

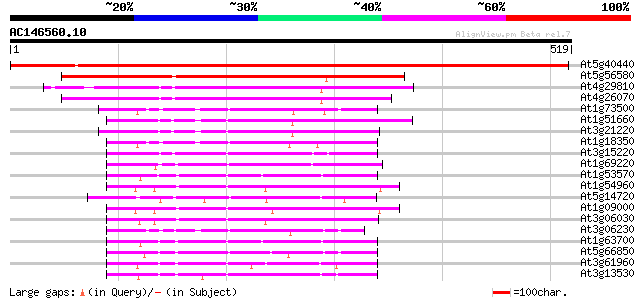
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146560.10 + phase: 0
(519 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g40440 MAP kinase kinase 3 (ATMKK3) 833 0.0
At5g56580 protein kinase MEK1 homolog 248 4e-66
At4g29810 MAP kinase kinase 2 234 7e-62
At4g26070 MEK1 221 6e-58
At1g73500 unknown protein 169 3e-42
At1g51660 unknown protein 163 2e-40
At3g21220 MAP kinase kinase 5 161 7e-40
At1g18350 MAP kinase kinase 5, putative 160 1e-39
At3g15220 putative MAP kinase 149 3e-36
At1g69220 serine/threonine kinase (SIK1) 147 1e-35
At1g53570 MEK kinase MAP3Ka, putative 142 3e-34
At1g54960 NPK1-related protein kinase 2 (ANP2) 141 1e-33
At5g14720 protein kinase -like protein 139 3e-33
At1g09000 NPK1-related protein kinase 1S (ANP1) 138 8e-33
At3g06030 NPK1-related protein kinase 3 135 5e-32
At3g06230 putative MAP kinase 127 1e-29
At1g63700 putative protein kinase 125 6e-29
At5g66850 MAP protein kinase like protein 122 5e-28
At3g61960 serine/threonine-protein kinase-like protein 122 6e-28
At3g13530 MAP3K epsilon protein kinase 122 6e-28
>At5g40440 MAP kinase kinase 3 (ATMKK3)
Length = 520
Score = 833 bits (2152), Expect = 0.0
Identities = 403/517 (77%), Positives = 453/517 (86%), Gaps = 1/517 (0%)
Query: 1 MSGLEELRKKLTPLFDAENGFSSTSSFDPNDSYTFSDGGTVNLLSRSYGVYNINELGLQK 60
M+ LEEL+KKL+PLFDAE GFSS+SS DPNDSY SDGGTVNLLSRSYGVYN NELGLQK
Sbjct: 1 MAALEELKKKLSPLFDAEKGFSSSSSLDPNDSYLLSDGGTVNLLSRSYGVYNFNELGLQK 60
Query: 61 CSTSRSVDESDDNEKTYRCGSHEMRIFGAIGSGASSVVQRAMHIPTHRVIALKKINIFEK 120
C TS VDES+ +E TY+C SHEMR+FGAIGSGASSVVQRA+HIP HR++ALKKINIFE+
Sbjct: 61 C-TSSHVDESESSETTYQCASHEMRVFGAIGSGASSVVQRAIHIPNHRILALKKINIFER 119
Query: 121 EKRQQLLTEIRTLCEAPCYEGLVEFHGAFYTPDSGQISIALEYMDGGSLADILRMHRTIP 180
EKRQQLLTEIRTLCEAPC+EGLV+FHGAFY+PDSGQISIALEYM+GGSLADIL++ + IP
Sbjct: 120 EKRQQLLTEIRTLCEAPCHEGLVDFHGAFYSPDSGQISIALEYMNGGSLADILKVTKKIP 179
Query: 181 EPILSSMFQKLLRGLSYLHGVRYLVHRDIKPANLLVNLKGEPKITDFGISAGLENSVAMC 240
EP+LSS+F KLL+GLSYLHGVR+LVHRDIKPANLL+NLKGEPKITDFGISAGLENS+AMC
Sbjct: 180 EPVLSSLFHKLLQGLSYLHGVRHLVHRDIKPANLLINLKGEPKITDFGISAGLENSMAMC 239
Query: 241 ATFVGTVTYMSPERIRNESYSYPADIWSLGLALLESGTGEFPYTANEGPVNLMLQILDDP 300
ATFVGTVTYMSPERIRN+SYSYPADIWSLGLAL E GTGEFPY ANEGPVNLMLQILDDP
Sbjct: 240 ATFVGTVTYMSPERIRNDSYSYPADIWSLGLALFECGTGEFPYIANEGPVNLMLQILDDP 299
Query: 301 SPSPSKEKFSPEFCSFVDACLQKDPDNRPTAEQLLLHPFITKYETAKVDLPGFVRSVFDP 360
SP+P K++FSPEFCSF+DACLQKDPD RPTA+QLL HPFITK+E +VDL FV+S+FDP
Sbjct: 300 SPTPPKQEFSPEFCSFIDACLQKDPDARPTADQLLSHPFITKHEKERVDLATFVQSIFDP 359
Query: 361 TQRMKDLADMLTIHYYLLFDGPDDSWQHTRNFYNENSIFSFSGKQHIGPNNIFTTLSSIR 420
TQR+KDLADMLTIHYY LFDG DD W H ++ Y E S+FSFSGK + G IF+ LS IR
Sbjct: 360 TQRLKDLADMLTIHYYSLFDGFDDLWHHAKSLYTETSVFSFSGKHNTGSTEIFSALSDIR 419
Query: 421 TTLIGDWPPEKLVHVVEKLQCRAHGEDGVAIRVSGSFIIGNQFLICGDGVQVEGLPNFKD 480
TL GD P EKLVHVVEKL C+ G GV IR GSFI+GNQFLICGDGVQ EGLP+FKD
Sbjct: 420 NTLTGDLPSEKLVHVVEKLHCKPCGSGGVIIRAVGSFIVGNQFLICGDGVQAEGLPSFKD 479
Query: 481 LGIDISSKRMGTFHEQFIVEPTSQIGCYTIVKQELYI 517
LG D++S+R+G F EQF+VE IG Y + KQELYI
Sbjct: 480 LGFDVASRRVGRFQEQFVVESGDLIGKYFLAKQELYI 516
>At5g56580 protein kinase MEK1 homolog
Length = 356
Score = 248 bits (634), Expect = 4e-66
Identities = 133/325 (40%), Positives = 199/325 (60%), Gaps = 11/325 (3%)
Query: 49 GVYNINELGLQKCSTSRSVDESDDNEKTYRCGSHEMRIFGAIGSGASSVVQRAMHIPTHR 108
G + +N+ GL+ S + +SD E + + ++ IG G+ VVQ H +
Sbjct: 35 GDFLLNQKGLRLTSDEKQSRQSDSKELDFEITAEDLETVKVIGKGSGGVVQLVRHKWVGK 94
Query: 109 VIALKKINI-FEKEKRQQLLTEIR-TLCEAPCYEGLVEFHGAFYTPDSGQISIALEYMDG 166
A+K I + ++E R+Q++ E++ + C +V +H ++ +G S+ LEYMD
Sbjct: 95 FFAMKVIQMNIQEEIRKQIVQELKINQASSQCPHVVVCYHSFYH---NGAFSLVLEYMDR 151
Query: 167 GSLADILRMHRTIPEPILSSMFQKLLRGLSYLHGVRYLVHRDIKPANLLVNLKGEPKITD 226
GSLAD++R +TI EP L+ + +++L GL YLH R+++HRDIKP+NLLVN KGE KI+D
Sbjct: 152 GSLADVIRQVKTILEPYLAVVCKQVLLGLVYLHNERHVIHRDIKPSNLLVNHKGEVKISD 211
Query: 227 FGISAGLENSVAMCATFVGTVTYMSPERIRNESYSYPADIWSLGLALLESGTGEFPYTAN 286
FG+SA L +S+ TFVGT YMSPERI +Y Y +DIWSLG+++LE G FPY +
Sbjct: 212 FGVSASLASSMGQRDTFVGTYNYMSPERISGSTYDYSSDIWSLGMSVLECAIGRFPYLES 271
Query: 287 EGPVN------LMLQILDDPSPSPSKEKFSPEFCSFVDACLQKDPDNRPTAEQLLLHPFI 340
E N L+ I+++P P+ ++FSPEFCSFV AC+QKDP R ++ LL HPFI
Sbjct: 272 EDQQNPPSFYELLAAIVENPPPTAPSDQFSPEFCSFVSACIQKDPPARASSLDLLSHPFI 331
Query: 341 TKYETAKVDLPGFVRSVFDPTQRMK 365
K+E +DL V ++ P ++
Sbjct: 332 KKFEDKDIDLGILVGTLEPPVNYLR 356
>At4g29810 MAP kinase kinase 2
Length = 363
Score = 234 bits (598), Expect = 7e-62
Identities = 135/349 (38%), Positives = 202/349 (57%), Gaps = 21/349 (6%)
Query: 32 SYTFSDGGTVNLLSRSYGVYNINELGLQKCSTSRSVDESDDNEKTYRCGSHEMRIFGAIG 91
S TF DG +L GV I++L + S + D+ + ++ + IG
Sbjct: 29 SGTFKDG---DLRVNKDGVRIISQLEPEVLSPIKPADD--------QLSLSDLDMVKVIG 77
Query: 92 SGASSVVQRAMHIPTHRVIALKKINI-FEKEKRQQLLTEIRTLCEAPCYEGLVEFHGAFY 150
G+S VVQ H T + ALK I + ++ R+ + E++ + C LV + +FY
Sbjct: 78 KGSSGVVQLVQHKWTGQFFALKVIQLNIDEAIRKAIAQELKINQSSQC-PNLVTSYQSFY 136
Query: 151 TPDSGQISIALEYMDGGSLADILRMHRTIPEPILSSMFQKLLRGLSYLHGVRYLVHRDIK 210
D+G IS+ LEYMDGGSLAD L+ + IP+ LS++F+++L+GL YLH R+++HRD+K
Sbjct: 137 --DNGAISLILEYMDGGSLADFLKSVKAIPDSYLSAIFRQVLQGLIYLHHDRHIIHRDLK 194
Query: 211 PANLLVNLKGEPKITDFGISAGLENSVAMCATFVGTVTYMSPERIRNESYSYPADIWSLG 270
P+NLL+N +GE KITDFG+S + N+ + TFVGT YMSPERI Y +DIWSLG
Sbjct: 195 PSNLLINHRGEVKITDFGVSTVMTNTAGLANTFVGTYNYMSPERIVGNKYGNKSDIWSLG 254
Query: 271 LALLESGTGEFPYTANE------GPVNLMLQILDDPSPSPSKEKFSPEFCSFVDACLQKD 324
L +LE TG+FPY LM I+D P P+ FSPE SF+ CLQKD
Sbjct: 255 LVVLECATGKFPYAPPNQEETWTSVFELMEAIVDQPPPALPSGNFSPELSSFISTCLQKD 314
Query: 325 PDNRPTAEQLLLHPFITKYETAKVDLPGFVRSVFDPTQRMKDLADMLTI 373
P++R +A++L+ HPF+ KY+ + ++L + P + +L+ ++
Sbjct: 315 PNSRSSAKELMEHPFLNKYDYSGINLASYFTDAGSPLATLGNLSGTFSV 363
>At4g26070 MEK1
Length = 354
Score = 221 bits (564), Expect = 6e-58
Identities = 125/312 (40%), Positives = 185/312 (59%), Gaps = 10/312 (3%)
Query: 49 GVYNINELGLQKCSTSRSVDESDDNEKTYRCGSHEMRIFGAIGSGASSVVQRAMHIPTHR 108
G +N+ G+Q S S + ++ + IG G+S VQ H T +
Sbjct: 33 GDLRVNKDGIQTVSLSEPGAPPPIEPLDNQLSLADLEVIKVIGKGSSGNVQLVKHKLTQQ 92
Query: 109 VIALKKINIFEKEKRQQLLT-EIRTLCEAPCYEGLVEFHGAFYTPDSGQISIALEYMDGG 167
ALK I + +E + ++ E+R + C LV + +FY +G +SI LE+MDGG
Sbjct: 93 FFALKVIQLNTEESTCRAISQELRINLSSQC-PYLVSCYQSFY--HNGLVSIILEFMDGG 149
Query: 168 SLADILRMHRTIPEPILSSMFQKLLRGLSYLHGVRYLVHRDIKPANLLVNLKGEPKITDF 227
SLAD+L+ +PE +LS++ +++LRGL Y+H R ++HRD+KP+NLL+N +GE KITDF
Sbjct: 150 SLADLLKKVGKVPENMLSAICKRVLRGLCYIHHERRIIHRDLKPSNLLINHRGEVKITDF 209
Query: 228 GISAGLENSVAMCATFVGTVTYMSPERIRNESYSYPADIWSLGLALLESGTGEFPYTANE 287
G+S L ++ ++ +FVGT YMSPERI YS +DIWSLGL LLE TG+FPYT E
Sbjct: 210 GVSKILTSTSSLANSFVGTYPYMSPERISGSLYSNKSDIWSLGLVLLECATGKFPYTPPE 269
Query: 288 ------GPVNLMLQILDDPSPSPSKEKFSPEFCSFVDACLQKDPDNRPTAEQLLLHPFIT 341
L+ I+++P P FSPEFCSF+ C+QKDP +R +A++LL H F+
Sbjct: 270 HKKGWSSVYELVDAIVENPPPCAPSNLFSPEFCSFISQCVQKDPRDRKSAKELLEHKFVK 329
Query: 342 KYETAKVDLPGF 353
+E + +L +
Sbjct: 330 MFEDSDTNLSAY 341
>At1g73500 unknown protein
Length = 310
Score = 169 bits (428), Expect = 3e-42
Identities = 109/272 (40%), Positives = 153/272 (56%), Gaps = 25/272 (9%)
Query: 83 EMRIFGAIGSGASSVVQRAMHIPTHRVIALKKIN-----IFEKEKRQQLLTEIRTLCEAP 137
++ +G G +V + H T + ALK +N IF ++ +++ EI ++P
Sbjct: 46 DLEKLNVLGCGNGGIVYKVRHKTTSEIYALKTVNGDMDPIFTRQLMREM--EILRRTDSP 103
Query: 138 CYEGLVEFHGAFYTPDSGQISIALEYMDGGSLADILRMHRTIPEPILSSMFQKLLRGLSY 197
+V+ HG F P G++SI +EYMDGG+L + + E L+ +++L+GLSY
Sbjct: 104 Y---VVKCHGIFEKPVVGEVSILMEYMDGGTLESL---RGGVTEQKLAGFAKQILKGLSY 157
Query: 198 LHGVRYLVHRDIKPANLLVNLKGEPKITDFGISAGLENSVAMCATFVGTVTYMSPERIRN 257
LH ++ +VHRDIKPANLL+N K E KI DFG+S L S+ C ++VGT YMSPER +
Sbjct: 158 LHALK-IVHRDIKPANLLLNSKNEVKIADFGVSKILVRSLDSCNSYVGTCAYMSPERFDS 216
Query: 258 ESYS-----YPADIWSLGLALLESGTGEFP-YTANEGP--VNLMLQI-LDDPSPSPSKEK 308
ES Y DIWS GL +LE G FP + P LM + +P +P E
Sbjct: 217 ESSGGSSDIYAGDIWSFGLMMLELLVGHFPLLPPGQRPDWATLMCAVCFGEPPRAP--EG 274
Query: 309 FSPEFCSFVDACLQKDPDNRPTAEQLLLHPFI 340
S EF SFV+ CL+KD R TA QLL HPF+
Sbjct: 275 CSEEFRSFVECCLRKDSSKRWTAPQLLAHPFL 306
>At1g51660 unknown protein
Length = 366
Score = 163 bits (412), Expect = 2e-40
Identities = 104/290 (35%), Positives = 157/290 (53%), Gaps = 15/290 (5%)
Query: 90 IGSGASSVVQRAMHIPTHRVIALKKI-NIFEKEKRQQLLTEIRTLCEAPCYEGLVEFHGA 148
IGSGA V + +H P+ R+ ALK I E+ R+Q+ EI L + + +V+ H
Sbjct: 85 IGSGAGGTVYKVIHRPSSRLYALKVIYGNHEETVRRQICREIEILRDVN-HPNVVKCHEM 143
Query: 149 FYTPDSGQISIALEYMDGGSLADILRMHRTIPEPILSSMFQKLLRGLSYLHGVRYLVHRD 208
F +G+I + LE+MD GSL E L+ + +++L GL+YLH R++VHRD
Sbjct: 144 F--DQNGEIQVLLEFMDKGSLEGA----HVWKEQQLADLSRQILSGLAYLHS-RHIVHRD 196
Query: 209 IKPANLLVNLKGEPKITDFGISAGLENSVAMCATFVGTVTYMSPERIRNESY-----SYP 263
IKP+NLL+N KI DFG+S L ++ C + VGT+ YMSPERI + Y
Sbjct: 197 IKPSNLLINSAKNVKIADFGVSRILAQTMDPCNSSVGTIAYMSPERINTDLNQGKYDGYA 256
Query: 264 ADIWSLGLALLESGTGEFPY-TANEGPVNLMLQILDDPSPSPSKEKFSPEFCSFVDACLQ 322
DIWSLG+++LE G FP+ + +G ++ + P + SPEF F+ CLQ
Sbjct: 257 GDIWSLGVSILEFYLGRFPFPVSRQGDWASLMCAICMSQPPEAPATASPEFRHFISCCLQ 316
Query: 323 KDPDNRPTAEQLLLHPFITKYETAKVDLPGFVRSVFDPTQRMKDLADMLT 372
++P R +A QLL HPFI + ++ P + + P + + + T
Sbjct: 317 REPGKRRSAMQLLQHPFILRASPSQNRSPQNLHQLLPPPRPLSSSSSPTT 366
>At3g21220 MAP kinase kinase 5
Length = 348
Score = 161 bits (408), Expect = 7e-40
Identities = 104/267 (38%), Positives = 145/267 (53%), Gaps = 15/267 (5%)
Query: 83 EMRIFGAIGSGASSVVQRAMHIPTHRVIALKKI-NIFEKEKRQQLLTEIRTLCEAPCYEG 141
E+ IGSGA V + +H PT R ALK I E R+Q+ EI L +
Sbjct: 69 ELERVNRIGSGAGGTVYKVIHTPTSRPFALKVIYGNHEDTVRRQICREIEILRSVD-HPN 127
Query: 142 LVEFHGAFYTPDSGQISIALEYMDGGSLADILRMHRTIPEPILSSMFQKLLRGLSYLHGV 201
+V+ H F +G+I + LE+MD GSL E L+ + +++L GL+YLH
Sbjct: 128 VVKCHDMF--DHNGEIQVLLEFMDQGSLEGA----HIWQEQELADLSRQILSGLAYLHR- 180
Query: 202 RYLVHRDIKPANLLVNLKGEPKITDFGISAGLENSVAMCATFVGTVTYMSPERIRNESY- 260
R++VHRDIKP+NLL+N KI DFG+S L ++ C + VGT+ YMSPERI +
Sbjct: 181 RHIVHRDIKPSNLLINSAKNVKIADFGVSRILAQTMDPCNSSVGTIAYMSPERINTDLNH 240
Query: 261 ----SYPADIWSLGLALLESGTGEFPY-TANEGPVNLMLQILDDPSPSPSKEKFSPEFCS 315
Y D+WSLG+++LE G FP+ + +G ++ + P + S EF
Sbjct: 241 GRYDGYAGDVWSLGVSILEFYLGRFPFAVSRQGDWASLMCAICMSQPPEAPATASQEFRH 300
Query: 316 FVDACLQKDPDNRPTAEQLLLHPFITK 342
FV CLQ DP R +A+QLL HPFI K
Sbjct: 301 FVSCCLQSDPPKRWSAQQLLQHPFILK 327
>At1g18350 MAP kinase kinase 5, putative
Length = 307
Score = 160 bits (406), Expect = 1e-39
Identities = 100/262 (38%), Positives = 146/262 (55%), Gaps = 20/262 (7%)
Query: 90 IGSGASSVVQRAMHIPTHRVIALKKIN-----IFEKEKRQQLLTEIRTLCEAPCYEGLVE 144
+G G+S +V + H T + ALK +N F ++ +++ EI ++P +V
Sbjct: 51 LGRGSSGIVYKVHHKTTGEIYALKSVNGDMSPAFTRQLAREM--EILRRTDSPY---VVR 105
Query: 145 FHGAFYTPDSGQISIALEYMDGGSLADILRMHRTIPEPILSSMFQKLLRGLSYLHGVRYL 204
G F P G++SI +EYMDGG+L + + E L+ +++L+GLSYLH ++ +
Sbjct: 106 CQGIFEKPIVGEVSILMEYMDGGNLESL---RGAVTEKQLAGFSRQILKGLSYLHSLK-I 161
Query: 205 VHRDIKPANLLVNLKGEPKITDFGISAGLENSVAMCATFVGTVTYMSPERIRN----ESY 260
VHRDIKPANLL+N + E KI DFG+S + S+ C ++VGT YMSPER + S
Sbjct: 162 VHRDIKPANLLLNSRNEVKIADFGVSKIITRSLDYCNSYVGTCAYMSPERFDSAAGENSD 221
Query: 261 SYPADIWSLGLALLESGTGEFPY--TANEGPVNLMLQILDDPSPSPSKEKFSPEFCSFVD 318
Y DIWS G+ +LE G FP ++ ++ P + E S EF SFVD
Sbjct: 222 VYAGDIWSFGVMILELFVGHFPLLPQGQRPDWATLMCVVCFGEPPRAPEGCSDEFRSFVD 281
Query: 319 ACLQKDPDNRPTAEQLLLHPFI 340
CL+K+ R TA QLL HPF+
Sbjct: 282 CCLRKESSERWTASQLLGHPFL 303
>At3g15220 putative MAP kinase
Length = 690
Score = 149 bits (377), Expect = 3e-36
Identities = 91/253 (35%), Positives = 149/253 (57%), Gaps = 8/253 (3%)
Query: 90 IGSGASSVVQRAMHIPTHRVIALKKINIFEKEKR-QQLLTEIRTLCEAPCYEGLVEFHGA 148
IG G+ V +A ++ +A+K I++ E E + + EI L + C + E++G+
Sbjct: 21 IGRGSFGDVYKAFDKDLNKEVAIKVIDLEESEDEIEDIQKEISVLSQCRC-PYITEYYGS 79
Query: 149 FYTPDSGQISIALEYMDGGSLADILRMHRTIPEPILSSMFQKLLRGLSYLHGVRYLVHRD 208
+ ++ I +EYM GGS+AD+L+ + + E ++ + + LL + YLH +HRD
Sbjct: 80 YL--HQTKLWIIMEYMAGGSVADLLQSNNPLDETSIACITRDLLHAVEYLHN-EGKIHRD 136
Query: 209 IKPANLLVNLKGEPKITDFGISAGLENSVAMCATFVGTVTYMSPERIRN-ESYSYPADIW 267
IK AN+L++ G+ K+ DFG+SA L +++ TFVGT +M+PE I+N E Y+ ADIW
Sbjct: 137 IKAANILLSENGDVKVADFGVSAQLTRTISRRKTFVGTPFWMAPEVIQNSEGYNEKADIW 196
Query: 268 SLGLALLESGTGEFPYTANEGPVNLMLQILDDPSPSPSKEKFSPEFCSFVDACLQKDPDN 327
SLG+ ++E GE P A+ P+ ++ I+ +P E FS + FV CL+K P
Sbjct: 197 SLGITVIEMAKGE-PPLADLHPMRVLF-IIPRETPPQLDEHFSRQVKEFVSLCLKKAPAE 254
Query: 328 RPTAEQLLLHPFI 340
RP+A++L+ H FI
Sbjct: 255 RPSAKELIKHRFI 267
>At1g69220 serine/threonine kinase (SIK1)
Length = 836
Score = 147 bits (371), Expect = 1e-35
Identities = 89/261 (34%), Positives = 153/261 (58%), Gaps = 12/261 (4%)
Query: 90 IGSGASSVVQRAMHIPTHRVIALKKINIFEKEKR-QQLLTEIRTL--CEAPCYEGLVEFH 146
+G G+ V +A + T ++A+K I++ E E+ +++ EI L C P +V +
Sbjct: 255 LGKGSYGSVYKARDLKTSEIVAVKVISLTEGEEGYEEIRGEIEMLQQCNHP---NVVRYL 311
Query: 147 GAFYTPDSGQISIALEYMDGGSLADILRM-HRTIPEPILSSMFQKLLRGLSYLHGVRYLV 205
G++ D + I +EY GGS+AD++ + + E ++ + ++ L+GL+YLH + Y V
Sbjct: 312 GSYQGED--YLWIVMEYCGGGSVADLMNVTEEALEEYQIAYICREALKGLAYLHSI-YKV 368
Query: 206 HRDIKPANLLVNLKGEPKITDFGISAGLENSVAMCATFVGTVTYMSPERIRNESYSYPAD 265
HRDIK N+L+ +GE K+ DFG++A L +++ TF+GT +M+PE I+ Y D
Sbjct: 369 HRDIKGGNILLTEQGEVKLGDFGVAAQLTRTMSKRNTFIGTPHWMAPEVIQENRYDGKVD 428
Query: 266 IWSLGLALLESGTGEFPYTANEGPVNLMLQILDDPSPS-PSKEKFSPEFCSFVDACLQKD 324
+W+LG++ +E G P ++ P+ ++ I +P+P KEK+S F FV CL K+
Sbjct: 429 VWALGVSAIEMAEG-LPPRSSVHPMRVLFMISIEPAPMLEDKEKWSLVFHDFVAKCLTKE 487
Query: 325 PDNRPTAEQLLLHPFITKYET 345
P RPTA ++L H F+ + +T
Sbjct: 488 PRLRPTAAEMLKHKFVERCKT 508
>At1g53570 MEK kinase MAP3Ka, putative
Length = 609
Score = 142 bits (359), Expect = 3e-34
Identities = 86/257 (33%), Positives = 152/257 (58%), Gaps = 12/257 (4%)
Query: 90 IGSGASSVVQRAMHIPTHRVIALKKINIFE-----KEKRQQLLTEIRTLCEAPCYEGLVE 144
+GSG V + ++ A+K++ + KE +QL EI L + C+ +V+
Sbjct: 220 LGSGTFGQVYLGFNSEKGKMCAIKEVKVISDDQTSKECLKQLNQEINLLNQL-CHPNIVQ 278
Query: 145 FHGAFYTPDSGQISIALEYMDGGSLADILRMHRTIPEPILSSMFQKLLRGLSYLHGVRYL 204
++G+ + ++ +S+ LEY+ GGS+ +L+ + + EP++ + +++L GL+YLHG R
Sbjct: 279 YYGSELSEET--LSVYLEYVSGGSIHKLLKDYGSFTEPVIQNYTRQILAGLAYLHG-RNT 335
Query: 205 VHRDIKPANLLVNLKGEPKITDFGISAGLENSVAMCATFVGTVTYMSPERIRNES-YSYP 263
VHRDIK AN+LV+ GE K+ DFG++ + + + +F G+ +M+PE + +++ Y++
Sbjct: 336 VHRDIKGANILVDPNGEIKLADFGMAKHV-TAFSTMLSFKGSPYWMAPEVVMSQNGYTHA 394
Query: 264 ADIWSLGLALLESGTGEFPYTANEGPVNLMLQILDDPSPSPSKEKFSPEFCSFVDACLQK 323
DIWSLG +LE T + P++ EG V + +I + + S + +F+ CLQ+
Sbjct: 395 VDIWSLGCTILEMATSKPPWSQFEG-VAAIFKIGNSKDTPEIPDHLSNDAKNFIRLCLQR 453
Query: 324 DPDNRPTAEQLLLHPFI 340
+P RPTA QLL HPF+
Sbjct: 454 NPTVRPTASQLLEHPFL 470
>At1g54960 NPK1-related protein kinase 2 (ANP2)
Length = 651
Score = 141 bits (355), Expect = 1e-33
Identities = 90/282 (31%), Positives = 152/282 (52%), Gaps = 14/282 (4%)
Query: 90 IGSGASSVVQRAMHIPTHRVIALKKI----NIFEKEKRQQLLTEIRT---LCEAPCYEGL 142
IG GA V M++ + ++A+K++ N KEK Q + E+ L + + +
Sbjct: 74 IGRGAFGTVYMGMNLDSGELLAVKQVLITSNCASKEKTQAHIQELEEEVKLLKNLSHPNI 133
Query: 143 VEFHGAFYTPDSGQISIALEYMDGGSLADILRMHRTIPEPILSSMFQKLLRGLSYLHGVR 202
V + G ++ ++I LE++ GGS++ +L PE ++ + +LL GL YLH
Sbjct: 134 VRYLGTVREDET--LNILLEFVPGGSISSLLEKFGAFPESVVRTYTNQLLLGLEYLHN-H 190
Query: 203 YLVHRDIKPANLLVNLKGEPKITDFGISAGLEN--SVAMCATFVGTVTYMSPERIRNESY 260
++HRDIK AN+LV+ +G K+ DFG S + +++ + GT +M+PE I +
Sbjct: 191 AIMHRDIKGANILVDNQGCIKLADFGASKQVAELATISGAKSMKGTPYWMAPEVILQTGH 250
Query: 261 SYPADIWSLGLALLESGTGEFPYTANEGPVNLMLQILDDPSPSPSKEKFSPEFCSFVDAC 320
S+ ADIWS+G ++E TG+ P++ + + I S P + S + F+ C
Sbjct: 251 SFSADIWSVGCTVIEMVTGKAPWSQQYKEIAAIFHIGTTKSHPPIPDNISSDANDFLLKC 310
Query: 321 LQKDPDNRPTAEQLLLHPFIT--KYETAKVDLPGFVRSVFDP 360
LQ++P+ RPTA +LL HPF+T + E+A DL F+ + P
Sbjct: 311 LQQEPNLRPTASELLKHPFVTGKQKESASKDLTSFMDNSCSP 352
>At5g14720 protein kinase -like protein
Length = 674
Score = 139 bits (351), Expect = 3e-33
Identities = 90/279 (32%), Positives = 151/279 (53%), Gaps = 19/279 (6%)
Query: 73 NEKTYRCGSHEMRIFGAIGSGASSVVQRAMHIPTHRVIALKKINIFEKEKRQQLLTEIRT 132
+EK + + + +++ IG G S+ V RA+ IP + V+A+K +++ EK L IR
Sbjct: 5 SEKKFPLNAKDYKLYEEIGDGVSATVHRALCIPLNVVVAIKVLDL---EKCNNDLDGIRR 61
Query: 133 LCEAPC---YEGLVEFHGAFYTPDSGQISIALEYMDGGSLADILRMHRT--IPEPILSSM 187
+ + +++ H +F T Q+ + + YM GGS I++ EP+++++
Sbjct: 62 EVQTMSLINHPNVLQAHCSFTT--GHQLWVVMPYMAGGSCLHIIKSSYPDGFEEPVIATL 119
Query: 188 FQKLLRGLSYLHGVRYLVHRDIKPANLLVNLKGEPKITDFGISAGLENS---VAMCATFV 244
++ L+ L YLH + +HRD+K N+L++ G K+ DFG+SA + ++ TFV
Sbjct: 120 LRETLKALVYLHAHGH-IHRDVKAGNILLDSNGAVKLADFGVSACMFDTGDRQRSRNTFV 178
Query: 245 GTVTYMSPERIRN-ESYSYPADIWSLGLALLESGTGEFPYTANEGPVNLMLQILDDPSPS 303
GT +M+PE ++ Y + AD+WS G+ LE G P++ P+ ++L L + P
Sbjct: 179 GTPCWMAPEVMQQLHGYDFKADVWSFGITALELAHGHAPFSKYP-PMKVLLMTLQNAPPG 237
Query: 304 PSKEK---FSPEFCSFVDACLQKDPDNRPTAEQLLLHPF 339
E+ FS F V CL KDP RPT+E+LL HPF
Sbjct: 238 LDYERDKRFSKAFKEMVGTCLVKDPKKRPTSEKLLKHPF 276
>At1g09000 NPK1-related protein kinase 1S (ANP1)
Length = 666
Score = 138 bits (347), Expect = 8e-33
Identities = 92/282 (32%), Positives = 148/282 (51%), Gaps = 14/282 (4%)
Query: 90 IGSGASSVVQRAMHIPTHRVIALKKI----NIFEKEKRQQLLTEIRT---LCEAPCYEGL 142
IG GA V M++ + ++A+K++ N KEK Q + E+ L + + +
Sbjct: 75 IGRGAFGTVYMGMNLDSGELLAVKQVLIAANFASKEKTQAHIQELEEEVKLLKNLSHPNI 134
Query: 143 VEFHGAFYTPDSGQISIALEYMDGGSLADILRMHRTIPEPILSSMFQKLLRGLSYLHGVR 202
V + G D+ ++I LE++ GGS++ +L PE ++ + ++LL GL YLH
Sbjct: 135 VRYLGTVREDDT--LNILLEFVPGGSISSLLEKFGPFPESVVRTYTRQLLLGLEYLHN-H 191
Query: 203 YLVHRDIKPANLLVNLKGEPKITDFGISAGLENSVAMCA--TFVGTVTYMSPERIRNESY 260
++HRDIK AN+LV+ KG K+ DFG S + M + GT +M+PE I +
Sbjct: 192 AIMHRDIKGANILVDNKGCIKLADFGASKQVAELATMTGAKSMKGTPYWMAPEVILQTGH 251
Query: 261 SYPADIWSLGLALLESGTGEFPYTANEGPVNLMLQILDDPSPSPSKEKFSPEFCSFVDAC 320
S+ ADIWS+G ++E TG+ P++ V + I S P + S + F+ C
Sbjct: 252 SFSADIWSVGCTVIEMVTGKAPWSQQYKEVAAIFFIGTTKSHPPIPDTLSSDAKDFLLKC 311
Query: 321 LQKDPDNRPTAEQLLLHPFI--TKYETAKVDLPGFVRSVFDP 360
LQ+ P+ RPTA +LL HPF+ E+A DL + ++ P
Sbjct: 312 LQEVPNLRPTASELLKHPFVMGKHKESASTDLGSVLNNLSTP 353
>At3g06030 NPK1-related protein kinase 3
Length = 651
Score = 135 bits (340), Expect = 5e-32
Identities = 88/261 (33%), Positives = 141/261 (53%), Gaps = 12/261 (4%)
Query: 90 IGSGASSVVQRAMHIPTHRVIALKKINIF----EKEKRQQLLTEIRT---LCEAPCYEGL 142
IG GA V M++ + ++A+K++ I KEK Q + E+ L + + +
Sbjct: 74 IGCGAFGRVYMGMNLDSGELLAIKQVLIAPSSASKEKTQGHIRELEEEVQLLKNLSHPNI 133
Query: 143 VEFHGAFYTPDSGQISIALEYMDGGSLADILRMHRTIPEPILSSMFQKLLRGLSYLHGVR 202
V + G DS ++I +E++ GGS++ +L + PEP++ ++LL GL YLH
Sbjct: 134 VRYLGTVRESDS--LNILMEFVPGGSISSLLEKFGSFPEPVIIMYTKQLLLGLEYLHN-N 190
Query: 203 YLVHRDIKPANLLVNLKGEPKITDFGISAGLEN--SVAMCATFVGTVTYMSPERIRNESY 260
++HRDIK AN+LV+ KG ++ DFG S + +V + GT +M+PE I +
Sbjct: 191 GIMHRDIKGANILVDNKGCIRLADFGASKKVVELATVNGAKSMKGTPYWMAPEVILQTGH 250
Query: 261 SYPADIWSLGLALLESGTGEFPYTANEGPVNLMLQILDDPSPSPSKEKFSPEFCSFVDAC 320
S+ ADIWS+G ++E TG+ P++ +L I + P E SPE F+ C
Sbjct: 251 SFSADIWSVGCTVIEMATGKPPWSEQYQQFAAVLHIGRTKAHPPIPEDLSPEAKDFLMKC 310
Query: 321 LQKDPDNRPTAEQLLLHPFIT 341
L K+P R +A +LL HPF+T
Sbjct: 311 LHKEPSLRLSATELLQHPFVT 331
>At3g06230 putative MAP kinase
Length = 293
Score = 127 bits (320), Expect = 1e-29
Identities = 89/249 (35%), Positives = 134/249 (53%), Gaps = 24/249 (9%)
Query: 90 IGSGASSVVQRAMHIPTHRVIALKKINI-FEKEKRQQLLTEIRTLCEAPCYEGLVEFHGA 148
+GSG V + T + ALKK+ ++ +++ EI + +P + + H
Sbjct: 59 LGSGNGGTVFKVKDKTTSEIYALKKVKENWDSTSLREI--EILRMVNSPY---VAKCHDI 113
Query: 149 FYTPDSGQISIALEYMDGGSLADILRMHRTIPEPILSSMFQKLLRGLSYLHGVRYLVHRD 208
F P SG++SI ++YMD GSL + R + E L+ M +++L G +YLH + +VHRD
Sbjct: 114 FQNP-SGEVSILMDYMDLGSLESL----RGVTEKQLALMSRQVLEGKNYLHEHK-IVHRD 167
Query: 209 IKPANLLVNLKGEPKITDFGISAGLENSVAMCATFVGTVTYMSPERIRNE---------S 259
IKPANLL + K E KI DFG+S + S+ C +FVGT YMSPER+ +E S
Sbjct: 168 IKPANLLRSSKEEVKIADFGVSKIVVRSLNKCNSFVGTFAYMSPERLDSEADGVTEEDKS 227
Query: 260 YSYPADIWSLGLALLESGTGEFPYTANEGPVNLMLQILDDPSPSPSKEKFSPEFCSFVDA 319
Y DIWS GL +LE G +P ++ + + +P +P E+ S + SF+D
Sbjct: 228 NVYAGDIWSFGLTMLEILVGYYPMLPDQAAI-VCAVCFGEPPKAP--EECSDDLKSFMDC 284
Query: 320 CLQKDPDNR 328
CL+K R
Sbjct: 285 CLRKKASER 293
>At1g63700 putative protein kinase
Length = 883
Score = 125 bits (314), Expect = 6e-29
Identities = 88/257 (34%), Positives = 142/257 (55%), Gaps = 12/257 (4%)
Query: 90 IGSGASSVVQRAMHIPTHRVIALKKINIFE-----KEKRQQLLTEIRTLCEAPCYEGLVE 144
+G G+ V + + + A+K++ + +E QQL EI L ++ +V+
Sbjct: 406 LGMGSFGHVYLGFNSESGEMCAMKEVTLCSDDPKSRESAQQLGQEISVLSRLR-HQNIVQ 464
Query: 145 FHGAFYTPDSGQISIALEYMDGGSLADILRMHRTIPEPILSSMFQKLLRGLSYLHGVRYL 204
++G+ D ++ I LEY+ GGS+ +L+ + E + + Q++L GL+YLH +
Sbjct: 465 YYGSETVDD--KLYIYLEYVSGGSIYKLLQEYGQFGENAIRNYTQQILSGLAYLHA-KNT 521
Query: 205 VHRDIKPANLLVNLKGEPKITDFGISAGLENSVAMCATFVGTVTYMSPERIRNESYSYPA 264
VHRDIK AN+LV+ G K+ DFG++ + + + +F G+ +M+PE I+N + S A
Sbjct: 522 VHRDIKGANILVDPHGRVKVADFGMAKHI-TAQSGPLSFKGSPYWMAPEVIKNSNGSNLA 580
Query: 265 -DIWSLGLALLESGTGEFPYTANEGPVNLMLQILDDPSPSPSKEKFSPEFCSFVDACLQK 323
DIWSLG +LE T + P++ EG V M +I + + S E FV CLQ+
Sbjct: 581 VDIWSLGCTVLEMATTKPPWSQYEG-VPAMFKIGNSKELPDIPDHLSEEGKDFVRKCLQR 639
Query: 324 DPDNRPTAEQLLLHPFI 340
+P NRPTA QLL H F+
Sbjct: 640 NPANRPTAAQLLDHAFV 656
>At5g66850 MAP protein kinase like protein
Length = 533
Score = 122 bits (306), Expect = 5e-28
Identities = 88/264 (33%), Positives = 139/264 (52%), Gaps = 21/264 (7%)
Query: 90 IGSGASSVVQRAMHIPTHRVIALKKINIFEKEKR-----QQLLTEIRTLCEAPCYEGLVE 144
IG G V A + T + A+K++ +F + + +QL EI+ L + +V+
Sbjct: 169 IGRGTFGSVYVASNSETGALCAMKEVELFPDDPKSAECIKQLEQEIKLLSNLQ-HPNIVQ 227
Query: 145 FHGAFYTPDSGQISIALEYMDGGSLADILRMH-RTIPEPILSSMFQKLLRGLSYLHGVRY 203
+ G+ D + I LEY+ GS+ +R H T+ E ++ + + +L GL+YLH +
Sbjct: 228 YFGSETVED--RFFIYLEYVHPGSINKYIRDHCGTMTESVVRNFTRHILSGLAYLHNKK- 284
Query: 204 LVHRDIKPANLLVNLKGEPKITDFGISAGLENSVAMCATFVGTVTYMSPERIR------- 256
VHRDIK ANLLV+ G K+ DFG++ L A + G+ +M+PE ++
Sbjct: 285 TVHRDIKGANLLVDASGVVKLADFGMAKHLTGQRADLS-LKGSPYWMAPELMQAVMQKDS 343
Query: 257 NESYSYPADIWSLGLALLESGTGEFPYTANEGPVNLMLQILDDPSPSPSKEKFSPEFCSF 316
N ++ DIWSLG ++E TG+ P++ EG M +++ D P P E SPE F
Sbjct: 344 NPDLAFAVDIWSLGCTIIEMFTGKPPWSEFEGAA-AMFKVMRDSPPIP--ESMSPEGKDF 400
Query: 317 VDACLQKDPDNRPTAEQLLLHPFI 340
+ C Q++P RPTA LL H F+
Sbjct: 401 LRLCFQRNPAERPTASMLLEHRFL 424
>At3g61960 serine/threonine-protein kinase-like protein
Length = 626
Score = 122 bits (305), Expect = 6e-28
Identities = 84/259 (32%), Positives = 131/259 (50%), Gaps = 14/259 (5%)
Query: 90 IGSGASSVVQRAMHIPTHRVIALKKIN--IFEKEKRQQLLTEIRTLCEAPCYEGLVEFHG 147
IGSG+ +VV A H + +A+K+I+ + + R LL EI L + ++ F+
Sbjct: 16 IGSGSFAVVWLAKHRSSGLEVAVKEIDKKLLSPKVRDNLLKEISILSTID-HPNIIRFYE 74
Query: 148 AFYTPDSGQISIALEYMDGGSLADILRMHRTIPEPILSSMFQKLLRGLSYLHGVRYLVHR 207
A T D +I + LEY GG LA + H +PE + ++L GL L ++ +HR
Sbjct: 75 AIETGD--RIFLVLEYCSGGDLAGYINRHGKVPEAVAKHFMRQLALGLQVLQE-KHFIHR 131
Query: 208 DIKPANLLVNLKGEP---KITDFGISAGLENSVAMCATFVGTVTYMSPERIRNESYSYPA 264
D+KP NLL++ K KI DFG + L +M TF G+ YM+PE IRN+ Y A
Sbjct: 132 DLKPQNLLLSSKEVTPLLKIGDFGFARSLTPE-SMAETFCGSPLYMAPEIIRNQKYDAKA 190
Query: 265 DIWSLGLALLESGTGEFPYTANEGPVNLMLQILDDPS---PSPSKEKFSPEFCSFVDACL 321
D+WS G L + TG+ P+ N + L I+ D P ++ + P+ + L
Sbjct: 191 DLWSAGAILFQLVTGKPPFDGN-NHIQLFHNIVRDTELKFPEDTRNEIHPDCVDLCRSLL 249
Query: 322 QKDPDNRPTAEQLLLHPFI 340
+++P R T + H F+
Sbjct: 250 RRNPIERLTFREFFNHMFL 268
>At3g13530 MAP3K epsilon protein kinase
Length = 1368
Score = 122 bits (305), Expect = 6e-28
Identities = 80/255 (31%), Positives = 139/255 (54%), Gaps = 10/255 (3%)
Query: 90 IGSGASSVVQRAMHIPTHRVIALKKINI--FEKEKRQQLLTEIRTLCEAPCYEGLVEFHG 147
IG GA V + + + +A+K++++ +E ++ EI L ++ +V++ G
Sbjct: 26 IGKGAYGRVYKGLDLENGDFVAIKQVSLENIVQEDLNTIMQEIDLLKNLN-HKNIVKYLG 84
Query: 148 AFYTPDSGQISIALEYMDGGSLADILRMHR--TIPEPILSSMFQKLLRGLSYLHGVRYLV 205
+ T + I LEY++ GSLA+I++ ++ PE +++ ++L GL YLH + ++
Sbjct: 85 SSKTKT--HLHIILEYVENGSLANIIKPNKFGPFPESLVAVYIAQVLEGLVYLHE-QGVI 141
Query: 206 HRDIKPANLLVNLKGEPKITDFGISAGLENSVAMCATFVGTVTYMSPERIRNESYSYPAD 265
HRDIK AN+L +G K+ DFG++ L + + VGT +M+PE I +D
Sbjct: 142 HRDIKGANILTTKEGLVKLADFGVATKLNEADVNTHSVVGTPYWMAPEVIEMSGVCAASD 201
Query: 266 IWSLGLALLESGTGEFPYTANEGPVNLMLQILDDPSPSPSKEKFSPEFCSFVDACLQKDP 325
IWS+G ++E T PY + P+ + +I+ D +P P + SP+ F+ C +KD
Sbjct: 202 IWSVGCTVIELLTCVPPYYDLQ-PMPALFRIVQDDNP-PIPDSLSPDITDFLRQCFKKDS 259
Query: 326 DNRPTAEQLLLHPFI 340
RP A+ LL HP+I
Sbjct: 260 RQRPDAKTLLSHPWI 274
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.138 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,398,778
Number of Sequences: 26719
Number of extensions: 563932
Number of successful extensions: 3575
Number of sequences better than 10.0: 992
Number of HSP's better than 10.0 without gapping: 797
Number of HSP's successfully gapped in prelim test: 195
Number of HSP's that attempted gapping in prelim test: 1351
Number of HSP's gapped (non-prelim): 1163
length of query: 519
length of database: 11,318,596
effective HSP length: 104
effective length of query: 415
effective length of database: 8,539,820
effective search space: 3544025300
effective search space used: 3544025300
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 62 (28.5 bits)
Medicago: description of AC146560.10


