

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146559.6 + phase: 0
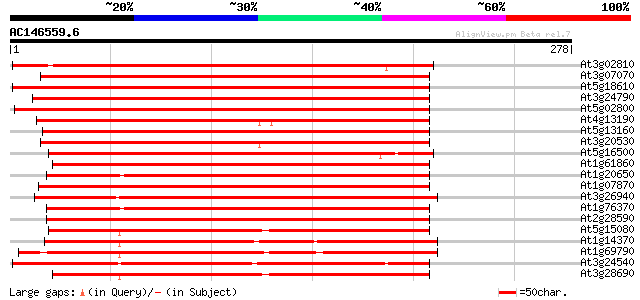
(278 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g02810 putative protein kinase 254 5e-68
At3g07070 putative protein kinase 253 1e-67
At5g18610 protein kinase -like protein 251 2e-67
At3g24790 protein kinase, putative 247 6e-66
At5g02800 protein kinase - like 240 7e-64
At4g13190 putative protein 237 6e-63
At5g13160 protein kinase-like 236 1e-62
At3g20530 protein kinase, putative 235 2e-62
At5g16500 protein kinase-like protein 232 2e-61
At1g61860 hypothetical protein 224 5e-59
At1g20650 unknown protein 223 7e-59
At1g07870 putative protein kinase (At1g07870) 218 3e-57
At3g26940 protein kinase, putative 216 1e-56
At1g76370 putative protein kinase 211 3e-55
At2g28590 putative protein kinase 209 1e-54
At5g15080 serine/threonine specific protein kinase -like 192 2e-49
At1g14370 putative protein 192 2e-49
At1g69790 protein kinase like protein 190 8e-49
At3g24540 protein kinase, putative 189 1e-48
At3g28690 protein kinase like protein 189 1e-48
>At3g02810 putative protein kinase
Length = 558
Score = 254 bits (648), Expect = 5e-68
Identities = 129/211 (61%), Positives = 153/211 (72%), Gaps = 4/211 (1%)
Query: 2 KKQRADEQMQGDPTNINAQNFTFRELATATKNFRQECLLSEGGFGRVYKGVIPATGQVVA 61
+ + +E Q + T++ FTFRELATATKNFRQECLL EGGFGRVYKG + +TGQVVA
Sbjct: 34 RAEETEEIEQSEGTSLKI--FTFRELATATKNFRQECLLGEGGFGRVYKGTLKSTGQVVA 91
Query: 62 VKQLDRHGTENSKEFLTEVSLLSHVHHENLVNLIGYCADGDQRLLVYEYFPGTTLEDRLF 121
VKQLD+HG +KEF EV L + H NLV LIGYCADGDQRLLVY+Y G +L+D L
Sbjct: 92 VKQLDKHGLHGNKEFQAEVLSLGQLDHPNLVKLIGYCADGDQRLLVYDYISGGSLQDHLH 151
Query: 122 ENKTDEPPLNWFDRMKVAEAASKGLEYLHDSANPPIIYRDFKAFNILLDVDLNAKLYDFG 181
E K D P++W RM++A AA++GL+YLHD ANPP+IYRD KA NILLD D + KL DFG
Sbjct: 152 EPKADSDPMDWTTRMQIAYAAAQGLDYLHDKANPPVIYRDLKASNILLDDDFSPKLSDFG 211
Query: 182 MVKF--SGGDKMNNAPPRVMGTYGYLAPEDT 210
+ K GDKM RVMGTYGY APE T
Sbjct: 212 LHKLGPGTGDKMMALSSRVMGTYGYSAPEYT 242
>At3g07070 putative protein kinase
Length = 414
Score = 253 bits (645), Expect = 1e-67
Identities = 123/193 (63%), Positives = 147/193 (75%)
Query: 16 NINAQNFTFRELATATKNFRQECLLSEGGFGRVYKGVIPATGQVVAVKQLDRHGTENSKE 75
NI AQ F+FRELATATKNFRQECL+ EGGFGRVYKG + TG +VAVKQLDR+G + +KE
Sbjct: 61 NIAAQTFSFRELATATKNFRQECLIGEGGFGRVYKGKLEKTGMIVAVKQLDRNGLQGNKE 120
Query: 76 FLTEVSLLSHVHHENLVNLIGYCADGDQRLLVYEYFPGTTLEDRLFENKTDEPPLNWFDR 135
F+ EV +LS +HH++LVNLIGYCADGDQRLLVYEY +LED L + D+ PL+W R
Sbjct: 121 FIVEVLMLSLLHHKHLVNLIGYCADGDQRLLVYEYMSRGSLEDHLLDLTPDQIPLDWDTR 180
Query: 136 MKVAEAASKGLEYLHDSANPPIIYRDFKAFNILLDVDLNAKLYDFGMVKFSGGDKMNNAP 195
+++A A+ GLEYLHD ANPP+IYRD KA NILLD + NAKL DFG+ K +
Sbjct: 181 IRIALGAAMGLEYLHDKANPPVIYRDLKAANILLDGEFNAKLSDFGLAKLGPVGDKQHVS 240
Query: 196 PRVMGTYGYLAPE 208
RVMGTYGY APE
Sbjct: 241 SRVMGTYGYCAPE 253
>At5g18610 protein kinase -like protein
Length = 513
Score = 251 bits (642), Expect = 2e-67
Identities = 125/207 (60%), Positives = 148/207 (71%)
Query: 2 KKQRADEQMQGDPTNINAQNFTFRELATATKNFRQECLLSEGGFGRVYKGVIPATGQVVA 61
+K+ +G +I AQ FTFRELA ATKNFR ECLL EGGFGRVYKG + TGQ+VA
Sbjct: 51 QKKELTAPKEGPTAHIAAQTFTFRELAAATKNFRPECLLGEGGFGRVYKGRLETTGQIVA 110
Query: 62 VKQLDRHGTENSKEFLTEVSLLSHVHHENLVNLIGYCADGDQRLLVYEYFPGTTLEDRLF 121
VKQLDR+G + ++EFL EV +LS +HH NLVNLIGYCADGDQRLLVYEY P +LED L
Sbjct: 111 VKQLDRNGLQGNREFLVEVLMLSLLHHPNLVNLIGYCADGDQRLLVYEYMPLGSLEDHLH 170
Query: 122 ENKTDEPPLNWFDRMKVAEAASKGLEYLHDSANPPIIYRDFKAFNILLDVDLNAKLYDFG 181
+ D+ PL+W RM +A A+KGLEYLHD ANPP+IYRD K+ NILL + KL DFG
Sbjct: 171 DLPPDKEPLDWSTRMTIAAGAAKGLEYLHDKANPPVIYRDLKSSNILLGDGYHPKLSDFG 230
Query: 182 MVKFSGGDKMNNAPPRVMGTYGYLAPE 208
+ K + RVMGTYGY APE
Sbjct: 231 LAKLGPVGDKTHVSTRVMGTYGYCAPE 257
>At3g24790 protein kinase, putative
Length = 379
Score = 247 bits (630), Expect = 6e-66
Identities = 122/197 (61%), Positives = 146/197 (73%)
Query: 12 GDPTNINAQNFTFRELATATKNFRQECLLSEGGFGRVYKGVIPATGQVVAVKQLDRHGTE 71
G N+ A+ FTFRELATATKNFRQECL+ EGGFGRVYKG + QVVAVKQLDR+G +
Sbjct: 41 GPSNNMGARIFTFRELATATKNFRQECLIGEGGFGRVYKGKLENPAQVVAVKQLDRNGLQ 100
Query: 72 NSKEFLTEVSLLSHVHHENLVNLIGYCADGDQRLLVYEYFPGTTLEDRLFENKTDEPPLN 131
+EFL EV +LS +HH NLVNLIGYCADGDQRLLVYEY P +LED L + + + PL+
Sbjct: 101 GQREFLVEVLMLSLLHHRNLVNLIGYCADGDQRLLVYEYMPLGSLEDHLLDLEPGQKPLD 160
Query: 132 WFDRMKVAEAASKGLEYLHDSANPPIIYRDFKAFNILLDVDLNAKLYDFGMVKFSGGDKM 191
W R+K+A A+KG+EYLHD A+PP+IYRD K+ NILLD + AKL DFG+ K
Sbjct: 161 WNTRIKIALGAAKGIEYLHDEADPPVIYRDLKSSNILLDPEYVAKLSDFGLAKLGPVGDT 220
Query: 192 NNAPPRVMGTYGYLAPE 208
+ RVMGTYGY APE
Sbjct: 221 LHVSSRVMGTYGYCAPE 237
>At5g02800 protein kinase - like
Length = 395
Score = 240 bits (612), Expect = 7e-64
Identities = 119/206 (57%), Positives = 142/206 (68%)
Query: 3 KQRADEQMQGDPTNINAQNFTFRELATATKNFRQECLLSEGGFGRVYKGVIPATGQVVAV 62
K E +I AQ FTF ELATAT+NFR+ECL+ EGGFGRVYKG + +T Q A+
Sbjct: 42 KSSLSESKSKGSDHIVAQTFTFSELATATRNFRKECLIGEGGFGRVYKGYLASTSQTAAI 101
Query: 63 KQLDRHGTENSKEFLTEVSLLSHVHHENLVNLIGYCADGDQRLLVYEYFPGTTLEDRLFE 122
KQLD +G + ++EFL EV +LS +HH NLVNLIGYCADGDQRLLVYEY P +LED L +
Sbjct: 102 KQLDHNGLQGNREFLVEVLMLSLLHHPNLVNLIGYCADGDQRLLVYEYMPLGSLEDHLHD 161
Query: 123 NKTDEPPLNWFDRMKVAEAASKGLEYLHDSANPPIIYRDFKAFNILLDVDLNAKLYDFGM 182
+ PL+W RMK+A A+KGLEYLHD PP+IYRD K NILLD D KL DFG+
Sbjct: 162 ISPGKQPLDWNTRMKIAAGAAKGLEYLHDKTMPPVIYRDLKCSNILLDDDYFPKLSDFGL 221
Query: 183 VKFSGGDKMNNAPPRVMGTYGYLAPE 208
K ++ RVMGTYGY APE
Sbjct: 222 AKLGPVGDKSHVSTRVMGTYGYCAPE 247
>At4g13190 putative protein
Length = 405
Score = 237 bits (604), Expect = 6e-63
Identities = 122/211 (57%), Positives = 148/211 (69%), Gaps = 16/211 (7%)
Query: 14 PTNINAQNFTFRELATATKNFRQECLLSEGGFGRVYKGVIPATGQVVAVKQLDRHGTENS 73
P NI A++F FRELATAT +FRQE L+ EGGFGRVYKG + TGQVVAVKQLDR+G + +
Sbjct: 51 PKNIKAKSFKFRELATATNSFRQEFLIGEGGFGRVYKGKMEKTGQVVAVKQLDRNGLQGN 110
Query: 74 KEFLTEVSLLSHVHHENLVNLIGYCADGDQRLLVYEYFPGTTLEDRLFE---------NK 124
+EFL E+ LS +HH NL NLIGYC DGDQRLLV+E+ P +LED L E N
Sbjct: 111 REFLVEIFRLSLLHHPNLANLIGYCLDGDQRLLVHEFMPLGSLEDHLLEFCTIVMELFNY 170
Query: 125 TDEP-------PLNWFDRMKVAEAASKGLEYLHDSANPPIIYRDFKAFNILLDVDLNAKL 177
EP PL+W R+++A A+KGLEYLH+ ANPP+IYRDFK+ NILL+VD +AKL
Sbjct: 171 LIEPDVVVGQQPLDWNSRIRIALGAAKGLEYLHEKANPPVIYRDFKSSNILLNVDFDAKL 230
Query: 178 YDFGMVKFSGGDKMNNAPPRVMGTYGYLAPE 208
DFG+ K N RV+GTYGY APE
Sbjct: 231 SDFGLAKLGSVGDTQNVSSRVVGTYGYCAPE 261
>At5g13160 protein kinase-like
Length = 456
Score = 236 bits (601), Expect = 1e-62
Identities = 117/192 (60%), Positives = 140/192 (71%)
Query: 17 INAQNFTFRELATATKNFRQECLLSEGGFGRVYKGVIPATGQVVAVKQLDRHGTENSKEF 76
I A F FRELA AT NF + L EGGFGRVYKG + +TGQVVAVKQLDR+G + ++EF
Sbjct: 69 IAAHTFAFRELAAATMNFHPDTFLGEGGFGRVYKGRLDSTGQVVAVKQLDRNGLQGNREF 128
Query: 77 LTEVSLLSHVHHENLVNLIGYCADGDQRLLVYEYFPGTTLEDRLFENKTDEPPLNWFDRM 136
L EV +LS +HH NLVNLIGYCADGDQRLLVYE+ P +LED L + D+ L+W RM
Sbjct: 129 LVEVLMLSLLHHPNLVNLIGYCADGDQRLLVYEFMPLGSLEDHLHDLPPDKEALDWNMRM 188
Query: 137 KVAEAASKGLEYLHDSANPPIIYRDFKAFNILLDVDLNAKLYDFGMVKFSGGDKMNNAPP 196
K+A A+KGLE+LHD ANPP+IYRDFK+ NILLD + KL DFG+ K ++
Sbjct: 189 KIAAGAAKGLEFLHDKANPPVIYRDFKSSNILLDEGFHPKLSDFGLAKLGPTGDKSHVST 248
Query: 197 RVMGTYGYLAPE 208
RVMGTYGY APE
Sbjct: 249 RVMGTYGYCAPE 260
>At3g20530 protein kinase, putative
Length = 386
Score = 235 bits (599), Expect = 2e-62
Identities = 118/194 (60%), Positives = 142/194 (72%), Gaps = 1/194 (0%)
Query: 16 NINAQNFTFRELATATKNFRQECLLSEGGFGRVYKGVIPATGQVVAVKQLDRHGTENSKE 75
NI+A FTFREL ATKNF + L EGGFGRVYKG I QVVAVKQLDR+G + ++E
Sbjct: 64 NISAHIFTFRELCVATKNFNPDNQLGEGGFGRVYKGQIETPEQVVAVKQLDRNGYQGNRE 123
Query: 76 FLTEVSLLSHVHHENLVNLIGYCADGDQRLLVYEYFPGTTLEDRLFE-NKTDEPPLNWFD 134
FL EV +LS +HH+NLVNL+GYCADGDQR+LVYEY +LED L E + + PL+W
Sbjct: 124 FLVEVMMLSLLHHQNLVNLVGYCADGDQRILVYEYMQNGSLEDHLLELARNKKKPLDWDT 183
Query: 135 RMKVAEAASKGLEYLHDSANPPIIYRDFKAFNILLDVDLNAKLYDFGMVKFSGGDKMNNA 194
RMKVA A++GLEYLH++A+PP+IYRDFKA NILLD + N KL DFG+ K +
Sbjct: 184 RMKVAAGAARGLEYLHETADPPVIYRDFKASNILLDEEFNPKLSDFGLAKVGPTGGETHV 243
Query: 195 PPRVMGTYGYLAPE 208
RVMGTYGY APE
Sbjct: 244 STRVMGTYGYCAPE 257
>At5g16500 protein kinase-like protein
Length = 636
Score = 232 bits (591), Expect = 2e-61
Identities = 116/193 (60%), Positives = 141/193 (72%), Gaps = 3/193 (1%)
Query: 20 QNFTFRELATATKNFRQECLLSEGGFGRVYKGVIPATGQVVAVKQLDRHGTENSKEFLTE 79
+ F FRELATATKNFRQECLL EGGFGRVYKG + +TGQ+VAVKQLD+HG +KEFL E
Sbjct: 60 KTFNFRELATATKNFRQECLLGEGGFGRVYKGTLQSTGQLVAVKQLDKHGLHGNKEFLAE 119
Query: 80 VSLLSHVHHENLVNLIGYCADGDQRLLVYEYFPGTTLEDRLFENKTDEPPLNWFDRMKVA 139
V L+ + H NLV LIGYCADGDQRLLV+EY G +L+D L+E K + P++W RMK+A
Sbjct: 120 VLSLAKLEHPNLVKLIGYCADGDQRLLVFEYVSGGSLQDHLYEQKPGQKPMDWITRMKIA 179
Query: 140 EAASKGLEYLHDSANPPIIYRDFKAFNILLDVDLNAKLYDFGM--VKFSGGDKMNNAPPR 197
A++GL+YLHD P +IYRD KA NILLD + KL DFG+ ++ GD + R
Sbjct: 180 FGAAQGLDYLHDKVTPAVIYRDLKASNILLDAEFYPKLCDFGLHNLEPGTGDSL-FLSSR 238
Query: 198 VMGTYGYLAPEDT 210
VM TYGY APE T
Sbjct: 239 VMDTYGYSAPEYT 251
>At1g61860 hypothetical protein
Length = 420
Score = 224 bits (570), Expect = 5e-59
Identities = 104/187 (55%), Positives = 134/187 (71%)
Query: 22 FTFRELATATKNFRQECLLSEGGFGRVYKGVIPATGQVVAVKQLDRHGTENSKEFLTEVS 81
F F+EL AT NF +C++ EGGFGRVYKG + + QVVAVK+LDR+G + ++EF EV
Sbjct: 73 FKFKELIAATDNFSMDCMIGEGGFGRVYKGFLTSLNQVVAVKRLDRNGLQGTREFFAEVM 132
Query: 82 LLSHVHHENLVNLIGYCADGDQRLLVYEYFPGTTLEDRLFENKTDEPPLNWFDRMKVAEA 141
+LS H NLVNLIGYC + +QR+LVYE+ P +LED LF+ P L+WF RM++
Sbjct: 133 VLSLAQHPNLVNLIGYCVEDEQRVLVYEFMPNGSLEDHLFDLPEGSPSLDWFTRMRIVHG 192
Query: 142 ASKGLEYLHDSANPPIIYRDFKAFNILLDVDLNAKLYDFGMVKFSGGDKMNNAPPRVMGT 201
A+KGLEYLHD A+PP+IYRDFKA NILL D N+KL DFG+ + + ++ RVMGT
Sbjct: 193 AAKGLEYLHDYADPPVIYRDFKASNILLQSDFNSKLSDFGLARLGPTEGKDHVSTRVMGT 252
Query: 202 YGYLAPE 208
YGY APE
Sbjct: 253 YGYCAPE 259
>At1g20650 unknown protein
Length = 381
Score = 223 bits (569), Expect = 7e-59
Identities = 109/190 (57%), Positives = 142/190 (74%), Gaps = 1/190 (0%)
Query: 19 AQNFTFRELATATKNFRQECLLSEGGFGRVYKGVIPATGQVVAVKQLDRHGTENSKEFLT 78
A++FTF+ELA AT+NFR+ LL EGGFGRVYKG + + GQVVA+KQL+ G + ++EF+
Sbjct: 63 ARSFTFKELAAATRNFREVNLLGEGGFGRVYKGRLDS-GQVVAIKQLNPDGLQGNREFIV 121
Query: 79 EVSLLSHVHHENLVNLIGYCADGDQRLLVYEYFPGTTLEDRLFENKTDEPPLNWFDRMKV 138
EV +LS +HH NLV LIGYC GDQRLLVYEY P +LED LF+ ++++ PL+W RMK+
Sbjct: 122 EVLMLSLLHHPNLVTLIGYCTSGDQRLLVYEYMPMGSLEDHLFDLESNQEPLSWNTRMKI 181
Query: 139 AEAASKGLEYLHDSANPPIIYRDFKAFNILLDVDLNAKLYDFGMVKFSGGDKMNNAPPRV 198
A A++G+EYLH +ANPP+IYRD K+ NILLD + + KL DFG+ K + RV
Sbjct: 182 AVGAARGIEYLHCTANPPVIYRDLKSANILLDKEFSPKLSDFGLAKLGPVGDRTHVSTRV 241
Query: 199 MGTYGYLAPE 208
MGTYGY APE
Sbjct: 242 MGTYGYCAPE 251
>At1g07870 putative protein kinase (At1g07870)
Length = 423
Score = 218 bits (555), Expect = 3e-57
Identities = 108/194 (55%), Positives = 130/194 (66%)
Query: 15 TNINAQNFTFRELATATKNFRQECLLSEGGFGRVYKGVIPATGQVVAVKQLDRHGTENSK 74
T AQ FTF+ELA AT NFR +C L EGGFG+V+KG I QVVA+KQLDR+G + +
Sbjct: 84 TGKKAQTFTFQELAEATGNFRSDCFLGEGGFGKVFKGTIEKLDQVVAIKQLDRNGVQGIR 143
Query: 75 EFLTEVSLLSHVHHENLVNLIGYCADGDQRLLVYEYFPGTTLEDRLFENKTDEPPLNWFD 134
EF+ EV LS H NLV LIG+CA+GDQRLLVYEY P +LED L + + PL+W
Sbjct: 144 EFVVEVLTLSLADHPNLVKLIGFCAEGDQRLLVYEYMPQGSLEDHLHVLPSGKKPLDWNT 203
Query: 135 RMKVAEAASKGLEYLHDSANPPIIYRDFKAFNILLDVDLNAKLYDFGMVKFSGGDKMNNA 194
RMK+A A++GLEYLHD PP+IYRD K NILL D KL DFG+ K +
Sbjct: 204 RMKIAAGAARGLEYLHDRMTPPVIYRDLKCSNILLGEDYQPKLSDFGLAKVGPSGDKTHV 263
Query: 195 PPRVMGTYGYLAPE 208
RVMGTYGY AP+
Sbjct: 264 STRVMGTYGYCAPD 277
>At3g26940 protein kinase, putative
Length = 432
Score = 216 bits (550), Expect = 1e-56
Identities = 107/200 (53%), Positives = 137/200 (68%), Gaps = 1/200 (0%)
Query: 13 DPTNINAQNFTFRELATATKNFRQECLLSEGGFGRVYKGVIPATGQVVAVKQLDRHGTEN 72
D + Q F++RELA AT +FR E L+ GGFG VYKG + +TGQ +AVK LD+ G +
Sbjct: 53 DSSRYRCQIFSYRELAIATNSFRNESLIGRGGFGTVYKGRL-STGQNIAVKMLDQSGIQG 111
Query: 73 SKEFLTEVSLLSHVHHENLVNLIGYCADGDQRLLVYEYFPGTTLEDRLFENKTDEPPLNW 132
KEFL EV +LS +HH NLV+L GYCA+GDQRL+VYEY P ++ED L++ + L+W
Sbjct: 112 DKEFLVEVLMLSLLHHRNLVHLFGYCAEGDQRLVVYEYMPLGSVEDHLYDLSEGQEALDW 171
Query: 133 FDRMKVAEAASKGLEYLHDSANPPIIYRDFKAFNILLDVDLNAKLYDFGMVKFSGGDKMN 192
RMK+A A+KGL +LH+ A PP+IYRD K NILLD D KL DFG+ KF D M+
Sbjct: 172 KTRMKIALGAAKGLAFLHNEAQPPVIYRDLKTSNILLDHDYKPKLSDFGLAKFGPSDDMS 231
Query: 193 NAPPRVMGTYGYLAPEDTGT 212
+ RVMGT+GY APE T
Sbjct: 232 HVSTRVMGTHGYCAPEYANT 251
>At1g76370 putative protein kinase
Length = 381
Score = 211 bits (538), Expect = 3e-55
Identities = 103/190 (54%), Positives = 136/190 (71%), Gaps = 1/190 (0%)
Query: 19 AQNFTFRELATATKNFRQECLLSEGGFGRVYKGVIPATGQVVAVKQLDRHGTENSKEFLT 78
A++FTF+ELA ATKNFR+ ++ +GGFG VYKG + + GQVVA+KQL+ G + ++EF+
Sbjct: 60 ARSFTFKELAAATKNFREGNIIGKGGFGSVYKGRLDS-GQVVAIKQLNPDGHQGNQEFIV 118
Query: 79 EVSLLSHVHHENLVNLIGYCADGDQRLLVYEYFPGTTLEDRLFENKTDEPPLNWFDRMKV 138
EV +LS HH NLV LIGYC G QRLLVYEY P +LED LF+ + D+ PL+W+ RMK+
Sbjct: 119 EVCMLSVFHHPNLVTLIGYCTSGAQRLLVYEYMPMGSLEDHLFDLEPDQTPLSWYTRMKI 178
Query: 139 AEAASKGLEYLHDSANPPIIYRDFKAFNILLDVDLNAKLYDFGMVKFSGGDKMNNAPPRV 198
A A++G+EYLH +P +IYRD K+ NILLD + + KL DFG+ K + RV
Sbjct: 179 AVGAARGIEYLHCKISPSVIYRDLKSANILLDKEFSVKLSDFGLAKVGPVGNRTHVSTRV 238
Query: 199 MGTYGYLAPE 208
MGTYGY APE
Sbjct: 239 MGTYGYCAPE 248
>At2g28590 putative protein kinase
Length = 424
Score = 209 bits (533), Expect = 1e-54
Identities = 102/190 (53%), Positives = 130/190 (67%)
Query: 19 AQNFTFRELATATKNFRQECLLSEGGFGRVYKGVIPATGQVVAVKQLDRHGTENSKEFLT 78
AQ FTF EL+ +T NF+ +C L EGGFG+VYKG I QVVA+KQLDR+G + +EF+
Sbjct: 83 AQTFTFEELSVSTGNFKSDCFLGEGGFGKVYKGFIEKINQVVAIKQLDRNGAQGIREFVV 142
Query: 79 EVSLLSHVHHENLVNLIGYCADGDQRLLVYEYFPGTTLEDRLFENKTDEPPLNWFDRMKV 138
EV LS H NLV LIG+CA+G QRLLVYEY P +L++ L + + + PL W RMK+
Sbjct: 143 EVLTLSLADHPNLVKLIGFCAEGVQRLLVYEYMPLGSLDNHLHDLPSGKNPLAWNTRMKI 202
Query: 139 AEAASKGLEYLHDSANPPIIYRDFKAFNILLDVDLNAKLYDFGMVKFSGGDKMNNAPPRV 198
A A++GLEYLHD+ PP+IYRD K NIL+D +AKL DFG+ K + RV
Sbjct: 203 AAGAARGLEYLHDTMKPPVIYRDLKCSNILIDEGYHAKLSDFGLAKVGPRGSETHVSTRV 262
Query: 199 MGTYGYLAPE 208
MGTYGY AP+
Sbjct: 263 MGTYGYCAPD 272
>At5g15080 serine/threonine specific protein kinase -like
Length = 493
Score = 192 bits (488), Expect = 2e-49
Identities = 101/198 (51%), Positives = 127/198 (64%), Gaps = 12/198 (6%)
Query: 20 QNFTFRELATATKNFRQECLLSEGGFGRVYKGVI---------PATGQVVAVKQLDRHGT 70
+ FTF +L +T+NFR E LL EGGFG V+KG I P TG VAVK L+ G
Sbjct: 128 RKFTFNDLKLSTRNFRPESLLGEGGFGCVFKGWIEENGTAPVKPGTGLTVAVKTLNPDGL 187
Query: 71 ENSKEFLTEVSLLSHVHHENLVNLIGYCADGDQRLLVYEYFPGTTLEDRLFENKTDEPPL 130
+ KE+L E++ L ++ H NLV L+GYC + DQRLLVYE+ P +LE+ LF PL
Sbjct: 188 QGHKEWLAEINFLGNLLHPNLVKLVGYCIEDDQRLLVYEFMPRGSLENHLFRRSL---PL 244
Query: 131 NWFDRMKVAEAASKGLEYLHDSANPPIIYRDFKAFNILLDVDLNAKLYDFGMVKFSGGDK 190
W RMK+A A+KGL +LH+ A P+IYRDFK NILLD D NAKL DFG+ K + +
Sbjct: 245 PWSIRMKIALGAAKGLSFLHEEALKPVIYRDFKTSNILLDADYNAKLSDFGLAKDAPDEG 304
Query: 191 MNNAPPRVMGTYGYLAPE 208
+ RVMGTYGY APE
Sbjct: 305 KTHVSTRVMGTYGYAAPE 322
>At1g14370 putative protein
Length = 426
Score = 192 bits (488), Expect = 2e-49
Identities = 107/204 (52%), Positives = 131/204 (63%), Gaps = 12/204 (5%)
Query: 18 NAQNFTFRELATATKNFRQECLLSEGGFGRVYKGVI---------PATGQVVAVKQLDRH 68
N + FTF EL ATKNFRQ+ LL EGGFG V+KG I P +G VVAVKQL
Sbjct: 70 NLKAFTFNELKNATKNFRQDNLLGEGGFGCVFKGWIDQTSLTASRPGSGIVVAVKQLKPE 129
Query: 69 GTENSKEFLTEVSLLSHVHHENLVNLIGYCADGDQRLLVYEYFPGTTLEDRLFENKTDEP 128
G + KE+LTEV+ L + H NLV L+GYCA+G+ RLLVYE+ P +LE+ LF +
Sbjct: 130 GFQGHKEWLTEVNYLGQLSHPNLVLLVGYCAEGENRLLVYEFMPKGSLENHLF--RRGAQ 187
Query: 129 PLNWFDRMKVAEAASKGLEYLHDSANPPIIYRDFKAFNILLDVDLNAKLYDFGMVKFSGG 188
PL W RMKVA A+KGL +LH+ A +IYRDFKA NILLD D NAKL DFG+ K
Sbjct: 188 PLTWAIRMKVAVGAAKGLTFLHE-AKSQVIYRDFKAANILLDADFNAKLSDFGLAKAGPT 246
Query: 189 DKMNNAPPRVMGTYGYLAPEDTGT 212
+ +V+GT+GY APE T
Sbjct: 247 GDNTHVSTKVIGTHGYAAPEYVAT 270
>At1g69790 protein kinase like protein
Length = 387
Score = 190 bits (482), Expect = 8e-49
Identities = 105/217 (48%), Positives = 137/217 (62%), Gaps = 17/217 (7%)
Query: 5 RADEQMQGDPTNINAQNFTFRELATATKNFRQECLLSEGGFGRVYKGVI---------PA 55
R++ ++ PT + FTF EL TAT+NF+ ++ EGGFG VYKG I P
Sbjct: 58 RSEGELLPSPT---LKAFTFNELKTATRNFKPNSMIGEGGFGCVYKGWIGERSLSPSKPG 114
Query: 56 TGQVVAVKQLDRHGTENSKEFLTEVSLLSHVHHENLVNLIGYCADGDQRLLVYEYFPGTT 115
+G VVAVK+L G + KE+LTEV L +HH NLV LIGYC +G++RLLVYEY P +
Sbjct: 115 SGMVVAVKKLKSEGFQGHKEWLTEVHYLGRLHHMNLVKLIGYCLEGEKRLLVYEYMPKGS 174
Query: 116 LEDRLFENKTDEPPLNWFDRMKVAEAASKGLEYLHDSANPPIIYRDFKAFNILLDVDLNA 175
LE+ LF + P+ W RMKVA +A++GL +LH++ +IYRDFKA NILLDVD NA
Sbjct: 175 LENHLFRRGAE--PIPWKTRMKVAFSAARGLSFLHEA---KVIYRDFKASNILLDVDFNA 229
Query: 176 KLYDFGMVKFSGGDKMNNAPPRVMGTYGYLAPEDTGT 212
KL DFG+ K + +V+GT GY APE T
Sbjct: 230 KLSDFGLAKAGPTGDRTHVTTQVIGTQGYAAPEYIAT 266
>At3g24540 protein kinase, putative
Length = 509
Score = 189 bits (481), Expect = 1e-48
Identities = 101/207 (48%), Positives = 132/207 (62%), Gaps = 4/207 (1%)
Query: 2 KKQRADEQMQGDPTNINAQNFTFRELATATKNFRQECLLSEGGFGRVYKGVIPATGQVVA 61
KK+ D++ P I+ FT+ ELA AT F + LL EGGFG VYKG++ G VA
Sbjct: 147 KKRPRDDKALPAPIGIHQSTFTYGELARATNKFSEANLLGEGGFGFVYKGILN-NGNEVA 205
Query: 62 VKQLDRHGTENSKEFLTEVSLLSHVHHENLVNLIGYCADGDQRLLVYEYFPGTTLEDRLF 121
VKQL + KEF EV+++S +HH NLV+L+GYC G QRLLVYE+ P TLE L
Sbjct: 206 VKQLKVGSAQGEKEFQAEVNIISQIHHRNLVSLVGYCIAGAQRLLVYEFVPNNTLEFHL- 264
Query: 122 ENKTDEPPLNWFDRMKVAEAASKGLEYLHDSANPPIIYRDFKAFNILLDVDLNAKLYDFG 181
+ P + W R+K+A ++SKGL YLH++ NP II+RD KA NIL+D AK+ DFG
Sbjct: 265 -HGKGRPTMEWSLRLKIAVSSSKGLSYLHENCNPKIIHRDIKAANILIDFKFEAKVADFG 323
Query: 182 MVKFSGGDKMNNAPPRVMGTYGYLAPE 208
+ K + D + RVMGT+GYLAPE
Sbjct: 324 LAKIA-LDTNTHVSTRVMGTFGYLAPE 349
>At3g28690 protein kinase like protein
Length = 376
Score = 189 bits (480), Expect = 1e-48
Identities = 100/196 (51%), Positives = 127/196 (64%), Gaps = 12/196 (6%)
Query: 22 FTFRELATATKNFRQECLLSEGGFGRVYKGVI---------PATGQVVAVKQLDRHGTEN 72
F F +L AT+NFR E LL EGGFG V+KG I P TG VAVK L+ G +
Sbjct: 14 FMFNDLKLATRNFRPESLLGEGGFGCVFKGWIEENGTAPVKPGTGLTVAVKTLNPDGLQG 73
Query: 73 SKEFLTEVSLLSHVHHENLVNLIGYCADGDQRLLVYEYFPGTTLEDRLFENKTDEPPLNW 132
KE+L E++ L ++ H +LV L+GYC + DQRLLVYE+ P +LE+ LF PL W
Sbjct: 74 HKEWLAEINFLGNLVHPSLVKLVGYCMEEDQRLLVYEFMPRGSLENHLFRRTL---PLPW 130
Query: 133 FDRMKVAEAASKGLEYLHDSANPPIIYRDFKAFNILLDVDLNAKLYDFGMVKFSGGDKMN 192
RMK+A A+KGL +LH+ A P+IYRDFK NILLD + NAKL DFG+ K + +K +
Sbjct: 131 SVRMKIALGAAKGLAFLHEEAEKPVIYRDFKTSNILLDGEYNAKLSDFGLAKDAPDEKKS 190
Query: 193 NAPPRVMGTYGYLAPE 208
+ RVMGTYGY APE
Sbjct: 191 HVSTRVMGTYGYAAPE 206
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.137 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,699,020
Number of Sequences: 26719
Number of extensions: 227894
Number of successful extensions: 2896
Number of sequences better than 10.0: 936
Number of HSP's better than 10.0 without gapping: 797
Number of HSP's successfully gapped in prelim test: 139
Number of HSP's that attempted gapping in prelim test: 566
Number of HSP's gapped (non-prelim): 966
length of query: 278
length of database: 11,318,596
effective HSP length: 98
effective length of query: 180
effective length of database: 8,700,134
effective search space: 1566024120
effective search space used: 1566024120
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 59 (27.3 bits)
Medicago: description of AC146559.6


