

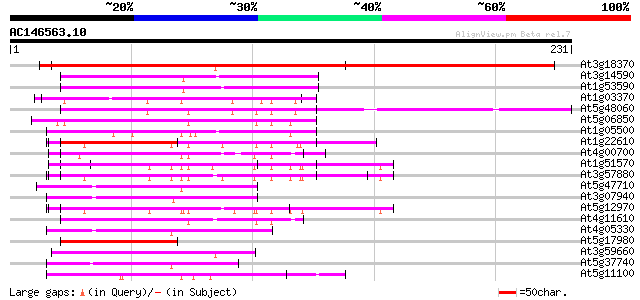
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146563.10 + phase: 0
(231 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g18370 unknown protein 280 5e-76
At3g14590 hypothetical protein 78 3e-15
At1g53590 Unknown protein (At1g53590) 74 7e-14
At1g03370 unknown protein 64 9e-11
At5g48060 phosphoribosylanthranilate transferase-like protein 61 6e-10
At5g06850 anthranilate phosphoribosyltransferase-like protein 59 3e-09
At1g05500 Ca2+-dependent lipid-binding protein, putative 57 6e-09
At1g22610 Highly similar to phosphoribosylanthranilate transferase 56 2e-08
At4g00700 putative phosphoribosylanthranilate transferase 55 2e-08
At1g51570 anthranilate phosphoribosyltransferase, putative (At1g... 55 4e-08
At3g57880 anthranilate phosphoribosyltransferase-like protein 53 1e-07
At5g47710 unknown protein 53 2e-07
At3g07940 putative GTPase activating protein 51 6e-07
At5g12970 anthranilate phosphoribosyltransferase -like protein 50 1e-06
At4g11610 phosphoribosylanthranilate transferase like protein 50 1e-06
At4g05330 unknown protein 50 1e-06
At5g17980 phosphoribosylanthranilate transferase-like protein 50 1e-06
At3g59660 unknown protein 50 1e-06
At5g37740 unknown protein 49 2e-06
At5g11100 CLB1 - like protein 49 2e-06
>At3g18370 unknown protein
Length = 815
Score = 280 bits (716), Expect = 5e-76
Identities = 138/212 (65%), Positives = 171/212 (80%)
Query: 13 SGNGWIELVLIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQTLEFPDD 72
S G IELVL+E RDLVAAD+RGTSDPYVRV YG K+RTKVIYKTL P+WNQT+EFPDD
Sbjct: 602 SSKGLIELVLVEARDLVAADIRGTSDPYVRVQYGEKKQRTKVIYKTLQPKWNQTMEFPDD 661
Query: 73 GSPLMLYVKDHNALLPTSSIGECVVEYQRLPPNQMADKWIPLQGVKRGEIHIQITRKVPE 132
GS L L+VKD+N LLPTSSIG CVVEYQ L PN+ ADKWI LQGVK GE+H+++TRKV E
Sbjct: 662 GSSLELHVKDYNTLLPTSSIGNCVVEYQGLKPNETADKWIILQGVKHGEVHVRVTRKVTE 721
Query: 133 MQKRQSMDSEPSLSKLHQIPTQIKQMMIKFRSQIEDGNLEGLSTTLSELETLEDTQEGYV 192
+Q+R S +K + Q+KQ+MIKF++ I+DG+LEGL+ L ELE+LED QE Y+
Sbjct: 722 IQRRASAGPGTPFNKALLLSNQMKQVMIKFQNLIDDGDLEGLAEALEELESLEDEQEQYL 781
Query: 193 AQLETEQMLLLSKIKELGQEIINSSPSPSLSR 224
QL+TEQ LL++KIK+LG+EI+NSSP+ + SR
Sbjct: 782 LQLQTEQSLLINKIKDLGKEILNSSPAQAPSR 813
Score = 57.4 bits (137), Expect = 6e-09
Identities = 37/122 (30%), Positives = 61/122 (49%), Gaps = 3/122 (2%)
Query: 18 IELVLIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQTLEFPDDGSPLM 77
I + ++ G++LV+ D G D V++ YG ++TK++ WNQ EF +
Sbjct: 483 IIVTVLAGKNLVSKDKSGKCDASVKLQYGKIIQKTKIV-NAAECVWNQKFEFEELAGEEY 541
Query: 78 LYVKDH-NALLPTSSIGECVVEYQRLPPNQMADKWIPLQGVKRGEIHIQITRKVPEMQKR 136
L VK + +L T +IG + Q + ++M W+PL+ V GEI + I PE +
Sbjct: 542 LKVKCYREEMLGTDNIGTATLSLQGINNSEM-HIWVPLEDVNSGEIELLIEALDPEYSEA 600
Query: 137 QS 138
S
Sbjct: 601 DS 602
>At3g14590 hypothetical protein
Length = 705
Score = 78.2 bits (191), Expect = 3e-15
Identities = 37/110 (33%), Positives = 62/110 (55%), Gaps = 5/110 (4%)
Query: 22 LIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQTLEFP----DDGSPLM 77
++E D+ +DL G +DPYV+ G ++ +TK+++KTL P+W + + P D + L
Sbjct: 257 VVEACDVKPSDLNGLADPYVKGQLGAYRFKTKILWKTLAPKWQEEFKIPICTWDSANILN 316
Query: 78 LYVKDHNALLPTSSIGECVVEYQRLPPNQMADKWIPLQGVKRGEIHIQIT 127
+ V+D + S+G+C V Q D W+PLQ +K G +H+ IT
Sbjct: 317 IEVQDKDR-FSDDSLGDCSVNIAEFRGGQRNDMWLPLQNIKMGRLHLAIT 365
>At1g53590 Unknown protein (At1g53590)
Length = 751
Score = 73.9 bits (180), Expect = 7e-14
Identities = 38/110 (34%), Positives = 61/110 (54%), Gaps = 5/110 (4%)
Query: 22 LIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQTLEFP----DDGSPLM 77
+ E DL +DL G +DPYV+ G ++ +TK+ KTL+P+W++ + P D S L
Sbjct: 288 VFEASDLKPSDLNGLADPYVKGKLGAYRFKTKIQKKTLSPKWHEEFKIPIFTWDSPSILN 347
Query: 78 LYVKDHNALLPTSSIGECVVEYQRLPPNQMADKWIPLQGVKRGEIHIQIT 127
+ V D + + ++GEC V + Q D W+ LQ +K G +H+ IT
Sbjct: 348 IEVGDKDRFV-DDTLGECSVNIEEFRGGQRNDMWLSLQNIKMGRLHLAIT 396
>At1g03370 unknown protein
Length = 1859
Score = 63.5 bits (153), Expect = 9e-11
Identities = 46/123 (37%), Positives = 65/123 (52%), Gaps = 11/123 (8%)
Query: 14 GNGWIELV-LIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVI-YKTLTPQWNQTLEF-- 69
G+GW+ V LIEG DL A D G DPY+ V N K RT I ++ PQWN+ EF
Sbjct: 1358 GDGWLLTVALIEGVDLAAVDPSGHCDPYI-VFTSNGKTRTSSIKFQKSNPQWNEIFEFDA 1416
Query: 70 -PDDGSPLMLYVKDHNALLPTS-SIGECVVEYQRLPPNQMADKWIPLQG----VKRGEIH 123
D S L + V D + + S+G V + R + +AD W+PLQG + ++H
Sbjct: 1417 MADPPSVLNVEVFDFDGPFDEAVSLGHAEVNFVRSNISDLADVWVPLQGKLAQACQSKLH 1476
Query: 124 IQI 126
++I
Sbjct: 1477 LRI 1479
Score = 63.2 bits (152), Expect = 1e-10
Identities = 39/115 (33%), Positives = 58/115 (49%), Gaps = 5/115 (4%)
Query: 11 SGSGNGWIELVLIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQTLEF- 69
SG G +++ ++E R+L A DL G SDPYVR+ G + RTKV+ K L P+W + F
Sbjct: 822 SGVGEMKLQVRVVEARNLPAMDLNGFSDPYVRLQLGKQRSRTKVVKKNLNPKWTEDFSFG 881
Query: 70 -PDDGSPLMLYVKDHNALLPTSSIGECVVEYQRL--PPNQ-MADKWIPLQGVKRG 120
D L++ V D + +G+ V + NQ + W PL K+G
Sbjct: 882 VDDLNDELVVSVLDEDKYFNDDFVGQVRVSVSLVFDAENQSLGTVWYPLNPKKKG 936
>At5g48060 phosphoribosylanthranilate transferase-like protein
Length = 1036
Score = 60.8 bits (146), Expect = 6e-10
Identities = 40/120 (33%), Positives = 61/120 (50%), Gaps = 15/120 (12%)
Query: 22 LIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVI-YKTLTPQWNQTLEFPDD---GSPLM 77
+++ ++L + G DPYV V GN+K RTK+ KT P+WNQ F + S L
Sbjct: 300 VVKAKELPPGSITGGCDPYVEVKLGNYKGRTKIFDRKTTIPEWNQVFAFTKERIQSSVLE 359
Query: 78 LYVKDHNALLPTSSIGECVVEYQ----RLPPNQ-MADKWIPLQG------VKRGEIHIQI 126
++VKD L +G+ V + R+PPN +A +W L+ V RGEI + +
Sbjct: 360 VFVKDKETLGRDDILGKVVFDLNEIPTRVPPNSPLAPQWYRLEDWRGEGKVVRGEIMLAV 419
Score = 43.1 bits (100), Expect = 1e-04
Identities = 53/230 (23%), Positives = 96/230 (41%), Gaps = 33/230 (14%)
Query: 22 LIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQTLEFPDDGSPLMLYVK 81
+++ + L+ D +G++ P+V V + N +T+ + K+L P WNQ L F D S +
Sbjct: 11 VVDAQYLMPRDGQGSASPFVEVDFLNQLSKTRTVPKSLNPVWNQKLYFDYDQS----VIN 66
Query: 82 DHNALLPTS-----------------SIGECVVEYQRLPPNQ---MADKWIPLQGVKRGE 121
HN + S I C + Y+ Q + KW+ L VK GE
Sbjct: 67 QHNQHIEVSVYHERRPIPGRSFLGRVKISLCNIVYKDDQVYQRFTLEKKWL-LSSVK-GE 124
Query: 122 IHIQITRKVPEMQKRQSMDSEPSLSKLHQIPTQIKQMMIKFRSQIEDGNLEGLSTTLSEL 181
I ++ E + + S+P S PTQ + + + S +E
Sbjct: 125 IGLKFYISSSEEDQTFPLPSKPYTS-----PTQASASGTEEDTADSETEDSLKSFASAEE 179
Query: 182 ETLEDTQEGYVAQLETEQMLLLSKIKELGQEIINSSPSPSLSRRISESVN 231
E L D+ V ++E+ + +++L ++ + + P+P S R+ N
Sbjct: 180 EDLADSVSECVEGKKSEE--VKEPVQKLHRQEVFARPAPMQSIRLRSREN 227
Score = 38.1 bits (87), Expect = 0.004
Identities = 41/164 (25%), Positives = 71/164 (43%), Gaps = 21/164 (12%)
Query: 16 GWIELVLIEGRDLVAADL---RGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQ--TLEFP 70
G +E+ ++ LV L RG+++ Y YG RT+ I TL+P+WN+ T E
Sbjct: 624 GMLEIGILGANGLVPMKLKDGRGSTNAYCVAKYGQKWVRTRTILDTLSPRWNEQYTWEVY 683
Query: 71 DDGSPLMLYVKDHNALLPTSS---------IGECVVEYQRLPPNQMADKWIPL-----QG 116
D + + L V D++ L S IG+ + L +++ PL G
Sbjct: 684 DPCTVITLGVFDNSHLGSAQSGTADSRDARIGKVRIRLSTLEAHKIYTHSFPLLVLQPHG 743
Query: 117 VKR-GEIHIQITRKVPEMQKRQSMDSEPSLSKLHQI-PTQIKQM 158
+K+ G++ I + + P L K+H + P + Q+
Sbjct: 744 LKKTGDLQISVRFTTLSLANIIYNYGHPLLPKMHYLFPFTVNQV 787
>At5g06850 anthranilate phosphoribosyltransferase-like protein
Length = 794
Score = 58.5 bits (140), Expect = 3e-09
Identities = 42/148 (28%), Positives = 67/148 (44%), Gaps = 31/148 (20%)
Query: 10 GSGSGNGWI---------ELV---------LIEGRDLVAADLRGTSDPYVRVHYGNFKKR 51
G G GWI +LV +++ +DL + DPYV V GN+K +
Sbjct: 30 GQRGGTGWIGSERAASTYDLVEQMFYLYVRVVKAKDLPPNPVTSNCDPYVEVKIGNYKGK 89
Query: 52 TKVIYKTLTPQWNQTLEFPDD---GSPLMLYVKDHNALLPTSSIGECVVEYQ----RLPP 104
TK K P+WNQ F D S + ++V+D + IG+ V + + R+PP
Sbjct: 90 TKHFEKRTNPEWNQVFAFSKDKVQSSTVEVFVRDKEMVTRDEYIGKVVFDMREVPTRVPP 149
Query: 105 NQ-MADKWIPL-----QGVKRGEIHIQI 126
+ +A +W L + KRGE+ + +
Sbjct: 150 DSPLAPQWYRLEDRRGESKKRGEVMVAV 177
Score = 36.6 bits (83), Expect = 0.012
Identities = 35/146 (23%), Positives = 63/146 (42%), Gaps = 19/146 (13%)
Query: 32 DLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQ--TLEFPDDGSPLMLYV--------- 80
D + T+DPY YG RT+ I + +P+WN+ T E D + + L V
Sbjct: 400 DGKATTDPYCVAKYGQKWVRTRTIIDSSSPKWNEQYTWEVYDPCTVITLGVFDNCHLGGS 459
Query: 81 -KDHNALLPTSSIGECVVEYQRLPPNQMADKWIPL-----QGVKR-GEIHIQITRKVPEM 133
K ++ S IG+ + L +++ PL +G+K+ GE+ + + +
Sbjct: 460 EKSNSGAKVDSRIGKVRIRLSTLEADRIYTHSYPLLVLQTKGLKKMGEVQLAVRFTCLSL 519
Query: 134 QKRQSMDSEPSLSKLHQI-PTQIKQM 158
+ P L K+H + P + Q+
Sbjct: 520 AHMIYLYGHPLLPKMHYLHPFTVNQL 545
Score = 32.3 bits (72), Expect = 0.22
Identities = 17/54 (31%), Positives = 29/54 (53%), Gaps = 1/54 (1%)
Query: 17 WIELVLIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVI-YKTLTPQWNQTLEF 69
++ + +IE +D+ +D +V+V GN +TK+ KT P WN+ L F
Sbjct: 218 YLRVNVIEAQDVEPSDRSQPPQAFVKVQVGNQILKTKLCPNKTTNPMWNEDLVF 271
>At1g05500 Ca2+-dependent lipid-binding protein, putative
Length = 560
Score = 57.4 bits (137), Expect = 6e-09
Identities = 35/116 (30%), Positives = 58/116 (49%), Gaps = 6/116 (5%)
Query: 16 GWIELVLIEGRDLVAADLRGTSDPYV--RVHYGNFKKRTKVIYKTLTPQWNQTLEF-PDD 72
G + + +I ++ DL G +DPYV + K +T+V+ +L P WNQT +F +D
Sbjct: 435 GVLSVTVISAEEIPIQDLMGKADPYVVLSMKKSGAKSKTRVVNDSLNPVWNQTFDFVVED 494
Query: 73 G--SPLMLYVKDHNALLPTSSIGECVVEYQRLPPNQMADKWIPLQGVKRGEIHIQI 126
G L+L V DH+ IG C++ R+ + W PL K G++ + +
Sbjct: 495 GLHDMLVLEVWDHDT-FGKDYIGRCILTLTRVIMEEEYKDWYPLDESKTGKLQLHL 549
Score = 54.3 bits (129), Expect = 5e-08
Identities = 35/125 (28%), Positives = 61/125 (48%), Gaps = 14/125 (11%)
Query: 16 GWIELVLIEGRDLVAADLRGTSDPYVRVHYGNFK---KRTKVIYKTLTPQWNQTLEF-PD 71
G +E+ L++ ++L DL G SDP+ ++ + KR+K I L P WN+ EF +
Sbjct: 262 GMLEVKLVQAKNLTNKDLVGKSDPFAKMFIRPLREKTKRSKTINNDLNPIWNEHFEFVVE 321
Query: 72 DGSP--LMLYVKDHNALLPTSSIGECVVEYQRLPPNQMADKWIPL--------QGVKRGE 121
D S L++ + D + + IG + L P ++ D W+ L RGE
Sbjct: 322 DASTQHLVVRIYDDEGVQASELIGCAQIRLCELEPGKVKDVWLKLVKDLEIQRDTKNRGE 381
Query: 122 IHIQI 126
+H+++
Sbjct: 382 VHLEL 386
>At1g22610 Highly similar to phosphoribosylanthranilate transferase
Length = 1029
Score = 55.8 bits (133), Expect = 2e-08
Identities = 37/123 (30%), Positives = 61/123 (49%), Gaps = 13/123 (10%)
Query: 17 WIELVLIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQTLEFPDD---G 73
++ + +++ RDL D+ G+ DPYV V GN+K TK + K P W Q F +
Sbjct: 295 YLYVSVVKARDLPVMDVSGSLDPYVEVKLGNYKGLTKHLEKNSNPIWKQIFAFSKERLQS 354
Query: 74 SPLMLYVKDHNALLPTSSIGECVVEYQ----RLPPNQ-MADKWIPLQGVK-----RGEIH 123
+ L + VKD + L +G ++ R+PP+ +A +W L+ K RGEI
Sbjct: 355 NLLEVTVKDKDLLTKDDFVGRVHIDLTEVPLRVPPDSPLAPQWYRLEDKKGMKTNRGEIM 414
Query: 124 IQI 126
+ +
Sbjct: 415 LAV 417
Score = 44.3 bits (103), Expect = 6e-05
Identities = 17/48 (35%), Positives = 31/48 (64%)
Query: 22 LIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQTLEF 69
+++ DL+ D +G++ P+V V + ++RT+ +K L PQWN+ L F
Sbjct: 8 IVDASDLMPKDGQGSASPFVEVEFDEQRQRTQTRFKDLNPQWNEKLVF 55
Score = 40.4 bits (93), Expect = 8e-04
Identities = 39/152 (25%), Positives = 65/152 (42%), Gaps = 17/152 (11%)
Query: 16 GWIELVLIEGRDLV---AADLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQ--TLEFP 70
G +EL ++ R+L+ D R T DPY YGN RT+ + L P+WN+ T E
Sbjct: 622 GILELGILSARNLMPMKGKDGRMT-DPYCVAKYGNKWVRTRTLLDALAPKWNEQYTWEVH 680
Query: 71 DDGSPLMLYVKDHNAL-----LPTSSIGECVVEYQRLPPNQMADKWIPLQGV------KR 119
D + + + V D++ + IG+ V L +++ + PL + K
Sbjct: 681 DPCTVITIGVFDNSHVNDGGDFKDQRIGKVRVRLSTLETDRVYTHFYPLLVLTPGGLKKN 740
Query: 120 GEIHIQITRKVPEMQKRQSMDSEPSLSKLHQI 151
GE+ + + + P L K+H I
Sbjct: 741 GELQLALRYTCTGFVNMMAQYGRPLLPKMHYI 772
Score = 34.3 bits (77), Expect = 0.057
Identities = 16/54 (29%), Positives = 31/54 (56%), Gaps = 1/54 (1%)
Query: 17 WIELVLIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVIY-KTLTPQWNQTLEF 69
++ + ++E +DLV +D D V++ GN + T+ +T+ PQW++ L F
Sbjct: 458 YLRIHVMEAQDLVPSDKGRVPDAIVKIQAGNQMRATRTPQMRTMNPQWHEELMF 511
>At4g00700 putative phosphoribosylanthranilate transferase
Length = 1006
Score = 55.5 bits (132), Expect = 2e-08
Identities = 41/110 (37%), Positives = 58/110 (52%), Gaps = 14/110 (12%)
Query: 22 LIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQTLEFPDD---GSPLML 78
+++ RDL DL G+ DPYV V GNFK T K P+WNQ F D + L +
Sbjct: 274 VVKARDLPNKDLTGSLDPYVVVKIGNFKGVTTHFNKNTDPEWNQVFAFAKDNLQSNFLEV 333
Query: 79 YVKDHNALLPTSSIGECVVEY------QRLPPNQ-MADKWIPLQGVKRGE 121
VKD + LL +G +V++ R+PP+ +A +W L+ KRGE
Sbjct: 334 MVKDKDILL-DDFVG--IVKFDLREVQSRVPPDSPLAPQWYRLEN-KRGE 379
Score = 40.8 bits (94), Expect = 6e-04
Identities = 29/123 (23%), Positives = 58/123 (46%), Gaps = 10/123 (8%)
Query: 17 WIELVLIEGRD-LVAADLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQTLEF----PD 71
++ + ++E +D ++ +D + +VRV GN RTK ++ P+W F P
Sbjct: 433 YLRVQILEAQDVIIVSDKSRVPEVFVRVKVGNQMLRTKFPQRSNNPKWGDEFTFVVAEPF 492
Query: 72 DGSPLMLYVKDHNALLPTSSIGECVVEY----QRLPPNQMADKWIPLQGVKRGEIHIQIT 127
+ + L+L V+DH A +G+ V+ +R+ D+W+ L+ + +
Sbjct: 493 EDN-LVLSVEDHTAPNRDEPVGKAVILMNDIEKRIDDKPFHDRWVHLEDSISDAMDVDKA 551
Query: 128 RKV 130
+KV
Sbjct: 552 KKV 554
Score = 38.5 bits (88), Expect = 0.003
Identities = 32/133 (24%), Positives = 57/133 (42%), Gaps = 15/133 (11%)
Query: 34 RGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQ--TLEFPDDGSPLMLYVKDHNALLP--- 88
+GTSD YV YG+ R++ + ++ P++N+ T E D + L + V D+
Sbjct: 617 KGTSDTYVVAKYGHKWVRSRTVINSMNPKYNEQYTWEVFDPATVLTICVFDNAHFAAGDG 676
Query: 89 ----TSSIGECVVEYQRLPPNQMADKWIPLQGV------KRGEIHIQITRKVPEMQKRQS 138
IG+ + L ++ PL + KRGE+H+ + +
Sbjct: 677 GNKRDQPIGKVRIRLSTLQTGRVYTHAYPLLVLQPTGLKKRGELHLAVRFTCTSVSSMLM 736
Query: 139 MDSEPSLSKLHQI 151
++P L K+H I
Sbjct: 737 KYTKPLLPKMHYI 749
>At1g51570 anthranilate phosphoribosyltransferase, putative
(At1g51570)
Length = 776
Score = 54.7 bits (130), Expect = 4e-08
Identities = 37/117 (31%), Positives = 60/117 (50%), Gaps = 13/117 (11%)
Query: 22 LIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQTLEFPDD---GSPLML 78
+++ ++L DL G+ DPYV V GN++ T+ K P+WNQ F D S L
Sbjct: 45 VVKAKELPGKDLTGSCDPYVEVKLGNYRGTTRHFEKKSNPEWNQVFAFSKDRVQASYLEA 104
Query: 79 YVKDHNALLPTSSIGECVVEY----QRLPPNQ-MADKWIPLQGVK----RGEIHIQI 126
VKD + L+ IG V + +R+PP+ +A +W L+ K +GE+ + +
Sbjct: 105 TVKDKD-LVKDDLIGRVVFDLNEIPKRVPPDSPLAPQWYRLEDGKGQKVKGELMLAV 160
Score = 44.7 bits (104), Expect = 4e-05
Identities = 41/143 (28%), Positives = 63/143 (43%), Gaps = 18/143 (12%)
Query: 34 RGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQ--TLEFPDDGSPLMLYVKD--------- 82
RGT+D Y YG RT+ I + TP+WN+ T E D + + + V D
Sbjct: 385 RGTTDAYCVAKYGQKWIRTRTIIDSFTPRWNEQYTWEVFDPCTVVTVGVFDNCHLHGGDK 444
Query: 83 HNALLPTSSIGECVVEYQRLPPNQMADKWIPL-----QGVKR-GEIHIQITRKVPEMQKR 136
+N S IG+ + L +++ PL GVK+ GEIH+ + +
Sbjct: 445 NNGGGKDSRIGKVRIRLSTLEADRVYTHSYPLLVLHPSGVKKMGEIHLAVRFTCSSLLNM 504
Query: 137 QSMDSEPSLSKLHQI-PTQIKQM 158
M S P L K+H + P + Q+
Sbjct: 505 MYMYSMPLLPKMHYLHPLTVSQL 527
Score = 42.4 bits (98), Expect = 2e-04
Identities = 28/90 (31%), Positives = 46/90 (51%), Gaps = 4/90 (4%)
Query: 17 WIELVLIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVIY-KTLTPQWNQTLEF---PDD 72
++ + +IE +DL+ +D + +V+V GN RT+V +++ P WN+ L F
Sbjct: 202 YLRVNVIEAQDLIPSDKGRYPEVFVKVIMGNQALRTRVSQSRSINPMWNEDLMFVVAEPF 261
Query: 73 GSPLMLYVKDHNALLPTSSIGECVVEYQRL 102
PL+L V+D A +G C V Q L
Sbjct: 262 EEPLILSVEDRVAPNKDEVLGRCAVPLQYL 291
>At3g57880 anthranilate phosphoribosyltransferase-like protein
Length = 773
Score = 53.1 bits (126), Expect = 1e-07
Identities = 36/117 (30%), Positives = 59/117 (49%), Gaps = 13/117 (11%)
Query: 22 LIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQTLEFPDD---GSPLML 78
+++ ++L D+ G+ DPYV V GN+K T+ K P+WNQ F D S L
Sbjct: 45 VVKAKELPGKDMTGSCDPYVEVKLGNYKGTTRHFEKKSNPEWNQVFAFSKDRIQASFLEA 104
Query: 79 YVKDHNALLPTSSIGECVVEY----QRLPPNQ-MADKWIPLQGVK----RGEIHIQI 126
VKD + + IG V + +R+PP+ +A +W L+ K +GE+ + +
Sbjct: 105 TVKDKD-FVKDDLIGRVVFDLNEVPKRVPPDSPLAPQWYRLEDRKGDKVKGELMLAV 160
Score = 49.3 bits (116), Expect = 2e-06
Identities = 47/163 (28%), Positives = 73/163 (43%), Gaps = 20/163 (12%)
Query: 16 GWIELVLIEGRDLV---AADLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQ--TLEFP 70
G +EL ++ L+ D RGT+D Y YG RT+ I + TP+WN+ T E
Sbjct: 362 GVLELGILNATGLMPMKTKDGRGTTDAYCVAKYGQKWIRTRTIIDSFTPRWNEQYTWEVF 421
Query: 71 DDGSPLMLYVKDHNAL--------LPTSSIGECVVEYQRLPPNQMADKWIPL-----QGV 117
D + + + V D+ L S IG+ + L +++ PL GV
Sbjct: 422 DPCTVVTVGVFDNCHLHGGEKIGGAKDSRIGKVRIRLSTLETDRVYTHSYPLLVLHPNGV 481
Query: 118 KR-GEIHIQITRKVPEMQKRQSMDSEPSLSKLHQI-PTQIKQM 158
K+ GEIH+ + + M S+P L K+H I P + Q+
Sbjct: 482 KKMGEIHLAVRFTCSSLLNMMYMYSQPLLPKMHYIHPLTVSQL 524
Score = 44.7 bits (104), Expect = 4e-05
Identities = 36/135 (26%), Positives = 59/135 (43%), Gaps = 19/135 (14%)
Query: 17 WIELVLIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVIY-KTLTPQWNQTLEF---PDD 72
++ + +IE +DL+ D + + YV+ GN RT+V +T+ P WN+ L F
Sbjct: 202 YLRVNVIEAQDLIPTDKQRYPEVYVKAIVGNQALRTRVSQSRTINPMWNEDLMFVAAEPF 261
Query: 73 GSPLMLYVKDHNALLPTSSIGECVVEYQRLPPNQMADKWIPLQGVKRGEIHIQITRKVPE 132
PL+L V+D A +G C IPLQ + R H + +
Sbjct: 262 EEPLILSVEDRVAPNKDEVLGRCA---------------IPLQYLDRRFDHKPVNSRWYN 306
Query: 133 MQKRQSMDSEPSLSK 147
++K +D E +K
Sbjct: 307 LEKHIMVDGEKKETK 321
>At5g47710 unknown protein
Length = 166
Score = 52.8 bits (125), Expect = 2e-07
Identities = 33/93 (35%), Positives = 47/93 (50%), Gaps = 3/93 (3%)
Query: 12 GSGNGWIELVLIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQTLEF-- 69
G G +++ +I+G+ LV D + +SDPYV V GN +TKVI L P WN+ L F
Sbjct: 2 GEPLGLLQVTVIQGKKLVIRDFK-SSDPYVIVKLGNESAKTKVINNCLNPVWNEELNFTL 60
Query: 70 PDDGSPLMLYVKDHNALLPTSSIGECVVEYQRL 102
D + L L V D + +G + Q L
Sbjct: 61 KDPAAVLALEVFDKDRFKADDKMGHASLSLQPL 93
>At3g07940 putative GTPase activating protein
Length = 385
Score = 50.8 bits (120), Expect = 6e-07
Identities = 30/89 (33%), Positives = 47/89 (52%), Gaps = 3/89 (3%)
Query: 16 GWIELVLIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQT--LEFPDDG 73
G I++ +++G +L D+ TSDPYV + G +T+VI L P WN+T L P+
Sbjct: 229 GLIKVNVVKGTNLAVRDVM-TSDPYVILALGQQSVKTRVIKNNLNPVWNETLMLSIPEPM 287
Query: 74 SPLMLYVKDHNALLPTSSIGECVVEYQRL 102
PL + V D + +GE ++ Q L
Sbjct: 288 PPLKVLVYDKDTFSTDDFMGEAEIDIQPL 316
>At5g12970 anthranilate phosphoribosyltransferase -like protein
Length = 769
Score = 50.1 bits (118), Expect = 1e-06
Identities = 31/102 (30%), Positives = 53/102 (51%), Gaps = 9/102 (8%)
Query: 22 LIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQTLEFPDD---GSPLML 78
+++ ++L D+ G+ DPYV V GN++ TK K P+W Q F + S L +
Sbjct: 46 VVKAKELPGKDVTGSCDPYVEVKLGNYRGMTKHFEKRSNPEWKQVFAFSKERIQASILEV 105
Query: 79 YVKDHNALLPTSSIGECVVEY----QRLPPNQ-MADKWIPLQ 115
VKD + +L IG + + +R+PP+ +A +W L+
Sbjct: 106 VVKDKDVVL-DDLIGRIMFDLNEIPKRVPPDSPLAPQWYRLE 146
Score = 45.1 bits (105), Expect = 3e-05
Identities = 31/107 (28%), Positives = 50/107 (45%), Gaps = 8/107 (7%)
Query: 17 WIELVLIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVIY-KTLTPQWNQTLEF---PDD 72
++ + +IE +DL+ D + YV+ GN RT++ KTL P WN+ L F
Sbjct: 202 YVRVNVIEAQDLIPHDKTKFPEVYVKAMLGNQTLRTRISQTKTLNPMWNEDLMFVVAEPF 261
Query: 73 GSPLMLYVKDHNALLPTSSIGECVVEYQ----RLPPNQMADKWIPLQ 115
L+L V+D A ++G C + Q RL + +W L+
Sbjct: 262 EEALILAVEDRVAPNKDETLGRCAIPLQNVQRRLDHRPLNSRWFNLE 308
Score = 43.9 bits (102), Expect = 7e-05
Identities = 41/159 (25%), Positives = 71/159 (43%), Gaps = 16/159 (10%)
Query: 16 GWIELVLIEGRDLV---AADLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQTLEFP-D 71
G +E+ +I L+ + D +GT+D Y YG RT+ I + TP+WN+ +
Sbjct: 362 GLLEVGIISAHGLMPMKSKDGKGTTDAYCVAKYGQKWIRTRTIVDSFTPKWNEQYTWEVF 421
Query: 72 DGSPLMLYVKDHNALLPTSS-----IGECVVEYQRLPPNQMADKWIPL-----QGVKR-G 120
D ++ + N +P S IG+ + L +++ PL G+K+ G
Sbjct: 422 DTCTVITFGAFDNGHIPGGSGKDLRIGKVRIRLSTLEADRIYTHSYPLLVFHPSGIKKTG 481
Query: 121 EIHIQITRKVPEMQKRQSMDSEPSLSKLHQI-PTQIKQM 158
EI + + + M S+P L K+H I P + Q+
Sbjct: 482 EIQLAVRFTCLSLINMLHMYSQPLLPKMHYIHPLSVLQL 520
>At4g11610 phosphoribosylanthranilate transferase like protein
Length = 1011
Score = 50.1 bits (118), Expect = 1e-06
Identities = 35/108 (32%), Positives = 56/108 (51%), Gaps = 10/108 (9%)
Query: 22 LIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQTLEFPDD---GSPLML 78
+++ R+L D+ G+ DP+V V GN+K T+ K P+WNQ F + S L +
Sbjct: 284 VVKARELPIMDITGSVDPFVEVRVGNYKGITRHFEKRQHPEWNQVFAFAKERMQASVLEV 343
Query: 79 YVKDHNALLPTSSIGECVVEYQ----RLPPNQ-MADKWIPLQGVKRGE 121
VKD + LL +G + R+PP+ +A +W L+ K+GE
Sbjct: 344 VVKDKD-LLKDDYVGFVRFDINDVPLRVPPDSPLAPQWYRLED-KKGE 389
Score = 36.2 bits (82), Expect = 0.015
Identities = 35/130 (26%), Positives = 56/130 (42%), Gaps = 12/130 (9%)
Query: 34 RGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQ--TLEFPDDGSPLMLYVKDHNALLPTSS 91
RGTSD + YG RT+ + L P++N+ T E D + L + V D+ L +
Sbjct: 625 RGTSDTFCVGKYGQKWVRTRTMVDNLCPKYNEQYTWEVFDPATVLTVGVFDNGQLGEKGN 684
Query: 92 ----IGECVVEYQRLPPNQMADKWIPL-----QGVKR-GEIHIQITRKVPEMQKRQSMDS 141
IG+ + L ++ PL GVK+ GE+H+ + S
Sbjct: 685 RDVKIGKIRIRLSTLETGRIYTHSYPLLVLHPTGVKKMGELHMAVRFTCISFANMLYQYS 744
Query: 142 EPSLSKLHQI 151
+P L K+H +
Sbjct: 745 KPLLPKMHYV 754
Score = 35.0 bits (79), Expect = 0.033
Identities = 16/48 (33%), Positives = 27/48 (55%)
Query: 22 LIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQTLEF 69
+I +L D +GTS+ YV +++ K RT + + L P WN++ F
Sbjct: 12 VIGAHNLFPKDGQGTSNAYVELYFDGQKHRTTIKDRDLNPVWNESFFF 59
Score = 30.0 bits (66), Expect = 1.1
Identities = 16/54 (29%), Positives = 27/54 (49%), Gaps = 1/54 (1%)
Query: 17 WIELVLIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVIY-KTLTPQWNQTLEF 69
++ + +IE +DL+ D D YV+ GN +T+ +TL WN+ F
Sbjct: 443 YVRVNVIEAQDLIPTDKTRFPDVYVKAQLGNQVMKTRPCQARTLGAVWNEDFLF 496
>At4g05330 unknown protein
Length = 336
Score = 50.1 bits (118), Expect = 1e-06
Identities = 30/95 (31%), Positives = 50/95 (52%), Gaps = 3/95 (3%)
Query: 16 GWIELVLIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQ--TLEFPDDG 73
G +++ + +G +L D+ +SDPYV ++ G K +T V+ L P WNQ L P+
Sbjct: 180 GLLKVTIKKGTNLAIRDMM-SSDPYVVLNLGKQKLQTTVMNSNLNPVWNQELMLSVPESY 238
Query: 74 SPLMLYVKDHNALLPTSSIGECVVEYQRLPPNQMA 108
P+ L V D++ +GE ++ Q L + MA
Sbjct: 239 GPVKLQVYDYDTFSADDIMGEADIDIQPLITSAMA 273
>At5g17980 phosphoribosylanthranilate transferase-like protein
Length = 1049
Score = 49.7 bits (117), Expect = 1e-06
Identities = 21/48 (43%), Positives = 31/48 (63%)
Query: 22 LIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQTLEF 69
+++ +DL D GTS PYV + Y ++RT+ I + L P WN+TLEF
Sbjct: 11 VVDAKDLTPKDGHGTSSPYVVLDYYGQRRRTRTIVRDLNPVWNETLEF 58
Score = 32.0 bits (71), Expect = 0.28
Identities = 16/53 (30%), Positives = 31/53 (58%), Gaps = 3/53 (5%)
Query: 16 GWIELVLIEGRDLV---AADLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQ 65
G +EL +I ++L+ + +G++D Y YG+ RT+ + +L P+WN+
Sbjct: 634 GIVELGIIGCKNLLPMKTVNGKGSTDAYTVAKYGSKWVRTRTVSDSLDPKWNE 686
>At3g59660 unknown protein
Length = 594
Score = 49.7 bits (117), Expect = 1e-06
Identities = 23/86 (26%), Positives = 47/86 (53%), Gaps = 2/86 (2%)
Query: 18 IELVLIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQTLEFPDDGSPLM 77
+++ L+ ++L+ A+L GTSDPY V+ G+ K+ + ++ + P W + FP D P
Sbjct: 83 VKVELLAAKNLIGANLNGTSDPYAIVNCGSEKRFSSMVPGSRNPMWGEEFNFPTDELPAK 142
Query: 78 LYVKDH--NALLPTSSIGECVVEYQR 101
+ V H + + ++ +G + +R
Sbjct: 143 INVTIHDWDIIWKSTVLGSVTINVER 168
>At5g37740 unknown protein
Length = 168
Score = 49.3 bits (116), Expect = 2e-06
Identities = 27/81 (33%), Positives = 44/81 (53%), Gaps = 3/81 (3%)
Query: 16 GWIELVLIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQ--TLEFPDDG 73
G + + + G +L D+ +SDPY+ VH G K +T+V+ ++ P+WN TL D
Sbjct: 6 GLLRIHVKRGVNLAIRDI-SSSDPYIVVHCGKQKLKTRVVKHSVNPEWNDDLTLSVTDPN 64
Query: 74 SPLMLYVKDHNALLPTSSIGE 94
P+ L V D++ L +GE
Sbjct: 65 LPIKLTVYDYDLLSADDKMGE 85
>At5g11100 CLB1 - like protein
Length = 574
Score = 48.9 bits (115), Expect = 2e-06
Identities = 33/128 (25%), Positives = 60/128 (46%), Gaps = 7/128 (5%)
Query: 16 GWIELVLIEGRDLVAADLRGTSDPYVRVHY--GNFKKRTKVIYKTLTPQWNQTLEFPDDG 73
G + + ++ DL A D G +D +V + K +T+V+ +L P WNQT +F +
Sbjct: 448 GVLSVTVVAAEDLPAVDFMGKADAFVVITLKKSETKSKTRVVPDSLNPVWNQTFDFVVED 507
Query: 74 S---PLMLYVKDHNALLPTSSIGECVVEYQRLPPNQMADKWIPLQGVKRGEIHIQITRKV 130
+ L L V DH+ IG ++ R+ +W L G K G++ + + +
Sbjct: 508 ALHDLLTLEVWDHDK-FGKDKIGRVIMTLTRVMLEGEFQEWFELDGAKSGKLCVHL-KWT 565
Query: 131 PEMQKRQS 138
P ++ R +
Sbjct: 566 PRLKLRDA 573
Score = 48.5 bits (114), Expect = 3e-06
Identities = 33/105 (31%), Positives = 52/105 (49%), Gaps = 6/105 (5%)
Query: 16 GWIELVLIEGRDLVAADLRGTSDPYVRVH---YGNFKKRTKVIYKTLTPQWNQTLEF-PD 71
G +++ +++ +DL D+ G SDPY V + K+TK I +L P WN+ EF +
Sbjct: 271 GKLDVKVVQAKDLANKDMIGKSDPYAIVFIRPLPDRTKKTKTISNSLNPIWNEHFEFIVE 330
Query: 72 DGSPLMLYVK--DHNALLPTSSIGECVVEYQRLPPNQMADKWIPL 114
D S L V+ D + + IG V L P ++ D W+ L
Sbjct: 331 DVSTQHLTVRVFDDEGVGSSQLIGAAQVPLNELVPGKVKDIWLKL 375
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.314 0.133 0.377
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,263,478
Number of Sequences: 26719
Number of extensions: 220160
Number of successful extensions: 645
Number of sequences better than 10.0: 80
Number of HSP's better than 10.0 without gapping: 61
Number of HSP's successfully gapped in prelim test: 19
Number of HSP's that attempted gapping in prelim test: 528
Number of HSP's gapped (non-prelim): 120
length of query: 231
length of database: 11,318,596
effective HSP length: 96
effective length of query: 135
effective length of database: 8,753,572
effective search space: 1181732220
effective search space used: 1181732220
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 58 (26.9 bits)
Medicago: description of AC146563.10


