

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146561.5 + phase: 2 /pseudo/partial
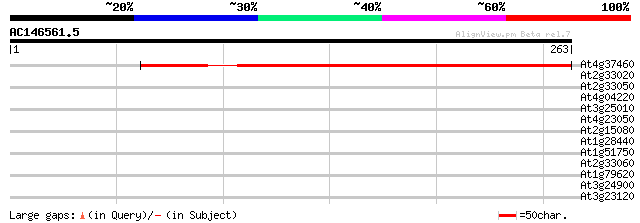
(263 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g37460 unknown protein 300 5e-82
At2g33020 putative leucine-rich repeat disease resistance protein 32 0.45
At2g33050 leucine-rich repeat disease resistance like protein 31 0.59
At4g04220 putative disease resistance protein, predicted GPI-anc... 30 1.3
At3g25010 disease resistance protein, putative 30 1.3
At4g23050 putative serine/threonine kinase 28 3.8
At2g15080 putative disease resistance protein 28 3.8
At1g28440 unknown protein 28 3.8
At1g51750 hypothetical protein 28 5.0
At2g33060 putative leucine-rich repeat disease resistance protein 28 6.6
At1g79620 unknown protein 28 6.6
At3g24900 leucine-rich repeat disease resistance protein, putative 27 8.6
At3g23120 disease resistance protein, putative 27 8.6
>At4g37460 unknown protein
Length = 869
Score = 300 bits (768), Expect = 5e-82
Identities = 135/202 (66%), Positives = 165/202 (80%), Gaps = 13/202 (6%)
Query: 62 EIMHAKSCSDLYKVVGEDFWSSTWCNSTAFEGKQLEGTRVTLVKMGQHGFDFAIRTPCTP 121
+IM A++C +L+ +VGEDFW +TWC+ST EG G+ G+DF+IRTPCTP
Sbjct: 680 QIMRAETCDELHNIVGEDFWVATWCDSTGSEG-------------GRLGYDFSIRTPCTP 726
Query: 122 ARWEDYDAEMAMAWEALCNAYCGENYGSTDFDVLENVRDAILRMTYYWYNFMPLSRGTAA 181
ARW D+D EM AWEALC AYCGENYGST+ D LE VRDAILRMTYYWYNFMPL+RGTA
Sbjct: 727 ARWSDFDEEMTSAWEALCTAYCGENYGSTELDALETVRDAILRMTYYWYNFMPLARGTAV 786
Query: 182 VGFAVMLGLLLAANMEFTGSIPQGFQVDWEAILNLDPNSFVDSVKSWLYPSLKVTTSWKD 241
GF V+LGLLLAANMEFT +IP+G Q+DWEAILN++P SFVDSVKSWLYPSLK+ TSW+D
Sbjct: 787 TGFVVLLGLLLAANMEFTETIPKGLQIDWEAILNVEPGSFVDSVKSWLYPSLKINTSWRD 846
Query: 242 YHDVASTFATTGSVVAALSSYD 263
+ +++S F+TTG+VVAALS+Y+
Sbjct: 847 HTEISSAFSTTGAVVAALSTYN 868
>At2g33020 putative leucine-rich repeat disease resistance protein
Length = 864
Score = 31.6 bits (70), Expect = 0.45
Identities = 21/58 (36%), Positives = 31/58 (53%), Gaps = 3/58 (5%)
Query: 155 LENVRDAILRMTYYWYNFMPLSRGTAAVGFAVMLGLLLAANMEFTGSIPQGFQVDWEA 212
L N+R LR ++ P +G +GF + +A NM FTGS+P F V+W+A
Sbjct: 586 LPNLRVLTLRSNKFYGPISPPHQGP--LGFPELRIFEIADNM-FTGSLPPSFFVNWKA 640
>At2g33050 leucine-rich repeat disease resistance like protein
Length = 800
Score = 31.2 bits (69), Expect = 0.59
Identities = 23/79 (29%), Positives = 37/79 (46%), Gaps = 6/79 (7%)
Query: 155 LENVRDAILRMTYYWYNFMPLSRGTAAVGFAVMLGLLLAANMEFTGSIPQGFQVDWEAI- 213
L N+ LR ++ + P RG A L +L ++ FTGS+P F V+W+A
Sbjct: 495 LPNLHVLTLRSNRFFGHLSPPDRGPLAFP---ELRILELSDNSFTGSLPPNFFVNWKASS 551
Query: 214 --LNLDPNSFVDSVKSWLY 230
+N D ++ K+ Y
Sbjct: 552 PKINEDGRIYMGDYKNAYY 570
>At4g04220 putative disease resistance protein, predicted
GPI-anchored protein
Length = 766
Score = 30.0 bits (66), Expect = 1.3
Identities = 20/68 (29%), Positives = 33/68 (48%), Gaps = 7/68 (10%)
Query: 156 ENVRDAILRMTYYWYNFMPLSRGT------AAVGFAVMLGLLLAANMEFTGSIPQGFQVD 209
+N + + +Y Y + LS+ ++G L +L +N EF+G IPQ F D
Sbjct: 585 KNSKQVLFDRNFYLYTLLDLSKNKLHGEIPTSLGNLKSLKVLNLSNNEFSGLIPQSFG-D 643
Query: 210 WEAILNLD 217
E + +LD
Sbjct: 644 LEKVESLD 651
>At3g25010 disease resistance protein, putative
Length = 881
Score = 30.0 bits (66), Expect = 1.3
Identities = 19/59 (32%), Positives = 34/59 (57%), Gaps = 3/59 (5%)
Query: 154 VLENVRDAILRMTYYWYNFMPLSRGTAAVGFAVMLGLLLAANMEFTGSIPQGFQVDWEA 212
VL ++ +L ++ P ++G+ +GF + L +A N + TGS+PQ F V+W+A
Sbjct: 599 VLPKLQVLLLSSNKFYGPLSPPNQGS--LGFPELRILEIAGN-KLTGSLPQDFFVNWKA 654
>At4g23050 putative serine/threonine kinase
Length = 736
Score = 28.5 bits (62), Expect = 3.8
Identities = 21/72 (29%), Positives = 34/72 (47%), Gaps = 6/72 (8%)
Query: 28 ERTLDIAKTVMKERSYVHGKTDQIIHLSNDGKLEEIMHAKSCSDLYKVVGEDFWSSTWCN 87
+R L +A V + +Y+H + I+H D K ++ K+ + V DF S W N
Sbjct: 564 KRRLRMALDVARGMNYLHRRNPPIVH--RDLKSSNLLVDKN----WNVKVGDFGLSKWKN 617
Query: 88 STAFEGKQLEGT 99
+T K +GT
Sbjct: 618 ATFLSTKSGKGT 629
>At2g15080 putative disease resistance protein
Length = 983
Score = 28.5 bits (62), Expect = 3.8
Identities = 10/23 (43%), Positives = 16/23 (69%)
Query: 197 EFTGSIPQGFQVDWEAILNLDPN 219
+F G++P F V+W A+ +LD N
Sbjct: 732 QFNGTLPANFFVNWTAMFSLDEN 754
>At1g28440 unknown protein
Length = 996
Score = 28.5 bits (62), Expect = 3.8
Identities = 12/24 (50%), Positives = 18/24 (75%)
Query: 181 AVGFAVMLGLLLAANMEFTGSIPQ 204
++G A L LL+ +N EFTGS+P+
Sbjct: 439 SIGGASNLSLLILSNNEFTGSLPE 462
>At1g51750 hypothetical protein
Length = 629
Score = 28.1 bits (61), Expect = 5.0
Identities = 14/42 (33%), Positives = 22/42 (52%), Gaps = 3/42 (7%)
Query: 206 FQVDWEAILNLDPNSFVDSVKSWLYP---SLKVTTSWKDYHD 244
F DW+ I+N + D ++S+L L V T WK+ +D
Sbjct: 540 FSTDWQTIVNYISETQTDRIRSFLSRYIFQLTVHTVWKERND 581
>At2g33060 putative leucine-rich repeat disease resistance protein
Length = 800
Score = 27.7 bits (60), Expect = 6.6
Identities = 18/58 (31%), Positives = 28/58 (48%), Gaps = 3/58 (5%)
Query: 155 LENVRDAILRMTYYWYNFMPLSRGTAAVGFAVMLGLLLAANMEFTGSIPQGFQVDWEA 212
L +++ LR + P RG A L +L ++ FTGS+P + V+WEA
Sbjct: 497 LPDLQALTLRSNKFHGPISPPDRGPLAFP---KLRILEISDNNFTGSLPPNYFVNWEA 551
>At1g79620 unknown protein
Length = 971
Score = 27.7 bits (60), Expect = 6.6
Identities = 34/133 (25%), Positives = 52/133 (38%), Gaps = 11/133 (8%)
Query: 120 TPARWEDYDAEMAMAWEAL-CNAYCGENYGSTDFDVLENVRDAILRMTYYWYNFMPLSRG 178
TP W D WE + CN G + + + I + + +RG
Sbjct: 50 TPPSWGGSDDPCGTPWEGVSCNNSRITALGLSTMGLKGRLSGDIGELAELRSLDLSFNRG 109
Query: 179 -----TAAVGFAVMLGLLLAANMEFTGSIPQ--GFQVDWEAILNLDPNSFVDSVKSWLYP 231
T+ +G L +L+ A FTG+IP G+ D + L L+ N+F + + L
Sbjct: 110 LTGSLTSRLGDLQKLNILILAGCGFTGTIPNELGYLKDL-SFLALNSNNFTGKIPASLGN 168
Query: 232 SLKVTTSWKDYHD 244
KV W D D
Sbjct: 169 LTKV--YWLDLAD 179
>At3g24900 leucine-rich repeat disease resistance protein, putative
Length = 884
Score = 27.3 bits (59), Expect = 8.6
Identities = 18/58 (31%), Positives = 32/58 (55%), Gaps = 3/58 (5%)
Query: 155 LENVRDAILRMTYYWYNFMPLSRGTAAVGFAVMLGLLLAANMEFTGSIPQGFQVDWEA 212
L ++ IL ++ P ++G+ +GF + L +A N +FTGS+P F +W+A
Sbjct: 601 LPKLQVLILHSNNFYGPLSPPNQGS--LGFPELRILEIAGN-KFTGSLPPDFFENWKA 655
>At3g23120 disease resistance protein, putative
Length = 784
Score = 27.3 bits (59), Expect = 8.6
Identities = 12/37 (32%), Positives = 21/37 (56%), Gaps = 1/37 (2%)
Query: 174 PLSRGTAAVGFAVMLGLLLAANMEFTGSIPQGFQVDW 210
P+ T +GF L ++ +N +F GS+PQ + +W
Sbjct: 510 PVYNSTTYLGFP-RLSIIDISNNDFVGSLPQDYFANW 545
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.134 0.419
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,152,471
Number of Sequences: 26719
Number of extensions: 256878
Number of successful extensions: 633
Number of sequences better than 10.0: 13
Number of HSP's better than 10.0 without gapping: 4
Number of HSP's successfully gapped in prelim test: 9
Number of HSP's that attempted gapping in prelim test: 621
Number of HSP's gapped (non-prelim): 20
length of query: 263
length of database: 11,318,596
effective HSP length: 97
effective length of query: 166
effective length of database: 8,726,853
effective search space: 1448657598
effective search space used: 1448657598
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 59 (27.3 bits)
Medicago: description of AC146561.5


