
Sbjct: 135 SLTLHRTVVFLLVCCVPISVLWFNVGKISVYLHQDPDIAKLAQTYLIFSLPDLLTNTLLH 194
Query: 86 -------------KLTLAMQIIGFIVRLSSSLLWIQIYRLGASYVDTAS 121
+TLA + G + L ++L + RLG + V AS
Sbjct: 195 PIRIYLRAQGIIHPVTLA-SLSGAVFHLPANLFLVSYLRLGLTGVAVAS 242
>At1g25480 hypothetical protein
Length = 548
Score = 28.5 bits (62), Expect = 2.0
Identities = 23/88 (26%), Positives = 41/88 (46%), Gaps = 17/88 (19%)
Query: 52 FCSILLDISWFILFT--HEIWNISSDDYAKFFIFSVKLTLAMQIIGFIVRLSSSL----- 104
FCS + SW L+ +++ + D K + FSVK+ +A+ + F++ L L
Sbjct: 38 FCSDGITASWKALYDIGAKLYEMGRSDRRKVY-FSVKMGMALALCSFVIYLKEPLRDASK 96
Query: 105 --LWIQI-------YRLGASYVDTASRA 123
+W + Y +GA+ V +RA
Sbjct: 97 YAVWAILTVVVVFEYSIGATLVKGFNRA 124
>At4g12460 putative SWH1 protein
Length = 694
Score = 27.7 bits (60), Expect = 3.4
Identities = 25/98 (25%), Positives = 41/98 (41%), Gaps = 14/98 (14%)
Query: 73 SSDDYAKFFIFSVKLTLAMQIIGFIVRLSSSLLWIQIYRLGASYVDTASRAADFDLRSSF 132
S D+ KF+IF+ TL ++ S W+Q V S DF SF
Sbjct: 117 SKSDHRKFYIFTATKTLHLRTDS----RSDRAAWLQALASTRGIVPLQSINGDF----SF 168
Query: 133 LSPVTPAVARQISNSNEILGGSIYDPAYYSSLFEDSQE 170
+SP + +S S E L +++ SL ++ ++
Sbjct: 169 VSP------KDLSISTERLKKRLFEEGMNESLVKECEQ 200
>At5g54830 unknown protein
Length = 907
Score = 27.3 bits (59), Expect = 4.4
Identities = 26/98 (26%), Positives = 42/98 (42%), Gaps = 8/98 (8%)
Query: 58 DISWFILFTHEIWNISSDDYAKFFIFSVKLTLAMQIIGFIVRLSSSLLWIQIYRLGASYV 117
++ W+ + + N+++D F I KL + FIVRL ++ W +LG V
Sbjct: 71 EVHWWGAMSSDFDNMTNDG---FVISDQKLNQTFKNSSFIVRLLGNVTW---DKLGVVSV 124
Query: 118 DTASRAADFD--LRSSFLSPVTPAVARQISNSNEILGG 153
A+DF L S+ T S SN++ G
Sbjct: 125 WDLPTASDFGHVLLSNATESDTSKAESPPSESNDVAPG 162
>At1g52360 coatomer complex subunit, putative
Length = 926
Score = 27.3 bits (59), Expect = 4.4
Identities = 18/44 (40%), Positives = 23/44 (51%), Gaps = 3/44 (6%)
Query: 129 RSSFLSPVTPAVA---RQISNSNEILGGSIYDPAYYSSLFEDSQ 169
RS S V+ VA + +S N S+ DP YS+LFED Q
Sbjct: 760 RSYLPSKVSEIVALWRKDLSKVNSKAAESLADPEEYSNLFEDWQ 803
>At5g04550 unknown protein
Length = 599
Score = 26.9 bits (58), Expect = 5.8
Identities = 16/70 (22%), Positives = 35/70 (49%)
Query: 37 ESSSQSLGRTYAFLLFCSILLDISWFILFTHEIWNISSDDYAKFFIFSVKLTLAMQIIGF 96
+++ +LG L + ++++ I F+ H I + + DD SV+ +L ++ +
Sbjct: 427 DAAPNTLGTACLALHYANVIIVIERFVASPHLIGDDARDDLYNMLPASVRTSLRERLKPY 486
Query: 97 IVRLSSSLLW 106
LSSS ++
Sbjct: 487 SKNLSSSTVY 496
>At5g08550 putative protein
Length = 908
Score = 26.6 bits (57), Expect = 7.6
Identities = 14/38 (36%), Positives = 21/38 (54%), Gaps = 4/38 (10%)
Query: 25 NLAISLFALVAIESSSQSLGRTYAFLL----FCSILLD 58
NL SL ++ A+ESS + G Y F+ F S++ D
Sbjct: 387 NLTASLMSITALESSLSAAGDKYVFMQKLRDFISVICD 424
>At4g13610 DNA (cytosine-5-)-methyltransferase - like protein
Length = 1404
Score = 26.2 bits (56), Expect = 9.9
Identities = 20/62 (32%), Positives = 31/62 (49%), Gaps = 2/62 (3%)
Query: 7 GCALLGSLGASYNGVSLINLAISLFALVAIESSSQSLGRTYAFLLFCSILLDISWFILFT 66
GC L S G GVS AI + A ++ Q+ +T F+ C+++L ISW L
Sbjct: 977 GCGGL-SYGLEKAGVSDTKWAIE-YEEPAAQAFKQNHPKTTVFVDNCNVILRISWLRLLI 1034
Query: 67 HE 68
++
Sbjct: 1035 ND 1036
>At4g00330 unknown protein
Length = 411
Score = 26.2 bits (56), Expect = 9.9
Identities = 11/38 (28%), Positives = 23/38 (59%)
Query: 112 LGASYVDTASRAADFDLRSSFLSPVTPAVARQISNSNE 149
+ ++Y+D + ++D ++RS S + +V R N+NE
Sbjct: 64 VSSNYIDNSKSSSDNNIRSRRSSTGSSSVQRSYGNANE 101
>At3g15980 putative coatomer complex subunit
Length = 921
Score = 26.2 bits (56), Expect = 9.9
Identities = 18/44 (40%), Positives = 22/44 (49%), Gaps = 3/44 (6%)
Query: 129 RSSFLSPVTPAVA---RQISNSNEILGGSIYDPAYYSSLFEDSQ 169
RS S V+ VA +S N S+ DP YS+LFED Q
Sbjct: 763 RSYLPSKVSEIVALWREDLSKVNPKAAESLADPEEYSNLFEDWQ 806
>At2g45430 putative AT-hook DNA-binding protein
Length = 317
Score = 26.2 bits (56), Expect = 9.9
Identities = 15/51 (29%), Positives = 23/51 (44%), Gaps = 5/51 (9%)
Query: 113 GASYVDTASRAADFDLRSSFLSPVTPAVARQIS-----NSNEILGGSIYDP 158
G+S V+ R L SFL P P A ++ +++GGS+ P
Sbjct: 168 GSSVVNLHGRFEILSLSGSFLPPPAPPAASGLTIYLAGGQGQVVGGSVVGP 218
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.324 0.137 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,522,232
Number of Sequences: 26719
Number of extensions: 124410
Number of successful extensions: 423
Number of sequences better than 10.0: 13
Number of HSP's better than 10.0 without gapping: 6
Number of HSP's successfully gapped in prelim test: 7
Number of HSP's that attempted gapping in prelim test: 414
Number of HSP's gapped (non-prelim): 13
length of query: 178
length of database: 11,318,596
effective HSP length: 93
effective length of query: 85
effective length of database: 8,833,729
effective search space: 750866965
effective search space used: 750866965
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 56 (26.2 bits)
Medicago: description of AC146567.1


