

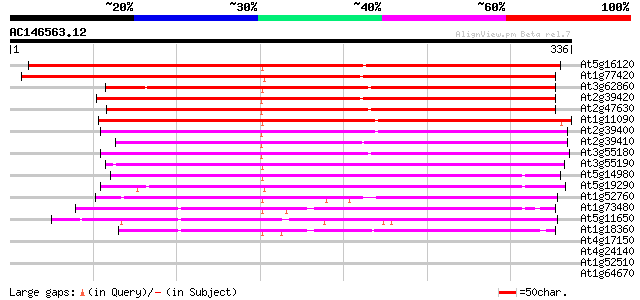
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146563.12 - phase: 0
(336 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g16120 lipase-like protein 424 e-119
At1g77420 lipase like protein 352 2e-97
At3g62860 unknown protein 249 2e-66
At2g39420 phospholipase like protein 246 2e-65
At2g47630 putative phospholipase 241 3e-64
At1g11090 lysophospholipase like protein 236 2e-62
At2g39400 phospholipase like protein 231 3e-61
At2g39410 putative phospholipase 227 6e-60
At3g55180 lipase -like protein 223 9e-59
At3g55190 lipase -like protein 211 3e-55
At5g14980 lysophospholipase -like protein 170 9e-43
At5g19290 phospholipase - like protein 167 1e-41
At1g52760 lipase like protein 167 1e-41
At1g73480 lysophospholipase like protein 123 2e-28
At5g11650 lysophospholipase - like protein 117 1e-26
At1g18360 lysophospholipase homolog, putative (At1g18360) 109 2e-24
At4g17150 unknown protein 38 0.007
At4g24140 unknown protein 37 0.012
At1g52510 unknown protein (At1g52510) 37 0.012
At1g64670 unknown protein 36 0.034
>At5g16120 lipase-like protein
Length = 351
Score = 424 bits (1089), Expect = e-119
Identities = 198/332 (59%), Positives = 264/332 (78%), Gaps = 14/332 (4%)
Query: 12 SEELQNIYLSNMDEAPARRLAREAFKDIQLAIDHCLFQLPADGVKMEEIYEVNSRGLKVF 71
+EEL+ + N+DEAP RR R++ KDIQL +DH LF+ P +G+K +E +EVNSRG+++F
Sbjct: 10 NEELRRLREVNIDEAPGRRRVRDSLKDIQLNLDHILFKTPENGIKTKESFEVNSRGVEIF 69
Query: 72 SKSWIPEKSPMKGIVYYCHGYADTCTFYFEGVARKLASSGFGVFALDYPGFGLSDGLHGY 131
SKSW+PE S + +V +CHGY DTCTF+FEG+AR+LA SG+GVFA+DYPGFGLS+GLHGY
Sbjct: 70 SKSWLPEASKPRALVCFCHGYGDTCTFFFEGIARRLALSGYGVFAMDYPGFGLSEGLHGY 129
Query: 132 IPSFENLVNDVIEHFSKIK-------------GESMGGAIALNIHFKQPTAWDGAALIAP 178
IPSF+ LV DVIEH+S IK G+SMGGA++L IH KQP AW GA L+AP
Sbjct: 130 IPSFDLLVQDVIEHYSNIKANPEFSSLPSFLFGQSMGGAVSLKIHLKQPNAWAGAVLLAP 189
Query: 179 LCKFAEDMIPHWLVKQILIGVAKVLPKTKLVPQKEEVKENIYRDARKRELAPYNVLFYKD 238
+CK A+D++P ++KQILIG+A VLPK KLVPQK ++ E +RD RKR++ PYN++ Y
Sbjct: 190 MCKIADDLVPPPVLKQILIGLANVLPKHKLVPQK-DLAEAGFRDIRKRDMTPYNMICYSG 248
Query: 239 KPRLGTALELLKATQELEQRLEEVSLPLLVMHGEADIITDPSASKALYQKAKVKDKKLCL 298
KPRL TA+E+L+ TQ++E++L+EVSLP+L++HGEAD +TDPS S+ LY+KAK DKK+ L
Sbjct: 249 KPRLRTAVEMLRTTQDIEKQLQEVSLPILILHGEADTVTDPSVSRELYEKAKSPDKKIVL 308
Query: 299 YKDAFHTLLEGEPDETIFHVLDDIISWLDDHS 330
Y++A+H+LLEGEPD+ I VL DIISWL+DHS
Sbjct: 309 YENAYHSLLEGEPDDMILRVLSDIISWLNDHS 340
>At1g77420 lipase like protein
Length = 382
Score = 352 bits (903), Expect = 2e-97
Identities = 167/334 (50%), Positives = 242/334 (72%), Gaps = 15/334 (4%)
Query: 8 IEGMSEELQNIYLSNMDEAPARRLAREAFKDIQLAIDHCLFQLPADGVKMEEIYEVNSRG 67
I+G+S+EL I N+D+APARRLAR AF D+QL +DHCLF+ G++ EE YE NS+G
Sbjct: 44 IDGVSDELNLIASQNLDQAPARRLARSAFVDLQLQLDHCLFKKAPSGIRTEEWYERNSKG 103
Query: 68 LKVFSKSWIPEKSP-MKGIVYYCHGYADTCTFYFEGVARKLASSGFGVFALDYPGFGLSD 126
+F KSW+P+ +K V +CHGY TCTF+F+G+A+++A G+GV+A+D+PGFGLSD
Sbjct: 104 EDIFCKSWLPKSGDEIKAAVCFCHGYGSTCTFFFDGIAKQIAGFGYGVYAIDHPGFGLSD 163
Query: 127 GLHGYIPSFENLVNDVIEHFSKIKG-------------ESMGGAIALNIHFKQPTAWDGA 173
GLHG+IPSF++L ++ IE F+K+KG +SMGGA+AL IH K+P AWDG
Sbjct: 164 GLHGHIPSFDDLADNAIEQFTKMKGRSELRNLPRFLLGQSMGGAVALKIHLKEPQAWDGL 223
Query: 174 ALIAPLCKFAEDMIPHWLVKQILIGVAKVLPKTKLVPQKEEVKENIYRDARKRELAPYNV 233
L+AP+CK +ED+ P LV + LI ++ + PK KL P K ++ + +RD KR+L Y+V
Sbjct: 224 ILVAPMCKISEDVKPPPLVLKTLILMSTLFPKAKLFP-KRDLSDFFFRDLSKRKLCEYDV 282
Query: 234 LFYKDKPRLGTALELLKATQELEQRLEEVSLPLLVMHGEADIITDPSASKALYQKAKVKD 293
+ Y D+ RL TA+ELL AT+++E ++++VSLPLL++HG+ D +TDP+ SK L++ A +D
Sbjct: 283 ICYDDQTRLKTAVELLNATRDIEMQVDKVSLPLLILHGDTDKVTDPTVSKFLHKHAVSQD 342
Query: 294 KKLCLYKDAFHTLLEGEPDETIFHVLDDIISWLD 327
K L LY +H +LEG+ DE IF V++DI++WLD
Sbjct: 343 KTLKLYPGGYHCILEGDTDENIFTVINDIVAWLD 376
>At3g62860 unknown protein
Length = 348
Score = 249 bits (635), Expect = 2e-66
Identities = 124/283 (43%), Positives = 178/283 (62%), Gaps = 15/283 (5%)
Query: 58 EEIYEVNSRGLKVFSKSWIPEKSPMKGIVYYCHGYADTCTFYFEGVARKLASSGFGVFAL 117
EE Y NSR +++F+ W+P SP + +V+ CHGY C+ + +LAS+G+ VF +
Sbjct: 9 EEEYIKNSRDVELFACRWLPSSSP-RALVFLCHGYGMECSSFMRECGIRLASAGYAVFGM 67
Query: 118 DYPGFGLSDGLHGYIPSFENLVNDVIEHFSKIK-------------GESMGGAIALNIHF 164
DY G G S G YI F N+VND ++++ I GESMGGA+AL +H
Sbjct: 68 DYEGHGRSKGARCYIKKFSNIVNDCFDYYTSISAQEEYKEKGRFLYGESMGGAVALLLHK 127
Query: 165 KQPTAWDGAALIAPLCKFAEDMIPHWLVKQILIGVAKVLPKTKLVPQKEEVKENIYRDAR 224
K P+ W+GA L+AP+CK +E + PH +V +L V ++PK K+VP K+ + + ++D
Sbjct: 128 KDPSFWNGALLVAPMCKISEKVKPHPVVINLLTRVEDIIPKWKIVPTKDVI-DAAFKDPV 186
Query: 225 KRELAPYNVLFYKDKPRLGTALELLKATQELEQRLEEVSLPLLVMHGEADIITDPSASKA 284
KRE N L Y+DKPRL TALE+L+ + +LE L E++LP V+HGEADI+TDP SKA
Sbjct: 187 KREEIRNNKLIYQDKPRLKTALEMLRTSMDLEDTLHEITLPFFVLHGEADIVTDPEISKA 246
Query: 285 LYQKAKVKDKKLCLYKDAFHTLLEGEPDETIFHVLDDIISWLD 327
L++KA +DK + LY +H L GEPD + V DI++WLD
Sbjct: 247 LFEKASTRDKTIKLYPGMWHGLTSGEPDANVDLVFADIVNWLD 289
>At2g39420 phospholipase like protein
Length = 317
Score = 246 bits (627), Expect = 2e-65
Identities = 125/288 (43%), Positives = 176/288 (60%), Gaps = 14/288 (4%)
Query: 53 DGVKMEEIYEVNSRGLKVFSKSWIPEKSPMKGIVYYCHGYADTCTFYFEGVARKLASSGF 112
+ +K EE + N+RG+K+F+ W+P K K +V+ CHGYA C+ AR+L +GF
Sbjct: 6 ENIKYEESFIKNTRGMKLFTCKWVPAKQEPKALVFICHGYAMECSITMNSTARRLVKAGF 65
Query: 113 GVFALDYPGFGLSDGLHGYIPSFENLVNDVIEHFSKI-------------KGESMGGAIA 159
V+ +DY G G SDGL Y+P+F++LV+DV H++ I GESMGGA+
Sbjct: 66 AVYGIDYEGHGKSDGLSAYVPNFDHLVDDVSTHYTSICEKEENKGKMRFLLGESMGGAVL 125
Query: 160 LNIHFKQPTAWDGAALIAPLCKFAEDMIPHWLVKQILIGVAKVLPKTKLVPQKEEVKENI 219
L +H K+P WDGA L+AP+CK AE+M P LV IL ++ V+P K++P +++ E
Sbjct: 126 LLLHRKKPQFWDGAVLVAPMCKIAEEMKPSPLVISILAKLSGVIPSWKIIP-GQDIIETA 184
Query: 220 YRDARKRELAPYNVLFYKDKPRLGTALELLKATQELEQRLEEVSLPLLVMHGEADIITDP 279
++ R+ N YK +PRL TA ELL+ + +LE+RL EVSLP +V+HGE D +TD
Sbjct: 185 FKQPEIRKQVRENPYCYKGRPRLKTAYELLRVSTDLEKRLNEVSLPFIVLHGEDDKVTDK 244
Query: 280 SASKALYQKAKVKDKKLCLYKDAFHTLLEGEPDETIFHVLDDIISWLD 327
+ S+ LY+ A DK LY +H LL GE E I V DII WLD
Sbjct: 245 AVSRQLYEVASSSDKTFKLYPGMWHGLLYGETPENIETVFADIIGWLD 292
>At2g47630 putative phospholipase
Length = 351
Score = 241 bits (616), Expect = 3e-64
Identities = 125/283 (44%), Positives = 174/283 (61%), Gaps = 15/283 (5%)
Query: 59 EIYEVNSRGLKVFSKSWIPEKSPM-KGIVYYCHGYADTCTFYFEGVARKLASSGFGVFAL 117
E Y NSRG+++F+ WIP S K +V+ CHGY C+ + +LAS+G+ VF +
Sbjct: 10 EEYVRNSRGVELFACRWIPSSSSSPKALVFLCHGYGMECSDSMKECGIRLASAGYAVFGM 69
Query: 118 DYPGFGLSDGLHGYIPSFENLVNDVIEHFSKI-------------KGESMGGAIALNIHF 164
DY G G S G YI F N+VND ++++ I GESMGGA+ L +H
Sbjct: 70 DYEGHGRSMGSRCYIKKFANVVNDCYDYYTSICAQEEYMDKGRFLYGESMGGAVTLLLHK 129
Query: 165 KQPTAWDGAALIAPLCKFAEDMIPHWLVKQILIGVAKVLPKTKLVPQKEEVKENIYRDAR 224
K P W+GA L+AP+CK +E + PH +V +L V +++PK K+VP K+ + + ++D
Sbjct: 130 KDPLFWNGAILVAPMCKISEKVKPHPIVINLLTRVEEIIPKWKIVPTKDVI-DAAFKDLV 188
Query: 225 KRELAPYNVLFYKDKPRLGTALELLKATQELEQRLEEVSLPLLVMHGEADIITDPSASKA 284
KRE N L Y+DKPRL TALE+L+ + LE L E+++P V+HGEAD +TDP SKA
Sbjct: 189 KREEVRNNKLIYQDKPRLKTALEMLRTSMNLEDTLHEITMPFFVLHGEADTVTDPEVSKA 248
Query: 285 LYQKAKVKDKKLCLYKDAFHTLLEGEPDETIFHVLDDIISWLD 327
LY+KA +DK L LY +H L GEPD + V DII+WLD
Sbjct: 249 LYEKASTRDKTLKLYPGMWHALTSGEPDCNVDLVFADIINWLD 291
>At1g11090 lysophospholipase like protein
Length = 324
Score = 236 bits (601), Expect = 2e-62
Identities = 121/300 (40%), Positives = 185/300 (61%), Gaps = 18/300 (6%)
Query: 54 GVKMEEIYEVNSRGLKVFSKSWIPEKS-PMKGIVYYCHGYADTCTFYFEGVARKLASSGF 112
G+ + + + RGL +F++SW+P S P +G+++ HGY + ++ F+ LA GF
Sbjct: 26 GIIGSKSFFTSPRGLNLFTRSWLPSSSSPPRGLIFMVHGYGNDVSWTFQSTPIFLAQMGF 85
Query: 113 GVFALDYPGFGLSDGLHGYIPSFENLVNDVIEHFSKIK-------------GESMGGAIA 159
FALD G G SDG+ Y+PS + +V+D+I F+ IK GESMGGAI
Sbjct: 86 ACFALDIEGHGRSDGVRAYVPSVDLVVDDIISFFNSIKQNPKFQGLPRFLFGESMGGAIC 145
Query: 160 LNIHFKQPTAWDGAALIAPLCKFAEDMIPHWLVKQILIGVAKVLPKTKLVPQKEEVKENI 219
L I F P +DGA L+AP+CK ++ + P W V Q LI +++ LP +VP ++ ++++I
Sbjct: 146 LLIQFADPLGFDGAVLVAPMCKISDKVRPKWPVDQFLIMISRFLPTWAIVPTEDLLEKSI 205
Query: 220 YRDARKRELAPYNVLFYKDKPRLGTALELLKATQELEQRLEEVSLPLLVMHGEADIITDP 279
+ K+ +A N + Y +KPRLGT +ELL+ T L ++L++VS+P +++HG AD +TDP
Sbjct: 206 -KVEEKKPIAKRNPMRYNEKPRLGTVMELLRVTDYLGKKLKDVSIPFIIVHGSADAVTDP 264
Query: 280 SASKALYQKAKVKDKKLCLYKDAFHTLLEGEPDETIFHVLDDIISWLDDH---SSTKNKV 336
S+ LY+ AK KDK L +Y H++L GEPD+ I V DI+SWL+D TK +V
Sbjct: 265 EVSRELYEHAKSKDKTLKIYDGMMHSMLFGEPDDNIEIVRKDIVSWLNDRCGGDKTKTQV 324
>At2g39400 phospholipase like protein
Length = 311
Score = 231 bits (590), Expect = 3e-61
Identities = 123/293 (41%), Positives = 177/293 (59%), Gaps = 14/293 (4%)
Query: 55 VKMEEIYEVNSRGLKVFSKSWIPEKSPMKGIVYYCHGYADTCTFYFEGVARKLASSGFGV 114
V EE + +NSRG+K+F+ W P K K +++ CHGYA + A +LA++GF V
Sbjct: 2 VMYEEDFVLNSRGMKLFTCVWKPVKQEPKALLFLCHGYAMESSITMNSAATRLANAGFAV 61
Query: 115 FALDYPGFGLSDGLHGYIPSFENLVNDVIEHFSKI-------------KGESMGGAIALN 161
+ +DY G G S+GL+GYI +F++LV+DV H+S I GESMGGA+ L
Sbjct: 62 YGMDYEGHGKSEGLNGYISNFDDLVDDVSNHYSTICEREENKGKMRFLLGESMGGAVVLL 121
Query: 162 IHFKQPTAWDGAALIAPLCKFAEDMIPHWLVKQILIGVAKVLPKTKLVPQKEEVKENIYR 221
+ K+P WDGA L+AP+CK A+++ PH +V ILI +AK +P K+VP + + I +
Sbjct: 122 LARKKPDFWDGAVLVAPMCKLADEIKPHPVVISILIKLAKFIPTWKIVPGNDIIDIAI-K 180
Query: 222 DARKRELAPYNVLFYKDKPRLGTALELLKATQELEQRLEEVSLPLLVMHGEADIITDPSA 281
+ R N YK +PRL TA +LL + +LE+ L +VS+P +V+HGE D +TD S
Sbjct: 181 EPHIRNQVRENKYCYKGRPRLNTAYQLLLVSLDLEKNLHQVSIPFIVLHGEDDKVTDKSI 240
Query: 282 SKALYQKAKVKDKKLCLYKDAFHTLLEGEPDETIFHVLDDIISWLDDHSSTKN 334
SK LY+ A DK LY +H LL GE +E V DII+WL+D ++ N
Sbjct: 241 SKMLYEVASSSDKTFKLYPKMWHALLYGETNENSEIVFGDIINWLEDRATDSN 293
>At2g39410 putative phospholipase
Length = 311
Score = 227 bits (579), Expect = 6e-60
Identities = 120/284 (42%), Positives = 168/284 (58%), Gaps = 14/284 (4%)
Query: 64 NSRGLKVFSKSWIPEKSPMKGIVYYCHGYADTCTFYFEGVARKLASSGFGVFALDYPGFG 123
N+RG K+F+ W+P + +V+ CHGY C+ AR+L +GF V+ +DY G G
Sbjct: 11 NTRGFKLFTCRWLPTNREPRALVFLCHGYGMECSITMNSTARRLVKAGFAVYGMDYEGHG 70
Query: 124 LSDGLHGYIPSFENLVNDVIEHFSKI-------------KGESMGGAIALNIHFKQPTAW 170
SDGL YI +F+ LV+DV H++ I GESMGGA+ L + K P W
Sbjct: 71 KSDGLSAYISNFDRLVDDVSTHYTAICEREENKWKMRFMLGESMGGAVVLLLGRKNPDFW 130
Query: 171 DGAALIAPLCKFAEDMIPHWLVKQILIGVAKVLPKTKLVPQKEEVKENIYRDARKRELAP 230
DGA L+AP+CK AE+M P V IL + ++PK K++P +++ E Y++ R+
Sbjct: 131 DGAILVAPMCKIAEEMKPSPFVISILTKLISIIPKWKIIPS-QDIIEISYKEPEIRKQVR 189
Query: 231 YNVLFYKDKPRLGTALELLKATQELEQRLEEVSLPLLVMHGEADIITDPSASKALYQKAK 290
N L K +PRL TA ELL+ + +LE+RL+EVSLP LV+HG+ D +TD + S+ LY+ A
Sbjct: 190 ENPLCSKGRPRLKTAYELLRISNDLEKRLQEVSLPFLVLHGDDDKVTDKAVSQELYKVAL 249
Query: 291 VKDKKLCLYKDAFHTLLEGEPDETIFHVLDDIISWLDDHSSTKN 334
DK L LY +H LL GE E I V D+ISWL+ S N
Sbjct: 250 SADKTLKLYPGMWHGLLTGETPENIEIVFADVISWLEKRSDYGN 293
>At3g55180 lipase -like protein
Length = 312
Score = 223 bits (569), Expect = 9e-59
Identities = 118/295 (40%), Positives = 175/295 (59%), Gaps = 15/295 (5%)
Query: 55 VKMEEIYEVNSRGLKVFSKSWIPEKSPM-KGIVYYCHGYADTCTFYFEGVARKLASSGFG 113
V +E Y NSRG+++F+ SW E+ K +++ CHGYA + A +LA++GF
Sbjct: 2 VMYKEDYVSNSRGIQLFTCSWKQEEQQEPKALIFLCHGYAMESSITMSSTAVRLANAGFS 61
Query: 114 VFALDYPGFGLSDGLHGYIPSFENLVNDVIEHFSKI-------------KGESMGGAIAL 160
V+ +DY G G S GL+GY+ F++LV DV H+S I GESMGGA+ L
Sbjct: 62 VYGMDYEGHGKSGGLNGYVKKFDDLVQDVSSHYSSICELEENKGKMRFLMGESMGGAVVL 121
Query: 161 NIHFKQPTAWDGAALIAPLCKFAEDMIPHWLVKQILIGVAKVLPKTKLVPQKEEVKENIY 220
+ K+P WDGA L+AP+CK AED+ PH +V L + + +P K+VP + + + +
Sbjct: 122 LLERKKPNFWDGAVLVAPMCKLAEDIKPHPMVISFLTKLTRFIPTWKIVPSNDII-DVAF 180
Query: 221 RDARKRELAPYNVLFYKDKPRLGTALELLKATQELEQRLEEVSLPLLVMHGEADIITDPS 280
++ R+ N YK +PRL TA +LL + +LE+ L++VS+P +V+HGE D +TD +
Sbjct: 181 KETHIRKQVRDNEYCYKGRPRLKTAHQLLMVSLDLEKNLDQVSMPFIVLHGEDDKVTDKN 240
Query: 281 ASKALYQKAKVKDKKLCLYKDAFHTLLEGEPDETIFHVLDDIISWLDDHSSTKNK 335
SK LY+ A DK LY + +H LL GE E + V DIISWL + +S N+
Sbjct: 241 VSKLLYEVASSSDKTFKLYPNMWHGLLYGESPENLEIVFSDIISWLKERASVTNQ 295
>At3g55190 lipase -like protein
Length = 319
Score = 211 bits (538), Expect = 3e-55
Identities = 112/288 (38%), Positives = 162/288 (55%), Gaps = 14/288 (4%)
Query: 58 EEIYEVNSRGLKVFSKSWIPEKSPMKGIVYYCHGYADTCTFYFEGVARKLASSGFGVFAL 117
EE E NSRG+++ + W P + ++++CHGYA C+ F+ +A K A GF V +
Sbjct: 12 EEFIE-NSRGMQLLTCKWFPVNQEPRALIFFCHGYAIDCSTTFKDIAPKFAKEGFAVHGI 70
Query: 118 DYPGFGLSDGLHGYIPSFENLVNDVIEHFSKIK-------------GESMGGAIALNIHF 164
+Y G G S GL YI +F+ L++DV HFSKI GESMGGA+ L +H
Sbjct: 71 EYEGHGRSSGLSVYIDNFDLLIDDVSSHFSKISEMGDNTKKKRFLMGESMGGAVVLLLHR 130
Query: 165 KQPTAWDGAALIAPLCKFAEDMIPHWLVKQILIGVAKVLPKTKLVPQKEEVKENIYRDAR 224
K+P WDG LIAP+CK AE+M P +V ++ V ++P K + ++ + +
Sbjct: 131 KKPEFWDGGILIAPMCKIAEEMKPSRMVISMINMVTNLIPSWKSIIHGPDILNSAIKLPE 190
Query: 225 KRELAPYNVLFYKDKPRLGTALELLKATQELEQRLEEVSLPLLVMHGEADIITDPSASKA 284
KR N Y PR+ T EL + + +LE RL EV++P +V+HGE D +TD SK
Sbjct: 191 KRHEIRTNPNCYNGWPRMKTMSELFRISLDLENRLNEVTMPFIVLHGEDDKVTDKGGSKL 250
Query: 285 LYQKAKVKDKKLCLYKDAFHTLLEGEPDETIFHVLDDIISWLDDHSST 332
LY+ A DK L LY + +H+LL GEP E V +DI+ W+ +T
Sbjct: 251 LYEVALSNDKTLKLYPEMWHSLLFGEPPENSEIVFNDIVQWMQTRITT 298
>At5g14980 lysophospholipase -like protein
Length = 327
Score = 170 bits (431), Expect = 9e-43
Identities = 88/283 (31%), Positives = 151/283 (53%), Gaps = 14/283 (4%)
Query: 61 YEVNSRGLKVFSKSWIPEKSPMKGIVYYCHGYADTCTFYFEGVARKLASSGFGVFALDYP 120
Y N GLK+F++ W P P G++ HG+ +++ + + A SG+ A+D+
Sbjct: 35 YVTNPTGLKLFTQWWTPLNRPPLGLIAVVHGFTGESSWFLQLTSVLFAKSGYLTCAIDHQ 94
Query: 121 GFGLSDGLHGYIPSFENLVNDVIEHFSKIK-------------GESMGGAIALNIHFKQP 167
G G SDGL +IP+ +V+D I F + ES+GGAIAL I +Q
Sbjct: 95 GHGFSDGLTAHIPNINLIVDDCISFFDDFRKRHASSFLPAFLYSESLGGAIALYITLRQK 154
Query: 168 TAWDGAALIAPLCKFAEDMIPHWLVKQILIGVAKVLPKTKLVPQKEEVKENIYRDARKRE 227
W+G L +C + P W ++ +L A ++P ++VP + + +++ KR+
Sbjct: 155 HQWNGLILSGAMCSISHKFKPPWPLQHLLTLAATLIPTWRVVPTRGSIAGVSFKEPWKRK 214
Query: 228 LAPYNVLFYKDKPRLGTALELLKATQELEQRLEEVSLPLLVMHGEADIITDPSASKALYQ 287
LA N KPR TA EL++ ++L+ R EEV +PL+++HG D++ DP++ + LY+
Sbjct: 215 LAYANPNRTVGKPRAATAYELVRVCEDLQNRFEEVEVPLMIVHGRDDVVCDPASVEELYR 274
Query: 288 KAKVKDKKLCLYKDAFHTLLEGEPDETIFHVLDDIISWLDDHS 330
+ +DK + +Y +H L+ GE +E + V D++ W+ S
Sbjct: 275 RCSSRDKTIKIYPGMWHQLI-GESEENVDLVFGDVLDWIKTRS 316
>At5g19290 phospholipase - like protein
Length = 330
Score = 167 bits (422), Expect = 1e-41
Identities = 92/294 (31%), Positives = 159/294 (53%), Gaps = 17/294 (5%)
Query: 55 VKMEEIYEVNSRGLKVFSKSW--IPEKSPMKGIVYYCHGYADTCTFYFEGVARKLASSGF 112
V + N RGLK+F++ W +P P+ GI+ HG+ +++ + + A SGF
Sbjct: 29 VSHSSAFITNPRGLKLFTQWWSPLPPTKPI-GIIAVVHGFTGESSWFLQLTSILFAKSGF 87
Query: 113 GVFALDYPGFGLSDGLHGYIPSFENLVNDVIEHFSKIKG-------------ESMGGAIA 159
A+D+ G G SDGL +IP +V+D I F + ES+GGAIA
Sbjct: 88 ITCAIDHQGHGFSDGLIAHIPDINPVVDDCISFFDDFRSRQTPSDLPCFLYSESLGGAIA 147
Query: 160 LNIHFKQPTAWDGAALIAPLCKFAEDMIPHWLVKQILIGVAKVLPKTKLVPQKEEVKENI 219
L I +Q WDG L +C ++ P W ++ +L VA ++P +++P + + +
Sbjct: 148 LYISLRQRGVWDGLILNGAMCGISDKFKPPWPLEHLLFVVANLIPTWRVIPTRGSIPDVS 207
Query: 220 YRDARKRELAPYNVLFYKDKPRLGTALELLKATQELEQRLEEVSLPLLVMHGEADIITDP 279
+++ KR+LA + KPR TA EL++ ++L+ R EEV +PLL++HG D++ D
Sbjct: 208 FKEPWKRKLAMASPRRTVAKPRAATAYELIRVCKDLQGRFEEVEVPLLIVHGGGDVVCDV 267
Query: 280 SASKALYQKAKVKDKKLCLYKDAFHTLLEGEPDETIFHVLDDIISWLDDHSSTK 333
+ + L+++A +DK + +Y + +H ++ GE +E + V D++SWL + K
Sbjct: 268 ACVEELHRRAISEDKTIKIYPELWHQMI-GESEEKVDLVYGDMLSWLKSRAERK 320
>At1g52760 lipase like protein
Length = 332
Score = 167 bits (422), Expect = 1e-41
Identities = 99/297 (33%), Positives = 155/297 (51%), Gaps = 28/297 (9%)
Query: 52 ADGVKMEEIYEVNSRGLKVFSKSWIPEKSPMKGIVYYCHGYADTCTFYFEGVARKLASSG 111
+ GV+ + Y G K+F++S++P +KG VY HGY ++ F+ + +S G
Sbjct: 34 SQGVRNSKSYFETPNG-KLFTQSFLPLDGEIKGTVYMSHGYGSDTSWMFQKICMSFSSWG 92
Query: 112 FGVFALDYPGFGLSDGLHGYIPSFENLVNDVIEHFSKIK-------------GESMGGAI 158
+ VFA D G G SDG+ Y+ E + + F ++ GESMGG +
Sbjct: 93 YAVFAADLLGHGRSDGIRCYMGDMEKVAATSLAFFKHVRCSDPYKDLPAFLFGESMGGLV 152
Query: 159 ALNIHFK-QPTAWDGAALIAPLCKFAEDMIP---HWLVKQILIGVAKV---LPKTKLVPQ 211
L ++F+ +P W G APL EDM P H +L G+A +P K+V +
Sbjct: 153 TLLMYFQSEPETWTGLMFSAPLFVIPEDMKPSKAHLFAYGLLFGLADTWAAMPDNKMVGK 212
Query: 212 KEEVKENIYRDARKRELAPYNVLFYKDKPRLGTALELLKATQELEQRLEEVSLPLLVMHG 271
+D K ++ N Y KPR+GT ELL+ TQ +++ +V++P+ HG
Sbjct: 213 A-------IKDPEKLKIIASNPQRYTGKPRVGTMRELLRKTQYVQENFGKVTIPVFTAHG 265
Query: 272 EADIITDPSASKALYQKAKVKDKKLCLYKDAFHTLLEGEPDETIFHVLDDIISWLDD 328
AD +T P++SK LY+KA DK L +Y+ +H+L++GEPDE VL D+ W+D+
Sbjct: 266 TADGVTCPTSSKLLYEKASSADKTLKIYEGMYHSLIQGEPDENAEIVLKDMREWIDE 322
>At1g73480 lysophospholipase like protein
Length = 463
Score = 123 bits (308), Expect = 2e-28
Identities = 95/302 (31%), Positives = 140/302 (45%), Gaps = 22/302 (7%)
Query: 40 QLAIDHCLFQLPADGVKMEEI-YEVNSRGLKVFSKSWIPEKSPMKGIVYYCHGYADTCTF 98
+LAI L DG + + RG +FS+SW P +G++ HG +
Sbjct: 167 ELAIKRVLEDEGGDGSSVRDYSLFTTKRGDTLFSQSWSPLSPNHRGLIVLLHGLNEHSGR 226
Query: 99 YFEGVARKLASSGFGVFALDYPGFGLSDGLHGYIPSFENLVNDVIEHFSKIK-------- 150
Y + A++L ++GF V+ +D+ G G SDGLH Y+PS + V D+ K+
Sbjct: 227 YSD-FAKQLNANGFKVYGIDWIGHGGSDGLHAYVPSLDYAVTDLKSFLEKVFTENPGLPC 285
Query: 151 ---GESMGGAIALNIHF--KQPTAWDGAALIAPLCKFAEDMIPHWLVKQILIGVAKVLPK 205
G S GGAI L K + G AL +P A + P + +L + L
Sbjct: 286 FCFGHSTGGAIILKAMLDPKIESRVSGIALTSP----AVGVQPSHPIFAVLAPIMAFLLP 341
Query: 206 TKLVPQKEEVKENIYRDARKRELAPYNVLFYKDKPRLGTALELLKATQELEQRLEEVSLP 265
+ + + RD + L + R+ T E+L+ T L+Q L +V +P
Sbjct: 342 RYQISAANKKGMPVSRDPAALIAKYSDPLVFTGSIRVKTGYEILRITAHLQQNLNKVKVP 401
Query: 266 LLVMHGEADIITDPSASKALYQKAKVKDKKLCLYKDAFHTLLEGEPDETIFHVLDDIISW 325
LVMHG D +TDPSASK LY++A DK L LY H LL EP+ I + I+ W
Sbjct: 402 FLVMHGTDDTVTDPSASKKLYEEAASSDKSLKLYDGLLHDLL-FEPEREI--IAGAILDW 458
Query: 326 LD 327
L+
Sbjct: 459 LN 460
>At5g11650 lysophospholipase - like protein
Length = 390
Score = 117 bits (292), Expect = 1e-26
Identities = 102/323 (31%), Positives = 157/323 (48%), Gaps = 26/323 (8%)
Query: 26 APARRLAREAFKDIQLAIDHCLFQLPADGVKMEEIYEVNS------RGLKVFSKSWIPEK 79
AP+RR R+ ++ D + A+GV+M E++ RG +FS+SW+P
Sbjct: 65 APSRRWRRKMAWKLEEE-DTARRRSLAEGVEMAGDGEISCSLFYGRRGNALFSRSWLPIS 123
Query: 80 SPMKGIVYYCHGYADTCTFYFEGVARKLASSGFGVFALDYPGFGLSDGLHGYIPSFENLV 139
++GI+ HG + Y + A++L +S GV+A+D+ G G SDGLHGY+PS + +V
Sbjct: 124 GELRGILIIIHGLNEHSGRYSQ-FAKQLNASNLGVYAMDWIGHGGSDGLHGYVPSLDYVV 182
Query: 140 NDVIEHFSKIKGESMGGAIALNIHFKQPTAWDGAALIAPLCKFAEDMI-------PHWLV 192
+D KI+ E+ G L H + L A EDM+ P V
Sbjct: 183 SDTEAFLEKIRSENPGVPCFLFGH----STGGAVVLKAASSPSIEDMLAGIVLTSPALRV 238
Query: 193 KQILIGVAKVLPKTKLVPQKEEVKENIYRD---ARKRE--LAPY-NVLFYKDKPRLGTAL 246
K V + P L+ + + K R +R E LA Y + L Y R+ T
Sbjct: 239 KPAHPIVGAIAPIFSLLAPRFQFKGANKRGIPVSRDPEALLAKYSDPLVYTGPIRVRTGY 298
Query: 247 ELLKATQELEQRLEEVSLPLLVMHGEADIITDPSASKALYQKAKVKDKKLCLYKDAFHTL 306
E+L+ T L + + V++P V+HG D +TDP AS+ LY +A K + LY H L
Sbjct: 299 EILRITAYLTRNFKSVTVPFFVLHGTEDKVTDPLASQDLYNQAPSVFKDIKLYDGFLHDL 358
Query: 307 L-EGEPDETIFHVLDDIISWLDD 328
L E E +E ++D +++ LDD
Sbjct: 359 LFEPEREEVGRDIIDWMMNRLDD 381
>At1g18360 lysophospholipase homolog, putative (At1g18360)
Length = 382
Score = 109 bits (272), Expect = 2e-24
Identities = 85/277 (30%), Positives = 133/277 (47%), Gaps = 24/277 (8%)
Query: 66 RGLKVFSKSWIP-EKSPMKGIVYYCHGYADTCTFYFEGVARKLASSGFGVFALDYPGFGL 124
RG +F++SW P + + +G+V HG + Y + A++L +GF V+ +D+ G G
Sbjct: 112 RGDTLFTQSWTPVDSAKNRGLVVLLHGLNEHSGRYSD-FAKQLNVNGFKVYGIDWIGHGG 170
Query: 125 SDGLHGYIPSFENLVNDVIEHFSKIK-----------GESMGGAIALN--IHFKQPTAWD 171
SDGLH Y+PS + V D+ K+ G S GGAI L + K
Sbjct: 171 SDGLHAYVPSLDYAVADLKSFIEKVIAENPGLPCFCIGHSTGGAIILKAMLDAKIEARVS 230
Query: 172 GAALIAPLCKFAEDMIPHWLVKQILIG-VAKVLPKTKLVPQKEEVKENIYRDARKRELAP 230
G L +P A + P + + ++ ++ ++P+ +L K+++ + RD
Sbjct: 231 GIVLTSP----AVGVQPTYPIFGVIAPFLSFLIPRYQLSAAKKKIMP-VSRDPEALLAKY 285
Query: 231 YNVLFYKDKPRLGTALELLKATQELEQRLEEVSLPLLVMHGEADIITDPSASKALYQKAK 290
+ L Y R T E+L+ L Q L + +P LVMHG AD +TDP ++ LY +A
Sbjct: 286 SDPLVYTGFIRARTGNEILRLGAHLLQNLNRIKVPFLVMHGTADTVTDPKGTQKLYNEAS 345
Query: 291 VKDKKLCLYKDAFHTLLEGEPDETIFHVLDDIISWLD 327
DK + LY H LL ETI V I+ WL+
Sbjct: 346 SSDKSIKLYDGLLHDLLFEPERETIAGV---ILDWLN 379
>At4g17150 unknown protein
Length = 502
Score = 38.1 bits (87), Expect = 0.007
Identities = 26/84 (30%), Positives = 39/84 (45%), Gaps = 6/84 (7%)
Query: 48 FQLPADGVKMEEIYEVNSRGLKVFSKSWIP----EKSPMKGIVYYCHGYADTCTFYFEGV 103
F L K +++ NSRG + ++P E +P+ ++Y CHG + C
Sbjct: 37 FSLGGTKCKRQDLELTNSRGHTLRCSHYVPSSSREDTPLPCVIY-CHGNSG-CRADANEA 94
Query: 104 ARKLASSGFGVFALDYPGFGLSDG 127
L S VF LD+ G GLS+G
Sbjct: 95 VMVLLPSNITVFTLDFSGSGLSEG 118
>At4g24140 unknown protein
Length = 498
Score = 37.4 bits (85), Expect = 0.012
Identities = 33/119 (27%), Positives = 53/119 (43%), Gaps = 10/119 (8%)
Query: 70 VFSKSWIPEKSPMKGIVYYCHGYADTCTFYFEGVARKL--ASSGFGVFALDYPGFGLS-- 125
+F K+ IP K V + HG+ + F+ E V L +SS +FA+D GFG S
Sbjct: 204 LFVKTQIPNGVTAKEDVLFIHGFISSSAFWTETVFPSLSASSSTHRLFAVDLLGFGKSPK 263
Query: 126 --DGLHGYIPSFENLVNDVIEHFS----KIKGESMGGAIALNIHFKQPTAWDGAALIAP 178
D L+ E + V+ ++ I S+G +AL++ + L+AP
Sbjct: 264 PADSLYTLREHVEMIEKSVLHKYNVKSFHIVAHSLGCILALSLAARHGGLIKSLTLLAP 322
>At1g52510 unknown protein (At1g52510)
Length = 380
Score = 37.4 bits (85), Expect = 0.012
Identities = 19/48 (39%), Positives = 28/48 (57%), Gaps = 1/48 (2%)
Query: 79 KSPMKGIVYYCHGYADTCTFYFEGVARKLASSGFGVFALDYPGFGLSD 126
K +G + + HG A T +F + V +L+ +GF FA D+ GFG SD
Sbjct: 121 KESRRGTIVFVHG-APTQSFSYRTVMSELSDAGFHCFAPDWIGFGFSD 167
>At1g64670 unknown protein
Length = 469
Score = 35.8 bits (81), Expect = 0.034
Identities = 32/127 (25%), Positives = 52/127 (40%), Gaps = 11/127 (8%)
Query: 63 VNSRGLKVFSKSWIPEKSPMKGIVYYCHGYADTCTFYFEGVARKL---ASSGFGVFALDY 119
++S +F P + + V + HG+ + TF+ E + A S + A+D
Sbjct: 162 LSSSNQSLFVNVQQPTDNKAQENVVFIHGFLSSSTFWTETLFPNFSDSAKSNYRFLAVDL 221
Query: 120 PGFGLS----DGLHGYIPSFENLVNDVIEHFS----KIKGESMGGAIALNIHFKQPTAWD 171
G+G S D L+ E + VI F + S+G +AL + K P A
Sbjct: 222 LGYGKSPKPNDSLYTLKEHLEMIERSVISQFRLKTFHLVAHSLGCILALALAVKHPGAIK 281
Query: 172 GAALIAP 178
L+AP
Sbjct: 282 SLTLLAP 288
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.138 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,730,463
Number of Sequences: 26719
Number of extensions: 339745
Number of successful extensions: 935
Number of sequences better than 10.0: 50
Number of HSP's better than 10.0 without gapping: 28
Number of HSP's successfully gapped in prelim test: 22
Number of HSP's that attempted gapping in prelim test: 866
Number of HSP's gapped (non-prelim): 52
length of query: 336
length of database: 11,318,596
effective HSP length: 100
effective length of query: 236
effective length of database: 8,646,696
effective search space: 2040620256
effective search space used: 2040620256
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC146563.12


