

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146563.9 + phase: 0
(620 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
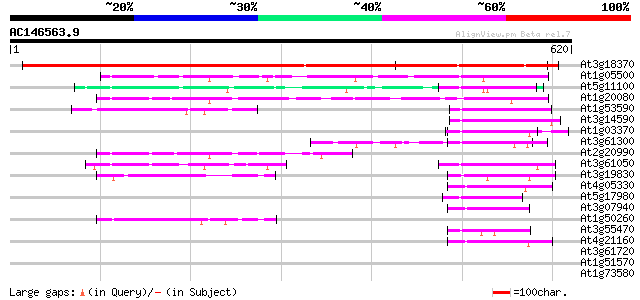
Score E
Sequences producing significant alignments: (bits) Value
At3g18370 unknown protein 686 0.0
At1g05500 Ca2+-dependent lipid-binding protein, putative 91 2e-18
At5g11100 CLB1 - like protein 89 7e-18
At1g20080 unknown protein 66 7e-11
At1g53590 Unknown protein (At1g53590) 62 9e-10
At3g14590 hypothetical protein 57 3e-08
At1g03370 unknown protein 55 2e-07
At3g61300 anthranilate phosphoribosyltransferase-like protein 54 2e-07
At2g20990 unknown protein 53 4e-07
At3g61050 CaLB protein 50 4e-06
At3g19830 unknown protein 49 6e-06
At4g05330 unknown protein 48 1e-05
At5g17980 phosphoribosylanthranilate transferase-like protein 47 3e-05
At3g07940 putative GTPase activating protein 47 3e-05
At1g50260 hypothetical protein 46 7e-05
At3g55470 elicitor responsive/phloem -like protein 45 2e-04
At4g21160 unknown protein 44 3e-04
At3g61720 putative protein 42 0.001
At1g51570 anthranilate phosphoribosyltransferase, putative (At1g... 41 0.002
At1g73580 hypothetical protein 40 0.003
>At3g18370 unknown protein
Length = 815
Score = 686 bits (1770), Expect = 0.0
Identities = 332/592 (56%), Positives = 444/592 (74%), Gaps = 3/592 (0%)
Query: 15 EVAVDFFNYVLQEKPKIPFFIPVILIACAVEKWVFSFSTWVPLALAVWATIQYGRYQRKL 74
E A +F N+++ E+ + +P++L A+E+WVF+FS WVPL +AVWA++QYG YQR L
Sbjct: 14 EAAREFINHLVAERHSLLLLVPLVLAFWAIERWVFAFSNWVPLVVAVWASLQYGSYQRAL 73
Query: 75 LVEDLDKKWKRIILNNSPITPLEHCEWLNKLLTEIWPNYFNPKLSSRLSAIVEARLKLRK 134
L EDL KKW++ + N S ITPLEHC+WLNKLL+EIW NY N KLS R S++VE RL+ R+
Sbjct: 74 LAEDLTKKWRQTVFNASTITPLEHCQWLNKLLSEIWLNYMNKKLSLRFSSMVEKRLRQRR 133
Query: 135 PRFLERVELQEFSLGSCPPSLALQGMRWSTIGDQRVMQLGFDWDTHEMSILLLAKLAKPL 194
R +E ++L EFSLGSCPP L L G WS G+Q++M+L F+WDT ++SILL AKL+ P
Sbjct: 134 SRLIENIQLLEFSLGSCPPLLGLHGTCWSKSGEQKIMRLDFNWDTTDLSILLQAKLSMPF 193
Query: 195 MGTARIVINSLHIKGDLIFTPILDGKALLYSFVSAPEVRVGVAFGSGGSQSLPATEWPGV 254
TARIV+NSL IKGD++ PIL+G+ALLYSFVS PEVR+GVAFG GG QSLPATE PGV
Sbjct: 194 NRTARIVVNSLCIKGDILIRPILEGRALLYSFVSNPEVRIGVAFGGGGGQSLPATELPGV 253
Query: 255 SSWLEKLFTDTLVKTMVEPRRRCFTLPAVDLRKKAVGGIIYVRVISANKLSSSSFKASRR 314
SSWL K+ T+TL K MVEPRR CF+LPA DL K A+GGIIYV V+S N L+ + S
Sbjct: 254 SSWLVKILTETLNKKMVEPRRGCFSLPATDLHKTAIGGIIYVTVVSGNNLNRRILRGSPS 313
Query: 315 QQSGSTNGSSEDVSDDKDLHTFVEVEIEELTRRTDVRLGSTPRWDAPFNMVLHDNTGTLR 374
+ S GSS + S K + TFVEVE+E+L+RRT+++ G P + + FNM+LHDNTGTL+
Sbjct: 314 KSSEIGEGSSGN-SSSKPVQTFVEVELEQLSRRTEMKSGPNPAYQSTFNMILHDNTGTLK 372
Query: 375 FNLYECIPNNVKCDYLGSCEIKLRHVEDDSTIMWAVGPDSGIIAKQAQFCGDEIEMVVPF 434
FNLYE P +V+ D L SCE+K+++V DDST+ WAVG D+G+IAK A+FCG EIEMVVPF
Sbjct: 373 FNLYENNPGSVRYDSLASCEVKMKYVGDDSTMFWAVGSDNGVIAKHAEFCGQEIEMVVPF 432
Query: 435 EGTNSGELKVSIVVKEWQFSDGTHSLNNLRNNSQQSLNGSSNIQLRTGKKLKITVVEGKD 494
EG +SGEL V +++KEW FSDG+HSLN++ ++S SL+ SS + +TG+K+ +TV+ GK+
Sbjct: 433 EGVSSGELTVRLLLKEWHFSDGSHSLNSVNSSSLHSLDSSSALLSKTGRKIIVTVLAGKN 492
Query: 495 LAAAKEKTGKFDPYIKLQYGKVMQKTKTSHTPNPVWNQTIEFDEVGGGEYLKLKVFTEEL 554
L +K+K+GK D +KLQYGK++QKTK + VWNQ EF+E+ G EYLK+K + EE+
Sbjct: 493 L-VSKDKSGKCDASVKLQYGKIIQKTKIVNAAECVWNQKFEFEELAGEEYLKVKCYREEM 551
Query: 555 FGDENIGSAQVNLEGLVDGSVRDVWIPLERVRSGEIRLKIEAIKVDDQEGST 606
G +NIG+A ++L+G ++ S +W+PLE V SGEI L IEA+ + E +
Sbjct: 552 LGTDNIGTATLSLQG-INNSEMHIWVPLEDVNSGEIELLIEALDPEYSEADS 602
Score = 80.1 bits (196), Expect = 3e-15
Identities = 52/169 (30%), Positives = 88/169 (51%), Gaps = 25/169 (14%)
Query: 427 EIEMVVPFEGTNSGELKVSIVVKEWQFSDGTHSLNNLRNNSQQSLNGSSNIQLRTGKKLK 486
E+ + VP E NSGE+++ I + ++S+ +S + L ++
Sbjct: 571 EMHIWVPLEDVNSGEIELLIEALDPEYSEA---------DSSKGL-------------IE 608
Query: 487 ITVVEGKDLAAAKEKTGKFDPYIKLQYGKVMQKTKTSH-TPNPVWNQTIEFDEVGGGEYL 545
+ +VE +DL AA + G DPY+++QYG+ Q+TK + T P WNQT+EF + G L
Sbjct: 609 LVLVEARDLVAADIR-GTSDPYVRVQYGEKKQRTKVIYKTLQPKWNQTMEFPDDGSSLEL 667
Query: 546 KLKVFTEELFGDENIGSAQVNLEGLVDGSVRDVWIPLERVRSGEIRLKI 594
+K + L +IG+ V +GL D WI L+ V+ GE+ +++
Sbjct: 668 HVKDYNT-LLPTSSIGNCVVEYQGLKPNETADKWIILQGVKHGEVHVRV 715
>At1g05500 Ca2+-dependent lipid-binding protein, putative
Length = 560
Score = 90.9 bits (224), Expect = 2e-18
Identities = 119/521 (22%), Positives = 223/521 (41%), Gaps = 69/521 (13%)
Query: 101 WLNKLLTEIWPNYFNPKLSSRLSAIVEARLKLRKPRFLERVELQEFSLGSCPPSLALQGM 160
WLN LT+IWP Y + S + A VE L+ +P + + + +LG+ P +
Sbjct: 73 WLNHHLTKIWP-YVDEAASELIKASVEPVLEQYRPAIVASLTFSKLTLGTVAPQFTGVSV 131
Query: 161 RWSTIGDQR--VMQLGFDWDTHEMSILLLAKLAKPLMGTAR-IVINSLHIKG--DLIFTP 215
GD+ ++L WD + +L + K L+G + I + ++ G LIF P
Sbjct: 132 ---IDGDKNGITLELDMQWDGNPNIVLGV----KTLVGVSLPIQVKNIGFTGVFRLIFRP 184
Query: 216 ILDG----KALLYSFVSAPEVRVGVAFGSGGSQSLPATEWPGVSSWLEKLFTDTLVKTMV 271
+++ A+ S ++ + G ++P G+S +E+ D + ++
Sbjct: 185 LVEDFPCFGAVSVSLREKKKLDFTLKVVGGDISAIP-----GLSEAIEETIRDAVEDSIT 239
Query: 272 EPRRRCFTLPAV-----DLRKKAVGGIIYVRVISANKLSSSSFKASRRQQSGSTNGSSED 326
P R+ +P + DL K VG ++ V+++ A L++
Sbjct: 240 WPVRK--VIPIIPGDYSDLELKPVG-MLEVKLVQAKNLTNKDLVGK-------------- 282
Query: 327 VSDDKDLHTFVEVEIEELTRRTDVRLGSTPRWDAPFNMVLHD-NTGTLRFNLYEC----I 381
D F+ E+ R + P W+ F V+ D +T L +Y+
Sbjct: 283 --SDPFAKMFIRPLREKTKRSKTINNDLNPIWNEHFEFVVEDASTQHLVVRIYDDEGVQA 340
Query: 382 PNNVKCDYLGSCEIKLRHVEDDSTIMWAVGPDSGIIAKQAQFCGD-EIEMVVPFEGTNSG 440
+ C + CE++ V+D +W I + + G+ +E++ G+ +G
Sbjct: 341 SELIGCAQIRLCELEPGKVKD----VWLKLVKDLEIQRDTKNRGEVHLELLYIPYGSGNG 396
Query: 441 ELKVSIVVKEWQFSDGTHSLNNLRNNSQQSLNGSSNIQLRTGKK--LKITVVEGKDLAAA 498
+V + S T L+N++ N SS + + L +TV+ +++
Sbjct: 397 ------IVNPFVTSSMTSLERVLKNDTTDEENASSRKRKDVIVRGVLSVTVISAEEIPI- 449
Query: 499 KEKTGKFDPYIKLQYGKVMQKTKT---SHTPNPVWNQTIEFD-EVGGGEYLKLKVFTEEL 554
++ GK DPY+ L K K+KT + + NPVWNQT +F E G + L L+V+ +
Sbjct: 450 QDLMGKADPYVVLSMKKSGAKSKTRVVNDSLNPVWNQTFDFVVEDGLHDMLVLEVWDHDT 509
Query: 555 FGDENIGSAQVNLEGLVDGSVRDVWIPLERVRSGEIRLKIE 595
FG + IG + L ++ W PL+ ++G+++L ++
Sbjct: 510 FGKDYIGRCILTLTRVIMEEEYKDWYPLDESKTGKLQLHLK 550
>At5g11100 CLB1 - like protein
Length = 574
Score = 89.0 bits (219), Expect = 7e-18
Identities = 133/557 (23%), Positives = 222/557 (38%), Gaps = 87/557 (15%)
Query: 72 RKLLVEDLDKKWKRIILNNSPITPLEHC--EWLNKLLTEIWPNYFNPKLSSRLSAIVEAR 129
RKLL D W ++ + C WLN L +IWP Y N S + + VE
Sbjct: 51 RKLLPGDFYPSW--VVFSQRQKLSYSKCLLNWLNLELEKIWP-YVNEAASELIKSSVEPV 107
Query: 130 LKLRKPRFLERVELQEFSLGSCPPSLALQGMRWSTIGDQRV-MQLGFDWDTHEMSILLLA 188
L+ P L ++ +F+LG+ P + S G + M+L WD + +L +
Sbjct: 108 LEQYTPAMLASLKFSKFTLGTVAPQFTGVSILESESGPNGITMELEMQWDGNPKIVLDV- 166
Query: 189 KLAKPLMGTA-RIVINSLHIKG--DLIFTPILDGKALLYSFVSAPEVRVGVAFG---SGG 242
K L+G + I + ++ G LIF P++D + + + G+ F GG
Sbjct: 167 ---KTLLGVSLPIEVKNIGFTGVFRLIFKPLVDEFPCFGALSYSLREKKGLDFTLKVIGG 223
Query: 243 SQSLPATEWPGVSSWLEKLFTDTLVKTMVEPRRRCF-TLPA--VDLRKKAVGGIIYVRVI 299
T PG+S +E+ D + ++ P R+ LP DL K VG + V+V+
Sbjct: 224 E----LTSIPGISDAIEETIRDAIEDSITWPVRKIIPILPGDYSDLELKPVGK-LDVKVV 278
Query: 300 SANKLSSSSFKASRRQQSGSTNGSSEDVSDDKDLHTFVEVE-IEELTRRTDVRLGS-TPR 357
A L +++D+ D + V + + + T++T S P
Sbjct: 279 QAKDL------------------ANKDMIGKSDPYAIVFIRPLPDRTKKTKTISNSLNPI 320
Query: 358 WDAPFNMVLHDNT--------------------GTLRFNLYECIPNNVKCDYLGSCEIKL 397
W+ F ++ D + G + L E +P VK +L +K
Sbjct: 321 WNEHFEFIVEDVSTQHLTVRVFDDEGVGSSQLIGAAQVPLNELVPGKVKDIWLKL--VKD 378
Query: 398 RHVEDDSTIMWAVGPDSGIIAKQAQFCGDEIEMVVPFEGTNSGELKVSIVVKEWQFSDGT 457
++ D+ + G + G E + PF S + ++ E + SD T
Sbjct: 379 LEIQRDT-------KNRGQLELLYCPLGKEGGLKNPFNPDYSLTILEKVLKPESEDSDAT 431
Query: 458 HSLNNLRNNSQQSLNGSSNIQLRTGKKLKITVVEGKDLAAAKEKTGKFDPYIKLQYGKVM 517
+ L S + L +TVV +DL A + GK D ++ + K
Sbjct: 432 ---------DMKKLVTSKKKDVIVRGVLSVTVVAAEDLPAV-DFMGKADAFVVITLKKSE 481
Query: 518 QKTKTSHTP---NPVWNQTIEF-DEVGGGEYLKLKVFTEELFGDENIGSAQVNLEGLVDG 573
K+KT P NPVWNQT +F E + L L+V+ + FG + IG + L ++
Sbjct: 482 TKSKTRVVPDSLNPVWNQTFDFVVEDALHDLLTLEVWDHDKFGKDKIGRVIMTLTRVMLE 541
Query: 574 SVRDVWIPLERVRSGEI 590
W L+ +SG++
Sbjct: 542 GEFQEWFELDGAKSGKL 558
Score = 68.2 bits (165), Expect = 1e-11
Identities = 44/114 (38%), Positives = 67/114 (58%), Gaps = 7/114 (6%)
Query: 475 SNIQLRTGKKLKITVVEGKDLAAAKEKTGKFDPYIKLQYGKVMQKTKT----SHTPNPVW 530
S+++L+ KL + VV+ KDLA K+ GK DPY + + +TK S++ NP+W
Sbjct: 263 SDLELKPVGKLDVKVVQAKDLAN-KDMIGKSDPYAIVFIRPLPDRTKKTKTISNSLNPIW 321
Query: 531 NQTIEFD-EVGGGEYLKLKVFTEELFGDEN-IGSAQVNLEGLVDGSVRDVWIPL 582
N+ EF E ++L ++VF +E G IG+AQV L LV G V+D+W+ L
Sbjct: 322 NEHFEFIVEDVSTQHLTVRVFDDEGVGSSQLIGAAQVPLNELVPGKVKDIWLKL 375
>At1g20080 unknown protein
Length = 535
Score = 65.9 bits (159), Expect = 7e-11
Identities = 104/514 (20%), Positives = 217/514 (41%), Gaps = 65/514 (12%)
Query: 97 EHCEWLNKLLTEIWPNYFNPKLSSRLSAIVEARLKLRKPRF-LERVELQEFSLGSCPPSL 155
+ +WLNKL+ +WP Y + + +I + + + P + ++ VE + +LGS PPS
Sbjct: 67 DRIDWLNKLIGHMWP-YMDKAICKMAKSIAKPIIAEQIPNYKIDSVEFEMLTLGSLPPS- 124
Query: 156 ALQGMR-WSTIGDQRVMQLGFDWDTHEMSILLLAKLAKPLMGTARIVINSLHIKGDLIFT 214
QGM+ ++T + +M+L W +I+++AK A L T +++ ++ +
Sbjct: 125 -FQGMKVYATDDKEIIMELSVKW-AGNPNIIVVAK-AFGLKATVQVIDLQVYATPRITLK 181
Query: 215 PILDG----KALLYSFVSAPEVRVGVAFGSGGSQSLPATEWPGVSSWLEKLFTDTLVKTM 270
P++ + S + P+V G+ ++ PG+ +++++ D +
Sbjct: 182 PLVPSFPCFANIFVSLMDKPQVDFGLKLLGADVMAI-----PGLYRFVQEIIKDQVANMY 236
Query: 271 VEPRRRCFTLPAVDLRKKAVGGIIYVRVISANKLSSSSFKASRRQQSGSTNGSSEDVSDD 330
+ P+ + K G++ V+VI A KL GS +S D
Sbjct: 237 LWPKTLNVQIMDPSKAMKKPVGLLSVKVIKAIKLKKKDL------LGGSDPYVKLTLSGD 290
Query: 331 KDLHTFVEVEIEELTRRTDVRLGSTPRWDAPFNMVLHD-NTGTLRFNLYECIPNNVKCDY 389
K V+ L P W+ F++V+ + + L+ +Y+ K D
Sbjct: 291 KVPGKKTVVKHSNL----------NPEWNEEFDLVVKEPESQELQLIVYDW-EQVGKHDK 339
Query: 390 LGSCEIKLRHVEDDSTIMWAVGPDSGIIAKQAQFCGDEIEMVVPFEGTNSGELKVSIVVK 449
+G I+L+ + + + + + K+ P + G+L V + K
Sbjct: 340 IGMNVIQLKDLTPEEPKLMTLELLKSMEPKE------------PVSEKSRGQLVVEVEYK 387
Query: 450 EWQFSDGTHSLNNLRNNSQQSLNGSSNIQLRTGKKLKITVVEGKDLAAAKEKTGKF--DP 507
++ D ++++ N +++ G+ + TG L + V E +DL GK+ +P
Sbjct: 388 PFKDDDIPENIDD-PNAVEKAPEGTPS----TGGLLVVIVHEAEDL------EGKYHTNP 436
Query: 508 YIKLQYGKVMQKTK-TSHTPNPVWNQTIEF--DEVGGGEYLKLKVFTEE---LFGDENIG 561
++L + +KTK P W++ +F DE + L ++V + + E +G
Sbjct: 437 SVRLLFRGEERKTKRVKKNREPRWDEDFQFPLDEPPINDKLHVEVISSSSRLIHPKETLG 496
Query: 562 SAQVNLEGLVDGSVRDVWIPLERVRSGEIRLKIE 595
+NL +V + L ++G I+++++
Sbjct: 497 YVVINLGDVVSNRRINDKYHLIDSKNGRIQIELQ 530
>At1g53590 Unknown protein (At1g53590)
Length = 751
Score = 62.0 bits (149), Expect = 9e-10
Identities = 57/218 (26%), Positives = 104/218 (47%), Gaps = 23/218 (10%)
Query: 69 RYQRKLLVEDLDKKWKRIILNNSPITPLEHCEWLNKLLTEIWPNYFNPKLSSR-LSAIVE 127
R +RKL E+ + +R +L++S E W+N + +IWP S + L I+
Sbjct: 47 RLKRKLQFEERKQANQRRVLSDS-----ESVRWMNYAVEKIWPICMEQIASQKILGPIIP 101
Query: 128 ARLKLRKPRFLERVELQEFSLGSCPPSLA-LQGMRWSTIGDQRVMQLGFDW-DTHEMSIL 185
L+ +P ++ +Q +G PP L ++ +R ST D V++LG ++ +MS +
Sbjct: 102 WFLEKYRPWTAKKAVIQHLYMGRNPPLLTDIRVLRQSTGDDHLVLELGMNFLAADDMSAI 161
Query: 186 LLAKLAKPL---MGTARIVINSLHIKGDLIFT-------PILDGKALLYSFVSAPEVRVG 235
L KL K L M T ++ + +H++G ++ P L + ++ ++ V
Sbjct: 162 LAVKLRKRLGFGMWT-KLHLTGMHVEGKVLIGVKFLRRWPFLGRLRVCFAEPPYFQMTVK 220
Query: 236 VAFGSGGSQSLPATEWPGVSSWLEKLFTDTLVKTMVEP 273
F G L PG++ WL+KL + +T+V+P
Sbjct: 221 PIFTHG----LDVAVLPGIAGWLDKLLSIAFEQTLVQP 254
Score = 52.0 bits (123), Expect = 1e-06
Identities = 32/115 (27%), Positives = 55/115 (47%), Gaps = 4/115 (3%)
Query: 487 ITVVEGKDLAAAKEKTGKFDPYIKLQYGKVMQKTKTSH-TPNPVWNQTIEFDEV--GGGE 543
+ V E DL + + G DPY+K + G KTK T +P W++ +
Sbjct: 286 VEVFEASDLKPS-DLNGLADPYVKGKLGAYRFKTKIQKKTLSPKWHEEFKIPIFTWDSPS 344
Query: 544 YLKLKVFTEELFGDENIGSAQVNLEGLVDGSVRDVWIPLERVRSGEIRLKIEAIK 598
L ++V ++ F D+ +G VN+E G D+W+ L+ ++ G + L I I+
Sbjct: 345 ILNIEVGDKDRFVDDTLGECSVNIEEFRGGQRNDMWLSLQNIKMGRLHLAITVIE 399
>At3g14590 hypothetical protein
Length = 705
Score = 57.0 bits (136), Expect = 3e-08
Identities = 38/131 (29%), Positives = 63/131 (48%), Gaps = 10/131 (7%)
Query: 487 ITVVEGKDLAAAKEKTGKFDPYIKLQYGKVMQKTKTS-HTPNPVWNQTIEFDEV--GGGE 543
+ VVE D+ + + G DPY+K Q G KTK T P W + +
Sbjct: 255 VEVVEACDVKPS-DLNGLADPYVKGQLGAYRFKTKILWKTLAPKWQEEFKIPICTWDSAN 313
Query: 544 YLKLKVFTEELFGDENIGSAQVNLEGLVDGSVRDVWIPLERVRSGEIRLKIEAI----KV 599
L ++V ++ F D+++G VN+ G D+W+PL+ ++ G + L I + K+
Sbjct: 314 ILNIEVQDKDRFSDDSLGDCSVNIAEFRGGQRNDMWLPLQNIKMGRLHLAITVLEDEAKL 373
Query: 600 DDQ--EGSTVC 608
+D EG T+C
Sbjct: 374 NDDPFEGVTIC 384
Score = 40.0 bits (92), Expect = 0.004
Identities = 52/216 (24%), Positives = 90/216 (41%), Gaps = 51/216 (23%)
Query: 69 RYQRKLLVEDLDKKWKRIILNNSPITPLEHCEWLNKLLTEIWPNYFNPKLSSRLSAIVEA 128
R ++KL E+ + +R +L++S E WLN + IWP S ++
Sbjct: 47 RLRKKLQFEERKQANQRRVLSDS-----ESVRWLNHAVERIWPICMEQIASQKI------ 95
Query: 129 RLKLRKPRFLERVELQEFSLGSCPPSLALQGMRWSTIGDQRVMQLGFDWDT-HEMSILLL 187
L+ P FL++ W+ V++LG ++ T +MS +L
Sbjct: 96 -LRPIIPWFLDKYR------------------PWTA-----VLELGMNFLTADDMSAILA 131
Query: 188 AKLAKPL---MGTARIVINSLHIKGDLIF-------TPILDGKALLYSFVSAPEVRVGVA 237
KL K L M T ++ + +H++G ++ P L L F P ++ V
Sbjct: 132 VKLRKRLGFGMWT-KLHLTGMHVEGKVLIGVKFLRRWPFLG--RLRVCFAEPPYFQMTVK 188
Query: 238 FGSGGSQSLPATEWPGVSSWLEKLFTDTLVKTMVEP 273
+ L PG++ WL+KL + +T+VEP
Sbjct: 189 --PITTHGLDVAVLPGIAGWLDKLLSVAFEQTLVEP 222
>At1g03370 unknown protein
Length = 1859
Score = 54.7 bits (130), Expect = 2e-07
Identities = 44/139 (31%), Positives = 65/139 (46%), Gaps = 14/139 (10%)
Query: 484 KLKITVVEGKDLAAAKEKTGKFDPYIKLQYGKVMQKTK-TSHTPNPVWNQTIEFDEVGGG 542
KL++ VVE ++L A + G DPY++LQ GK +TK NP W + F
Sbjct: 828 KLQVRVVEARNLPAM-DLNGFSDPYVRLQLGKQRSRTKVVKKNLNPKWTEDFSFGVDDLN 886
Query: 543 EYLKLKVFTEE-LFGDENIGSAQVNLEGLVDG---SVRDVWIPLERVRSGEIRLKIEAIK 598
+ L + V E+ F D+ +G +V++ + D S+ VW PL + G K
Sbjct: 887 DELVVSVLDEDKYFNDDFVGQVRVSVSLVFDAENQSLGTVWYPLNPKKKGS--------K 938
Query: 599 VDDQEGSTVCFFSQKLSIL 617
D E FSQK S+L
Sbjct: 939 KDCGEILLKICFSQKNSVL 957
Score = 51.2 bits (121), Expect = 2e-06
Identities = 36/106 (33%), Positives = 53/106 (49%), Gaps = 5/106 (4%)
Query: 482 GKKLKITVVEGKDLAAAKEKTGKFDPYIKL-QYGKVMQKTKTSHTPNPVWNQTIEFDEVG 540
G L + ++EG DLAA + +G DPYI GK + NP WN+ EFD +
Sbjct: 1360 GWLLTVALIEGVDLAAV-DPSGHCDPYIVFTSNGKTRTSSIKFQKSNPQWNEIFEFDAMA 1418
Query: 541 GG-EYLKLKVFTEELFGDE--NIGSAQVNLEGLVDGSVRDVWIPLE 583
L ++VF + DE ++G A+VN + DVW+PL+
Sbjct: 1419 DPPSVLNVEVFDFDGPFDEAVSLGHAEVNFVRSNISDLADVWVPLQ 1464
>At3g61300 anthranilate phosphoribosyltransferase-like protein
Length = 972
Score = 54.3 bits (129), Expect = 2e-07
Identities = 66/258 (25%), Positives = 112/258 (42%), Gaps = 42/258 (16%)
Query: 333 LHTFVEVEIEELTRRTD-VRLGSTPRWDAPFNMVLHDNTGTLRFNLYECI---PNNVKCD 388
L ++EV++ T +T P W N V + + N+ E I + VK D
Sbjct: 270 LDPYIEVKLGNYTGKTKHFEKNQNPVW----NEVFAFSKSNQQSNVLEVIVMDKDMVKDD 325
Query: 389 YLGSCEIKLRHVEDDSTIMWAVGPDSGIIAKQAQFC---GDEIEMVVPFEGTNSGELKVS 445
++G L + V PDS + + + G EI + V F GT + E
Sbjct: 326 FVGLIRFDLNQIPT------RVAPDSPLAPEWYRVNNEKGGEIMLAVWF-GTQADEA--- 375
Query: 446 IVVKEWQFSDGTHS--LNNLRNNSQQS-LNGSSNIQLRTGKKLKITVVEGKDLAAAKEKT 502
FSD T+S LN + +S +S + S + L++ V+E +DL ++T
Sbjct: 376 -------FSDATYSDALNAVNKSSLRSKVYHSPRLWY-----LRVNVIEAQDLVIVPDRT 423
Query: 503 GKFDPYIKLQYGKVMQKTKTSHTPNPVWNQTIEFDEVGGGEYLKLKVFTEELFG---DEN 559
+PY+K++ + +TK SH+ NP WN+ EF V + L + E+ +E
Sbjct: 424 RLPNPYVKIRLNNQVVRTKPSHSLNPRWNE--EFTLVAAEPFEDLIISIEDRVAPNREET 481
Query: 560 IGSAQVNLEGLVDGSVRD 577
+G + + G +D + D
Sbjct: 482 LGEVHIPI-GTIDKRIDD 498
Score = 45.4 bits (106), Expect = 9e-05
Identities = 31/117 (26%), Positives = 56/117 (47%), Gaps = 8/117 (6%)
Query: 485 LKITVVEGKDLAAAKEKTGKFDPYIKLQYGKVMQKTK-TSHTPNPVWNQTIEFDEVG-GG 542
L I +V+ ++L + + TG DPYI+++ G KTK NPVWN+ F +
Sbjct: 251 LFIKIVKARNLPSM-DLTGSLDPYIEVKLGNYTGKTKHFEKNQNPVWNEVFAFSKSNQQS 309
Query: 543 EYLKLKVFTEELFGDENIGSAQVNLEGL-----VDGSVRDVWIPLERVRSGEIRLKI 594
L++ V +++ D+ +G + +L + D + W + + GEI L +
Sbjct: 310 NVLEVIVMDKDMVKDDFVGLIRFDLNQIPTRVAPDSPLAPEWYRVNNEKGGEIMLAV 366
>At2g20990 unknown protein
Length = 541
Score = 53.1 bits (126), Expect = 4e-07
Identities = 62/296 (20%), Positives = 129/296 (42%), Gaps = 45/296 (15%)
Query: 97 EHCEWLNKLLTEIWPNYFNPKLSSRLSAIVEARLKLRKPRF-LERVELQEFSLGSCPPSL 155
+ +W+N+ L +WP Y + + I + ++ + P++ ++ VE + +LGS PP+
Sbjct: 69 DRVDWINRFLEYMWP-YLDKAICKTAKNIAKPIIEEQIPKYKIDSVEFETLTLGSLPPT- 126
Query: 156 ALQGMR-WSTIGDQRVMQLGFDWDTHEMSILLLAKLAKPLMGTARIVINSLHIKGDLIFT 214
QGM+ + T + +M+ W + +L+A A L T ++V + + +
Sbjct: 127 -FQGMKVYLTDEKELIMEPCLKWAANPN--ILVAIKAFGLKATVQVVDLQVFAQPRITLK 183
Query: 215 PILDG----KALLYSFVSAPEVRVGVAFGSGGSQSLPATEWPGVSSWLEKLFTDTLVKTM 270
P++ + S + P V G+ G S+ PG+ ++++ D +
Sbjct: 184 PLVPSFPCFANIYVSLMEKPHVDFGLKLGGADLMSI-----PGLYRFVQEQIKDQVANMY 238
Query: 271 VEPRRRCFTLPAVDLRK--KAVGGIIYVRVISANKLSSSSFKASRRQQSGSTNGSSEDVS 328
+ P + +P +D K + GI++V+V+ A L +D+
Sbjct: 239 LWP--KTLVVPILDPAKAFRRPVGIVHVKVVRAVGL------------------RKKDLM 278
Query: 329 DDKDLHTFVEVEIEE---LTRRTDVR-LGSTPRWDAPFNMVLHD-NTGTLRFNLYE 379
D FV++++ E +++T V+ P W+ F + D T L F++Y+
Sbjct: 279 GGAD--PFVKIKLSEDKIPSKKTTVKHKNLNPEWNEEFKFSVRDPQTQVLEFSVYD 332
>At3g61050 CaLB protein
Length = 510
Score = 50.1 bits (118), Expect = 4e-06
Identities = 42/142 (29%), Positives = 72/142 (50%), Gaps = 14/142 (9%)
Query: 475 SNIQLRTGKKLKITVVEGKDLAAAKEKTGKFDPYIKLQYGKVMQ-KTKT-SHTPNPVWNQ 532
S+++L+ KL +TVV+ +L KE GK DPY + V + KTK + NPVW+Q
Sbjct: 255 SDLELKPQGKLIVTVVKATNLKN-KELIGKSDPYATIYIRPVFKYKTKAIENNLNPVWDQ 313
Query: 533 TIE-FDEVGGGEYLKLKVFTEELFGDENIGSAQVNLEGLVDGSVRDVWIPL--------- 582
T E E + L ++VF +++ DE +G ++ L L G +++ + L
Sbjct: 314 TFELIAEDKETQSLTVEVFDKDVGQDERLGLVKLPLSSLEAGVTKELELNLLSSLDTLKV 373
Query: 583 -ERVRSGEIRLKIEAIKVDDQE 603
++ G I LK+ + + +E
Sbjct: 374 KDKKDRGSITLKVHYHEFNKEE 395
Score = 42.4 bits (98), Expect = 8e-04
Identities = 59/245 (24%), Positives = 107/245 (43%), Gaps = 40/245 (16%)
Query: 84 KRIILNNSP----ITPLEHCEWLNKLLTEIWPNYFNPKLSSRLSAIVEARLKLRKPRFLE 139
K+I +N P E +WLNKLL+++WP Y + + VE L+ +P +
Sbjct: 51 KKICGDNFPQWISFPAFEQVKWLNKLLSKMWP-YIAEAATMVIRDSVEPLLEDYRPPGIT 109
Query: 140 RVELQEFSLGSCPPSLALQGMRWSTIGDQRV-MQLGFDWDTHEMSILLLAKLAKPLMGTA 198
++ + +LG+ P ++G+R + + +V M + W +L + TA
Sbjct: 110 SLKFSKLTLGNVAPK--IEGIRVQSFKEGQVTMDVDLRWGGDPNIVLGV---------TA 158
Query: 199 RIVINSLHIKGDLIFT----------PILDGKALLYSFVSAPEVRVGVAFGS-GGSQSLP 247
+ + +K +FT I A++ + ++ P+ R+ + GGS
Sbjct: 159 LVASIPIQLKDLQVFTVARVIFQLADEIPCISAVVVALLAEPKPRIDYTLKAVGGS---- 214
Query: 248 ATEWPGVSSWLEKLFTDTLVKTMVE-PRRRCFTLPAV-----DLRKKAVGGIIYVRVISA 301
T PG+S ++ DT+VK M++ P R + + DL K G +I V V+ A
Sbjct: 215 LTAIPGLSDMIDDT-VDTIVKDMLQWPHRIVVPIGGIPVDLSDLELKPQGKLI-VTVVKA 272
Query: 302 NKLSS 306
L +
Sbjct: 273 TNLKN 277
>At3g19830 unknown protein
Length = 666
Score = 49.3 bits (116), Expect = 6e-06
Identities = 50/203 (24%), Positives = 84/203 (40%), Gaps = 42/203 (20%)
Query: 97 EHCEWLNKLLTEIWPNY---FNPKLSSRLSAIVEARLKLRKPRFLERVELQEFSLGSCPP 153
E EW+N +L ++W Y L L +++ L+KP +++RVE+++FSLG P
Sbjct: 202 ESVEWVNMVLVKLWKVYRGGIENWLVGLLQPVID---DLKKPDYVQRVEIKQFSLGDEPL 258
Query: 154 SLALQGMRWSTIGDQRVMQLGFDWDTHEMSILLLAKLAKPLMGTARIVINSLHIKGDLIF 213
S+ R S + Q+G + +L+L+ + + I I G+L
Sbjct: 259 SVRNVERRTSRRVNDLQYQIGLRYTGGARMLLMLSLKFGIIPVVVPVGIRDFDIDGELWV 318
Query: 214 TPILDGKALLYSFVSAPEVRVGVAFGSGGSQSLPATEWPGVSSW--LEKLFTDTLVKTMV 271
L +P+ W G +SW L KL T+ L + V
Sbjct: 319 KLRL----------------------------IPSAPWVGAASWAFLTKLLTEDLPRLFV 350
Query: 272 EPRRRCFTLPAVDLRK-KAVGGI 293
P++ +D +K KAVG +
Sbjct: 351 RPKK-----IVLDFQKGKAVGPV 368
Score = 43.1 bits (100), Expect = 5e-04
Identities = 42/136 (30%), Positives = 66/136 (47%), Gaps = 18/136 (13%)
Query: 484 KLKITVVEGKDLAAAKEKTGKFDPYIKLQYG-KVMQKTKTSHTP------NPVWNQTIEF 536
+L +T+V + L +GK DPY+ L+ G +V++ K S T P+WNQ +F
Sbjct: 387 ELSVTLVNAQKLPYMF--SGKTDPYVILRIGDQVIRSKKNSQTTVIGAPGQPIWNQDFQF 444
Query: 537 DEVGGGE-YLKLKVFTEELFGDENIGSAQVNLEGLVDG-------SVRDVWIPLERVRSG 588
E L+++V F D IG +V+LE L D S+R W + +G
Sbjct: 445 LVSNPREQVLQIEVNDCLGFADMAIGIGEVDLESLPDTVPTDRFVSLRGGWSLFGKGSTG 504
Query: 589 EIRLKIE-AIKVDDQE 603
EI L++ V+D+E
Sbjct: 505 EILLRLTYKAYVEDEE 520
>At4g05330 unknown protein
Length = 336
Score = 48.1 bits (113), Expect = 1e-05
Identities = 41/156 (26%), Positives = 69/156 (43%), Gaps = 42/156 (26%)
Query: 485 LKITVVEGKDLAAAKEKTGKFDPYIKLQYGKV-MQKTKTSHTPNPVWNQTIEFDEVGGGE 543
LK+T+ +G +LA + DPY+ L GK +Q T + NPVWNQ +
Sbjct: 182 LKVTIKKGTNLAIRDMMSS--DPYVVLNLGKQKLQTTVMNSNLNPVWNQELMLSVPESYG 239
Query: 544 YLKLKVFTEELF-GDENIGSAQVNLE---------------------------------- 568
+KL+V+ + F D+ +G A ++++
Sbjct: 240 PVKLQVYDYDTFSADDIMGEADIDIQPLITSAMAFGDPEMFGDMQIGKWLKSHDNPLIDD 299
Query: 569 ---GLVDGSVR-DVWIPLERVRSGEIRLKIEAIKVD 600
+VDG V+ +V I L+ V SGE+ L++E + +D
Sbjct: 300 SIINIVDGKVKQEVQIKLQNVESGELELEMEWLPLD 335
>At5g17980 phosphoribosylanthranilate transferase-like protein
Length = 1049
Score = 47.0 bits (110), Expect = 3e-05
Identities = 30/90 (33%), Positives = 47/90 (51%), Gaps = 3/90 (3%)
Query: 479 LRTGKKLKITVVEGKDLAAAKEKTGKFDPYIKLQYGKVMQKTKTS-HTPNPVWNQTIEFD 537
+ T +KL + VV+ KDL K+ G PY+ L Y ++T+T NPVWN+T+EF
Sbjct: 1 MATTRKLVVEVVDAKDLTP-KDGHGTSSPYVVLDYYGQRRRTRTIVRDLNPVWNETLEFS 59
Query: 538 EVGGGEY-LKLKVFTEELFGDENIGSAQVN 566
+ L V +++ D+N G + N
Sbjct: 60 LAKRPSHQLFTDVLELDMYHDKNFGQTRRN 89
Score = 37.0 bits (84), Expect = 0.033
Identities = 35/109 (32%), Positives = 51/109 (46%), Gaps = 12/109 (11%)
Query: 485 LKITVVEGKDLAAAKEKTGKFDPY-IKLQYGKVMQKTKTSHTPN--PVWNQTIEFDEVGG 541
L+ TV+E +DL + K + +K Q G +QKTK++ T N P WN+ + F V
Sbjct: 479 LRATVIEAQDLLPPQLTAFKEASFQLKAQLGSQVQKTKSAVTRNGAPSWNEDLLF--VAA 536
Query: 542 GEYLKLKVFTEEL---FGDENIGSAQVNLEGLV----DGSVRDVWIPLE 583
+ VFT E G +G A+V L + D V W+ LE
Sbjct: 537 EPFSDQLVFTLEYRTSKGPVTVGMARVPLSAIERRVDDRLVASRWLGLE 585
>At3g07940 putative GTPase activating protein
Length = 385
Score = 47.0 bits (110), Expect = 3e-05
Identities = 29/92 (31%), Positives = 51/92 (54%), Gaps = 4/92 (4%)
Query: 485 LKITVVEGKDLAAAKEKTGKFDPYIKLQYGKVMQKTKT-SHTPNPVWNQTIEFDEVGGGE 543
+K+ VV+G +LA T DPY+ L G+ KT+ + NPVWN+T+
Sbjct: 231 IKVNVVKGTNLAVRDVMTS--DPYVILALGQQSVKTRVIKNNLNPVWNETLMLSIPEPMP 288
Query: 544 YLKLKVFTEELFG-DENIGSAQVNLEGLVDGS 574
LK+ V+ ++ F D+ +G A+++++ LV +
Sbjct: 289 PLKVLVYDKDTFSTDDFMGEAEIDIQPLVSAA 320
>At1g50260 hypothetical protein
Length = 778
Score = 45.8 bits (107), Expect = 7e-05
Identities = 56/211 (26%), Positives = 99/211 (46%), Gaps = 24/211 (11%)
Query: 97 EHCEWLNKLLTEIWPNYFNPKLSSRLSAIVEARLK-LRKPRFLERVELQEFSLGSCPPSL 155
E EW+N +L ++W Y + + L +++ + L+KP +++RVE+++FSLG P S+
Sbjct: 190 ESVEWVNMVLGKLWKVY-RAGIENWLVGLLQPVIDDLKKPDYVQRVEIKQFSLGDEPLSV 248
Query: 156 ALQGMRWSTIGDQRVMQLGFDWDTHEMSILLLAKLAKPLMGTARIVINSLHIKGD----- 210
+ S + Q+G + +L+L+ + + + I G+
Sbjct: 249 RNVERKTSRRANDLQYQIGLRYTGGARMLLMLSLKFGVIPIVVPVGVRDFDIDGELWVKL 308
Query: 211 -LIFTPILDGKALLYSFVSAPEVRVGVA----FGSGGSQ-SLPATEWPGVSSWLEKLFTD 264
LI T G A+ SFVS P+V +A F G + S + +P +L KL T
Sbjct: 309 RLIPTQPWVG-AVSCSFVSLPKVTFQLAAFRLFNLMGMRLSWSSDPFP----FLTKLLTV 363
Query: 265 TLVKTMVEPRRRCFTLPAVDLRK-KAVGGII 294
L + V P++ +D +K KAVG ++
Sbjct: 364 DLPRLFVRPKK-----IVLDFQKGKAVGPVL 389
>At3g55470 elicitor responsive/phloem -like protein
Length = 156
Score = 44.7 bits (104), Expect = 2e-04
Identities = 31/99 (31%), Positives = 52/99 (52%), Gaps = 9/99 (9%)
Query: 485 LKITVVEGKDLAAAKEKTGKFDPYIKLQYGKVMQKT---KTSHTPNPVWNQTI----EFD 537
L+++++ GK L + + GK DPY+++QY +K+ K NP WN + EF
Sbjct: 6 LEVSLISGKGLKRS-DFLGKIDPYVEIQYKGQTRKSSVAKEDGGRNPTWNDKLKWRAEFP 64
Query: 538 EVGGGEYLKLKVFTEELF-GDENIGSAQVNLEGLVDGSV 575
G L +KV + F D+ IG A V+++ L++ V
Sbjct: 65 GSGADYKLIVKVMDHDTFSSDDFIGEATVHVKELLEMGV 103
>At4g21160 unknown protein
Length = 337
Score = 43.9 bits (102), Expect = 3e-04
Identities = 38/156 (24%), Positives = 68/156 (43%), Gaps = 42/156 (26%)
Query: 485 LKITVVEGKDLAAAKEKTGKFDPYIKLQYGKVM-QKTKTSHTPNPVWNQTIEFDEVGGGE 543
LK+T+ +G ++A + DPY+ L G+ Q T NPVWN+ +
Sbjct: 183 LKVTIKKGTNMAIRDMMSS--DPYVVLTLGQQKAQSTVVKSNLNPVWNEELMLSVPHNYG 240
Query: 544 YLKLKVFTEELF-GDENIGSAQVNLEGLV------------------------------- 571
+KL+VF + F D+ +G A+++++ L+
Sbjct: 241 SVKLQVFDYDTFSADDIMGEAEIDIQPLITSAMAFGDPEMFGDMQIGKWLKSHDNALIED 300
Query: 572 ------DGSVR-DVWIPLERVRSGEIRLKIEAIKVD 600
DG V+ +V I L+ V SGE+ L++E + ++
Sbjct: 301 SIINIADGKVKQEVQIKLQNVESGELELEMEWLPLE 336
>At3g61720 putative protein
Length = 795
Score = 42.0 bits (97), Expect = 0.001
Identities = 24/66 (36%), Positives = 36/66 (54%), Gaps = 4/66 (6%)
Query: 485 LKITVVEGKDLAAAKEKTGKFDPYIKLQYGKVMQKTKTSHTPNPVWNQTIEF---DEVGG 541
+++T+V G DL + K+K Y+ GKV KTK S NP WNQ + F + + G
Sbjct: 201 VRVTIVSGHDLIS-KDKNKTPSVYVTATLGKVALKTKVSSGTNPSWNQDLIFVASEPLEG 259
Query: 542 GEYLKL 547
Y++L
Sbjct: 260 TVYIRL 265
Score = 39.7 bits (91), Expect = 0.005
Identities = 30/105 (28%), Positives = 49/105 (46%), Gaps = 9/105 (8%)
Query: 484 KLKITVVEGKDLAAAKEKTGKFDPYIKLQYGKVMQKTKT-SHTPNPVWNQTIEFDEVGGG 542
KL+I ++ L + EK D Y+ +YG +T+T ++ +P WN+ +D
Sbjct: 359 KLEIGILGATGLKGSDEKKQTIDSYVVAKYGNKWARTRTVVNSVSPKWNEQYSWD----- 413
Query: 543 EYLKLKVFTEELFGDENIGSAQVNLEGLVDGSVRDVWIPLERVRS 587
Y K V T ++ + I + + G VR IPL RV+S
Sbjct: 414 VYEKCTVLTLGIYDNRQILEDKNKANDVPIGKVR---IPLNRVQS 455
>At1g51570 anthranilate phosphoribosyltransferase, putative
(At1g51570)
Length = 776
Score = 40.8 bits (94), Expect = 0.002
Identities = 33/115 (28%), Positives = 53/115 (45%), Gaps = 10/115 (8%)
Query: 485 LKITVVEGKDLAAAKEKTGKFDPYIKLQYGKVMQKTK-TSHTPNPVWNQTIEF--DEVGG 541
L + VV+ K+L K+ TG DPY++++ G T+ NP WNQ F D V
Sbjct: 41 LYVRVVKAKELPG-KDLTGSCDPYVEVKLGNYRGTTRHFEKKSNPEWNQVFAFSKDRV-Q 98
Query: 542 GEYLKLKVFTEELFGDENIGSAQVNLEGL-----VDGSVRDVWIPLERVRSGEIR 591
YL+ V ++L D+ IG +L + D + W LE + +++
Sbjct: 99 ASYLEATVKDKDLVKDDLIGRVVFDLNEIPKRVPPDSPLAPQWYRLEDGKGQKVK 153
Score = 28.9 bits (63), Expect = 8.9
Identities = 15/55 (27%), Positives = 33/55 (59%), Gaps = 5/55 (9%)
Query: 485 LKITVVEGKDLAAAKEKTGKF-DPYIKLQYGKVMQKTKTSHTP--NPVWNQTIEF 536
L++ V+E +DL + + G++ + ++K+ G +T+ S + NP+WN+ + F
Sbjct: 203 LRVNVIEAQDLIPSDK--GRYPEVFVKVIMGNQALRTRVSQSRSINPMWNEDLMF 255
>At1g73580 hypothetical protein
Length = 168
Score = 40.4 bits (93), Expect = 0.003
Identities = 25/90 (27%), Positives = 44/90 (48%), Gaps = 4/90 (4%)
Query: 485 LKITVVEGKDLAAAKEKTGKFDPYIKLQYGKVMQKTK-TSHTPNPVWNQTIEFDEVGGGE 543
L++ V G +LA + DPY+ L+ G+ KTK NP W + + F
Sbjct: 11 LRVRVQRGVNLAVRDVSSS--DPYVVLKLGRQKLKTKVVKQNVNPQWQEDLSFTVTDPNL 68
Query: 544 YLKLKVFTEELFG-DENIGSAQVNLEGLVD 572
L L V+ + F D+ +G A+++L+ ++
Sbjct: 69 PLTLIVYDHDFFSKDDKMGDAEIDLKPYIE 98
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.137 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,380,693
Number of Sequences: 26719
Number of extensions: 633554
Number of successful extensions: 1771
Number of sequences better than 10.0: 60
Number of HSP's better than 10.0 without gapping: 14
Number of HSP's successfully gapped in prelim test: 46
Number of HSP's that attempted gapping in prelim test: 1696
Number of HSP's gapped (non-prelim): 93
length of query: 620
length of database: 11,318,596
effective HSP length: 105
effective length of query: 515
effective length of database: 8,513,101
effective search space: 4384247015
effective search space used: 4384247015
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 63 (28.9 bits)
Medicago: description of AC146563.9


