

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146559.11 - phase: 0
(367 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
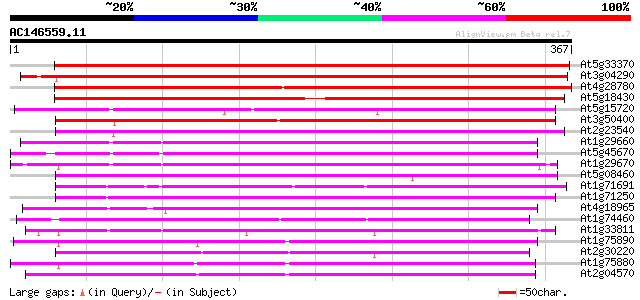
Score E
Sequences producing significant alignments: (bits) Value
At5g33370 unknown protein 511 e-145
At3g04290 GDSL-motif lipase/acylhydrolase like protein 504 e-143
At4g28780 Proline-rich APG - like protein 463 e-131
At5g18430 putative protein 437 e-123
At5g15720 unknown protein 243 1e-64
At3g50400 unknown protein 239 2e-63
At2g23540 putative GDSL-motif lipase/hydrolase 234 7e-62
At1g29660 lipase/hydrolase like protein 234 7e-62
At5g45670 GDSL-motif lipase/hydrolase-like protein 233 2e-61
At1g29670 lipase/hydrolase-like protein 230 8e-61
At5g08460 GDSL-motif lipase/acylhydrolase-like protein 229 2e-60
At1g71691 unknown protein 226 1e-59
At1g71250 putative GDSL-motif lipase/acylhydrolase 225 3e-59
At4g18965 unknown protein 223 2e-58
At1g74460 putative lipase/acylhydrolase 220 1e-57
At1g33811 unknown protein 220 1e-57
At1g75890 family II lipase EXL2 (EXL2) 213 1e-55
At2g30220 putative GDSL-motif lipase/hydrolase 213 1e-55
At1g75880 family II extracellular lipase 1 like protein (EXL1) 212 3e-55
At2g04570 putative GDSL-motif lipase/hydrolase 211 5e-55
>At5g33370 unknown protein
Length = 366
Score = 511 bits (1316), Expect = e-145
Identities = 244/337 (72%), Positives = 284/337 (83%)
Query: 30 RAFFVFGDSVADNGNNNFLTTTARADAPPYGIDFPTHEPTGRFSNGLNIPDLTSERLGLE 89
RAF VFGDS+ DNGNN+FL TTARAD PYGIDFPTH PTGRFSNGLNIPDL SE LG E
Sbjct: 29 RAFLVFGDSLVDNGNNDFLATTARADNYPYGIDFPTHRPTGRFSNGLNIPDLISEHLGQE 88
Query: 90 PSLPYLSPLLVGEKLLVGANFASAGVGILNDTGFQFLQIIHIGKQLDLFNQYQQKLSAQI 149
+PYLSP+L +KLL GANFASAG+GILNDTG QFL II I KQL+ F QY+ ++S +
Sbjct: 89 SPMPYLSPMLKKDKLLRGANFASAGIGILNDTGIQFLNIIRITKQLEYFEQYKVRVSGLV 148
Query: 150 GAEGAKQLVNKAIVLIMLGGNDFVNNYYLVPFSARSRQFSLPNYVTYLISEYKKILQRLY 209
G E +LVN A+VLI LGGNDFVNNYYLVPFSARSRQFSLP+YV ++ISEY+K+L+++Y
Sbjct: 149 GEEEMNRLVNGALVLITLGGNDFVNNYYLVPFSARSRQFSLPDYVVFVISEYRKVLRKMY 208
Query: 210 DLGARRVLVTGTGPMGCAPAELALKSRNGDCDAELMRAASLYNPQLVQMITQLNREIGDD 269
DLGARRVLVTGTGPMGC PAELA +SRNG+C EL RAASL+NPQL+QMIT LN E+G
Sbjct: 209 DLGARRVLVTGTGPMGCVPAELAQRSRNGECATELQRAASLFNPQLIQMITDLNNEVGSS 268
Query: 270 VFIAVNAHKMHMDFITNPKAFGFVTAKDACCGQGRFNGIGLCTPISKLCPNRNLYAFWDA 329
FIA N +MHMDFI++P+A+GFVT+K ACCGQG +NGIGLCTP+S LCPNR+L+AFWD
Sbjct: 269 AFIAANTQQMHMDFISDPQAYGFVTSKVACCGQGPYNGIGLCTPLSNLCPNRDLFAFWDP 328
Query: 330 FHPSEKASRIIVQQMFIGSNLYMNPMNLSTVLAMDSM 366
FHPSEKASRII QQ+ GS YM+PMNLST+L +DSM
Sbjct: 329 FHPSEKASRIIAQQILNGSPEYMHPMNLSTILTVDSM 365
>At3g04290 GDSL-motif lipase/acylhydrolase like protein
Length = 366
Score = 504 bits (1299), Expect = e-143
Identities = 243/361 (67%), Positives = 296/361 (81%), Gaps = 5/361 (1%)
Query: 8 CCSYILMINLFVGFDLAYAQPK---RAFFVFGDSVADNGNNNFLTTTARADAPPYGIDFP 64
C +I+LF F + + P+ RAFFVFGDS+ DNGNN++L TTARAD PYGID+P
Sbjct: 5 CSPLGFLISLF--FIVTFLAPQVKSRAFFVFGDSLVDNGNNDYLVTTARADNYPYGIDYP 62
Query: 65 THEPTGRFSNGLNIPDLTSERLGLEPSLPYLSPLLVGEKLLVGANFASAGVGILNDTGFQ 124
T PTGRFSNGLNIPD+ SE +G+ +LPYLSP L GE LLVGANFASAG+GILNDTG Q
Sbjct: 63 TRRPTGRFSNGLNIPDIISEAIGMPSTLPYLSPHLTGENLLVGANFASAGIGILNDTGIQ 122
Query: 125 FLQIIHIGKQLDLFNQYQQKLSAQIGAEGAKQLVNKAIVLIMLGGNDFVNNYYLVPFSAR 184
F+ II I KQ++ F QYQ ++SA IG E +QLVN+A+VLI LGGNDFVNNYYL+PFSAR
Sbjct: 123 FVNIIRISKQMEYFEQYQLRVSALIGPEATQQLVNQALVLITLGGNDFVNNYYLIPFSAR 182
Query: 185 SRQFSLPNYVTYLISEYKKILQRLYDLGARRVLVTGTGPMGCAPAELALKSRNGDCDAEL 244
SRQ++LP+YV YLISEY KIL++LY+LGARRVLVTGTG MGCAPAELA SRNG+C L
Sbjct: 183 SRQYALPDYVVYLISEYGKILRKLYELGARRVLVTGTGAMGCAPAELAQHSRNGECYGAL 242
Query: 245 MRAASLYNPQLVQMITQLNREIGDDVFIAVNAHKMHMDFITNPKAFGFVTAKDACCGQGR 304
AA+L+NPQLV +I +N EIG DVF+A NA++M+MD+++NP+ FGFVT+K ACCGQG
Sbjct: 243 QTAAALFNPQLVDLIASVNAEIGQDVFVAANAYQMNMDYLSNPEQFGFVTSKVACCGQGP 302
Query: 305 FNGIGLCTPISKLCPNRNLYAFWDAFHPSEKASRIIVQQMFIGSNLYMNPMNLSTVLAMD 364
+NGIGLCTP+S LCPNR+LYAFWDAFHP+EKA+RIIV Q+ GS+ YM+PMNLST + +D
Sbjct: 303 YNGIGLCTPVSNLCPNRDLYAFWDAFHPTEKANRIIVNQILTGSSKYMHPMNLSTAMLLD 362
Query: 365 S 365
S
Sbjct: 363 S 363
>At4g28780 Proline-rich APG - like protein
Length = 367
Score = 463 bits (1191), Expect = e-131
Identities = 223/339 (65%), Positives = 273/339 (79%), Gaps = 2/339 (0%)
Query: 30 RAFFVFGDSVADNGNNNFLTTTARADAPPYGIDFPTHEPTGRFSNGLNIPDLTSERLGLE 89
RAFFVFGDS+ D+GNNN+L TTARAD+PPYGID+PT PTGRFSNGLN+PD+ SE++G E
Sbjct: 30 RAFFVFGDSLVDSGNNNYLVTTARADSPPYGIDYPTGRPTGRFSNGLNLPDIISEQIGSE 89
Query: 90 PSLPYLSPLLVGEKLLVGANFASAGVGILNDTGFQFLQIIHIGKQLDLFNQYQQKLSAQI 149
P+LP LSP L GEKLL+GANFASAG+GILNDTG QFL I+ IG+Q +LF +YQ+++S I
Sbjct: 90 PTLPILSPELTGEKLLIGANFASAGIGILNDTGVQFLNILRIGRQFELFQEYQERVSEII 149
Query: 150 GAEGAKQLVNKAIVLIMLGGNDFVNNYYLVPFSARSRQFSLPNYVTYLISEYKKILQRLY 209
G++ +QLVN A+VL+ LGGNDFVNNY+ P S R RQ SL + LISEYKKIL LY
Sbjct: 150 GSDKTQQLVNGALVLMTLGGNDFVNNYFF-PISTRRRQSSLGEFSQLLISEYKKILTSLY 208
Query: 210 DLGARRVLVTGTGPMGCAPAELALK-SRNGDCDAELMRAASLYNPQLVQMITQLNREIGD 268
+LGARRV+VTGTGP+GC PAELA S NG+C E +AA+++NP LVQM+ LNREIG
Sbjct: 209 ELGARRVMVTGTGPLGCVPAELASSGSVNGECAPEAQQAAAIFNPLLVQMLQGLNREIGS 268
Query: 269 DVFIAVNAHKMHMDFITNPKAFGFVTAKDACCGQGRFNGIGLCTPISKLCPNRNLYAFWD 328
DVFI NA + DFI NP+ FGFVT+K ACCGQG +NG G+CTP+S LC +RN YAFWD
Sbjct: 269 DVFIGANAFNTNADFINNPQRFGFVTSKVACCGQGAYNGQGVCTPLSTLCSDRNAYAFWD 328
Query: 329 AFHPSEKASRIIVQQMFIGSNLYMNPMNLSTVLAMDSMV 367
FHP+EKA+R+IVQQ+ GS YMNPMNLST++A+DS +
Sbjct: 329 PFHPTEKATRLIVQQIMTGSVEYMNPMNLSTIMALDSRI 367
>At5g18430 putative protein
Length = 349
Score = 437 bits (1123), Expect = e-123
Identities = 216/336 (64%), Positives = 263/336 (77%), Gaps = 15/336 (4%)
Query: 30 RAFFVFGDSVADNGNNNFLTTTARADAPPYGIDFPTHEPTGRFSNGLNIPDLTSERLGLE 89
RAFFVFGDS+ D+GNNN+L TTARAD+PPYGIDFPT PTGRFSNGLNIPDL S +G E
Sbjct: 27 RAFFVFGDSLVDSGNNNYLVTTARADSPPYGIDFPTRRPTGRFSNGLNIPDLISYAIGNE 86
Query: 90 -PSLPYLSPLLVGEKLLVGANFASAGVGILNDTGFQFLQIIHIGKQLDLFNQYQQKLSAQ 148
P LPYLSP L G LL GANFASAG+GILNDTGFQF+ II + +QLD F QYQQ++S
Sbjct: 87 EPPLPYLSPELRGRSLLNGANFASAGIGILNDTGFQFINIIRMYQQLDYFQQYQQRVSRL 146
Query: 149 IGAEGAKQLVNKAIVLIMLGGNDFVNNYYLVPFSARSRQFSLPNYVTYLISEYKKILQRL 208
IG ++LV++A+VLI +GGNDFVNNY+L P+SARSRQF+LP+Y RL
Sbjct: 147 IGKPQTQRLVSQALVLITVGGNDFVNNYFLFPYSARSRQFTLPDY-------------RL 193
Query: 209 YDLGARRVLVTGTGPMGCAPAELALK-SRNGDCDAELMRAASLYNPQLVQMITQLNREIG 267
LG RVLVTG GP+GCAPAELA + NG C AEL RAASLY+PQL+QMI +LN++IG
Sbjct: 194 NSLGVGRVLVTGAGPLGCAPAELARSGTSNGRCSAELQRAASLYDPQLLQMINELNKKIG 253
Query: 268 DDVFIAVNAHKMHMDFITNPKAFGFVTAKDACCGQGRFNGIGLCTPISKLCPNRNLYAFW 327
+VFIA N ++M DF++ P+ +GFVT+K ACCGQG +NG+GLCT +S LCPNR LY FW
Sbjct: 254 RNVFIAANTNQMQEDFLSTPRRYGFVTSKVACCGQGPYNGMGLCTVLSNLCPNRELYVFW 313
Query: 328 DAFHPSEKASRIIVQQMFIGSNLYMNPMNLSTVLAM 363
DAFHP+EKA+R+IV+ + G+ YMNPMNLS+ LA+
Sbjct: 314 DAFHPTEKANRMIVRHILTGTTKYMNPMNLSSALAL 349
>At5g15720 unknown protein
Length = 364
Score = 243 bits (620), Expect = 1e-64
Identities = 133/359 (37%), Positives = 203/359 (56%), Gaps = 8/359 (2%)
Query: 4 SLMLCCSYILMINLFVGFDLAYAQPKRAFFVFGDSVADNGNNNFLTTTARADAPPYGIDF 63
SL++C + ++ L G + AFFVFGDS+ D+GNNN++ T ARA+ PYGIDF
Sbjct: 3 SLLICLVLLELVWLGNGQSRDHQPLAPAFFVFGDSLVDSGNNNYIPTLARANYFPYGIDF 62
Query: 64 PTHEPTGRFSNGLNIPDLTSERLGLEPSLPYLSPLLVGEKLLVGANFASAGVGILNDTGF 123
PTGRF NG + D + LGL PYLSPL +G+ L G N+ASA GIL++TG
Sbjct: 63 GF--PTGRFCNGRTVVDYGATYLGLPLVPPYLSPLSIGQNALRGVNYASAAAGILDETGR 120
Query: 124 QFLQIIHIGKQLDLFN---QYQQKLSAQIGAEGAKQLVNKAIVLIMLGGNDFVNNYYLVP 180
+ Q+ F + + + Q A+ K L K+I+ I +G ND++NNY +
Sbjct: 121 HYGARTTFNGQISQFEITIELRLRRFFQNPADLRKYLA-KSIIGINIGSNDYINNYLMPE 179
Query: 181 FSARSRQFSLPNYVTYLISEYKKILQRLYDLGARRVLVTGTGPMGCAPAELALKSRNGD- 239
+ S+ +S +Y LI + RLY+LGAR++++ G+GP+GC P++L++ + N
Sbjct: 180 RYSTSQTYSGEDYADLLIKTLSAQISRLYNLGARKMVLAGSGPLGCIPSQLSMVTGNNTS 239
Query: 240 -CDAELMRAASLYNPQLVQMITQLNREIGDDVFIAVNAHKMHMDFITNPKAFGFVTAKDA 298
C ++ S++N +L + LN + F+ N + D + NP +G V + +A
Sbjct: 240 GCVTKINNMVSMFNSRLKDLANTLNTTLPGSFFVYQNVFDLFHDMVVNPSRYGLVVSNEA 299
Query: 299 CCGQGRFNGIGLCTPISKLCPNRNLYAFWDAFHPSEKASRIIVQQMFIGSNLYMNPMNL 357
CCG GR+ G C P+ + C +RN Y FWDAFHP+E A++II F S Y P+++
Sbjct: 300 CCGNGRYGGALTCLPLQQPCLDRNQYVFWDAFHPTETANKIIAHNTFSKSANYSYPISV 358
>At3g50400 unknown protein
Length = 374
Score = 239 bits (610), Expect = 2e-63
Identities = 135/336 (40%), Positives = 203/336 (60%), Gaps = 10/336 (2%)
Query: 31 AFFVFGDSVADNGNNNFLTTTARADAPPYGIDFPTHE--PTGRFSNGLNIPDLTSERLGL 88
A FVFGDS+ D GNNN+L T +RA++PP GIDF PTGRF+NG I D+ E+LG
Sbjct: 34 ASFVFGDSLVDAGNNNYLQTLSRANSPPNGIDFKPSRGNPTGRFTNGRTIADIVGEKLGQ 93
Query: 89 EP-SLPYLSPLLVGEKLLVGANFASAGVGILNDTGFQFLQIIHIGKQLDLFNQYQQKLSA 147
+ ++PYL+P GE LL G N+AS G GILN TG F+ + + Q+D F +++
Sbjct: 94 QSYAVPYLAPNASGEALLNGVNYASGGGGILNATGSVFVNRLGMDIQVDYFTNTRKQFDK 153
Query: 148 QIGAEGAKQLVNK-AIVLIMLGGNDFVNNYYLVPF-SARSRQFSLP-NYVTYLISEYKKI 204
+G + A+ + K ++ +++G NDF+NN YLVPF +A++R P +V +IS +
Sbjct: 154 LLGQDKARDYIRKRSLFSVVIGSNDFLNN-YLVPFVAAQARLTQTPETFVDDMISHLRNQ 212
Query: 205 LQRLYDLGARRVLVTGTGPMGCAPAELALKSRNG-DCDAELMRAASLYNPQLVQMIT-QL 262
L+RLYD+ AR+ +V P+GC P + ++ N C + A YN +L ++T +L
Sbjct: 213 LKRLYDMDARKFVVGNVAPIGCIPYQKSINQLNDKQCVDLANKLAIQYNARLKDLLTVEL 272
Query: 263 NREIGDDVFIAVNAHKMHMDFITNPKAFGFVTAKDACC-GQGRFNGIGLCTPISKLCPNR 321
+ D F+ N + + MD I N K +GF TA +ACC +GR GI C P S LC +R
Sbjct: 273 KDSLKDAHFVYANVYDLFMDLIVNFKDYGFRTASEACCETRGRLAGILPCGPTSSLCTDR 332
Query: 322 NLYAFWDAFHPSEKASRIIVQQMFIGSNLYMNPMNL 357
+ + FWDA+HP+E A+ +I ++ G + ++ P NL
Sbjct: 333 SKHVFWDAYHPTEAANLLIADKLLYGDSKFVTPFNL 368
>At2g23540 putative GDSL-motif lipase/hydrolase
Length = 387
Score = 234 bits (596), Expect = 7e-62
Identities = 129/340 (37%), Positives = 198/340 (57%), Gaps = 7/340 (2%)
Query: 31 AFFVFGDSVADNGNNNFLTTTARADAPPYGIDFPTH--EPTGRFSNGLNIPDLTSERLG- 87
A F+FGDS+ D GNNN+L+T +RA+ P GIDF PTGRF+NG I D+ E LG
Sbjct: 48 ASFIFGDSLVDAGNNNYLSTLSRANMKPNGIDFKASGGTPTGRFTNGRTIGDIVGEELGS 107
Query: 88 LEPSLPYLSPLLVGEKLLVGANFASAGVGILNDTGFQFLQIIHIGKQLDLFNQYQQKLSA 147
++P+L+P G+ LL G N+AS G GI+N TG F+ + + Q+D FN +++
Sbjct: 108 ANYAIPFLAPDAKGKALLAGVNYASGGGGIMNATGRIFVNRLGMDVQVDFFNTTRKQFDD 167
Query: 148 QIGAEGAKQLV-NKAIVLIMLGGNDFVNNYYLVPFSARSRQFSLPN-YVTYLISEYKKIL 205
+G E AK + K+I I +G NDF+NNY S +R P+ ++ ++ + L
Sbjct: 168 LLGKEKAKDYIAKKSIFSITIGANDFLNNYLFPLLSVGTRFTQTPDDFIGDMLEHLRDQL 227
Query: 206 QRLYDLGARRVLVTGTGPMGCAPAELALKSRN-GDCDAELMRAASLYNPQLVQMITQLNR 264
RLY L AR+ ++ GP+GC P + + + +C + A+ YN +L ++ +LN+
Sbjct: 228 TRLYQLDARKFVIGNVGPIGCIPYQKTINQLDENECVDLANKLANQYNVRLKSLLEELNK 287
Query: 265 EIGDDVFIAVNAHKMHMDFITNPKAFGFVTAKDACCGQG-RFNGIGLCTPISKLCPNRNL 323
++ +F+ N + + M+ ITN +GF +A ACCG G ++ GI C P S LC R+
Sbjct: 288 KLPGAMFVHANVYDLVMELITNYDKYGFKSATKACCGNGGQYAGIIPCGPTSSLCEERDK 347
Query: 324 YAFWDAFHPSEKASRIIVQQMFIGSNLYMNPMNLSTVLAM 363
Y FWD +HPSE A+ II +Q+ G ++P+NLS + M
Sbjct: 348 YVFWDPYHPSEAANVIIAKQLLYGDVKVISPVNLSKLRDM 387
>At1g29660 lipase/hydrolase like protein
Length = 364
Score = 234 bits (596), Expect = 7e-62
Identities = 131/340 (38%), Positives = 191/340 (55%), Gaps = 5/340 (1%)
Query: 8 CCSYILMINLFVGFDLAYAQPKRAFFVFGDSVADNGNNNFLTTTARADAPPYGIDFPTHE 67
C + ++ L +GF + +F+FGDS+ DNGNNN L + ARAD PYGIDF
Sbjct: 9 CLVSVWVLLLGLGFKVKAEPQVPCYFIFGDSLVDNGNNNRLRSIARADYFPYGIDFGG-- 66
Query: 68 PTGRFSNGLNIPDLTSERLGLEPSLPYLSPLLVGEKLLVGANFASAGVGILNDTGFQFLQ 127
PTGRFSNG D+ +E LG + +P S + G+++L G N+ASA GI +TG Q Q
Sbjct: 67 PTGRFSNGRTTVDVLTELLGFDNYIPAYSTVS-GQEILQGVNYASAAAGIREETGAQLGQ 125
Query: 128 IIHIGKQLDLFNQYQQKLSAQIGAE-GAKQLVNKAIVLIMLGGNDFVNNYYLVPFSARSR 186
I Q++ + ++ +G E A + + I + +G ND++NNY++ F + SR
Sbjct: 126 RITFSGQVENYKNTVAQVVEILGDEYTAADYLKRCIYSVGMGSNDYLNNYFMPQFYSTSR 185
Query: 187 QFSLPNYVTYLISEYKKILQRLYDLGARRVLVTGTGPMGCAPAELALKSRNGDCDAELMR 246
Q++ Y LIS Y+ L LY+ GAR+ + G G +GC+P LA S++G E +
Sbjct: 186 QYTPEQYADDLISRYRDQLNALYNYGARKFALVGIGAIGCSPNALAQGSQDGTTCVERIN 245
Query: 247 AAS-LYNPQLVQMITQLNREIGDDVFIAVNAHKMHMDFITNPKAFGFVTAKDACCGQGRF 305
+A+ ++N +L+ M+ QLN D F +NA+ D I NP A+GF ACCG GR
Sbjct: 246 SANRIFNNRLISMVQQLNNAHSDASFTYINAYGAFQDIIANPSAYGFTNTNTACCGIGRN 305
Query: 306 NGIGLCTPISKLCPNRNLYAFWDAFHPSEKASRIIVQQMF 345
G C P C NR+ Y FWDAFHPS A+ I ++ +
Sbjct: 306 GGQLTCLPGEPPCLNRDEYVFWDAFHPSAAANTAIAKRSY 345
>At5g45670 GDSL-motif lipase/hydrolase-like protein
Length = 362
Score = 233 bits (593), Expect = 2e-61
Identities = 136/348 (39%), Positives = 194/348 (55%), Gaps = 11/348 (3%)
Query: 1 MASSLMLCCSYILMINLFVGFDLAYAQPKRAFFVFGDSVADNGNNNFLTTTARADAPPYG 60
M+ +M+ + MIN+ +A +F+FGDS+ DNGNNN L + ARA+ PYG
Sbjct: 4 MSLMIMMIMVAVTMINIAKSDPIA-----PCYFIFGDSLVDNGNNNQLQSLARANYFPYG 58
Query: 61 IDFPTHEPTGRFSNGLNIPDLTSERLGLEPSL-PYLSPLLVGEKLLVGANFASAGVGILN 119
IDF PTGRFSNGL D+ ++ LG E + PY S G+ +L G N+ASA GI +
Sbjct: 59 IDFAAG-PTGRFSNGLTTVDVIAQLLGFEDYITPYASAR--GQDILRGVNYASAAAGIRD 115
Query: 120 DTGFQFL-QIIHIGKQLDLFNQYQQKLSAQIGAEGAKQLVNKAIVLIMLGGNDFVNNYYL 178
+TG Q +I G+ + N Q ++ A ++K I I LG ND++NNY++
Sbjct: 116 ETGRQLGGRIAFAGQVANHVNTVSQVVNILGDQNEASNYLSKCIYSIGLGSNDYLNNYFM 175
Query: 179 VPFSARSRQFSLPNYVTYLISEYKKILQRLYDLGARRVLVTGTGPMGCAPAELALKSRNG 238
F + QFS +Y L++ Y + L+ LY GAR+ + G G +GC+P ELA SR+G
Sbjct: 176 PTFYSTGNQFSPESYADDLVARYTEQLRVLYTNGARKFALIGVGAIGCSPNELAQNSRDG 235
Query: 239 -DCDAELMRAASLYNPQLVQMITQLNREIGDDVFIAVNAHKMHMDFITNPKAFGFVTAKD 297
CD + A ++N +L+ ++ N+ D F +NA+ + D ITNP +GF
Sbjct: 236 RTCDERINSANRIFNSKLISIVDAFNQNTPDAKFTYINAYGIFQDIITNPARYGFRVTNA 295
Query: 298 ACCGQGRFNGIGLCTPISKLCPNRNLYAFWDAFHPSEKASRIIVQQMF 345
CCG GR NG C P C NRN Y FWDAFHP E A+ +I ++ F
Sbjct: 296 GCCGVGRNNGQITCLPGQAPCLNRNEYVFWDAFHPGEAANIVIGRRSF 343
>At1g29670 lipase/hydrolase-like protein
Length = 363
Score = 230 bits (587), Expect = 8e-61
Identities = 141/365 (38%), Positives = 199/365 (53%), Gaps = 14/365 (3%)
Query: 1 MASSLMLCCSYILMINLFVGFDLAYAQPKR---AFFVFGDSVADNGNNNFLTTTARADAP 57
M S L C ++++ L GF + AQ + FFVFGDS+ DNGNNN L + AR++
Sbjct: 1 MESYLTKWC--VVLVLLCFGFSVVKAQAQAQVPCFFVFGDSLVDNGNNNGLISIARSNYF 58
Query: 58 PYGIDFPTHEPTGRFSNGLNIPDLTSERLGLEPSLPYLSPLLVGEKLLVGANFASAGVGI 117
PYGIDF PTGRFSNG D+ +E LG +P + + G ++L G N+ASA GI
Sbjct: 59 PYGIDFGG--PTGRFSNGKTTVDVIAELLGFNGYIPAYNTVS-GRQILSGVNYASAAAGI 115
Query: 118 LNDTGFQFLQIIHIGKQLDLFNQYQQKLSAQIGAEG-AKQLVNKAIVLIMLGGNDFVNNY 176
+TG Q Q I Q+ + ++ +G E A + + I + LG ND++NNY
Sbjct: 116 REETGRQLGQRISFSGQVRNYQTTVSQVVQLLGDETRAADYLKRCIYSVGLGSNDYLNNY 175
Query: 177 YLVPFSARSRQFSLPNYVTYLISEYKKILQRLYDLGARRVLVTGTGPMGCAPAELALKSR 236
++ F + SRQF+ Y LIS Y L LY+ GAR+ ++G G +GC+P LA
Sbjct: 176 FMPTFYSSSRQFTPEQYANDLISRYSTQLNALYNYGARKFALSGIGAVGCSPNALAGSPD 235
Query: 237 NGDCDAELMRAASLYNPQLVQMITQLNREIGDDVFIAVNAHKMHMDFITNPKAFGFVTAK 296
C + A ++N +L ++ QLN D FI +NA+ + D ITNP FGF
Sbjct: 236 GRTCVDRINSANQIFNNKLRSLVDQLNNNHPDAKFIYINAYGIFQDMITNPARFGFRVTN 295
Query: 297 DACCGQGRFNGIGLCTPISKLCPNRNLYAFWDAFHPSEKASRIIVQQMF---IGSNLYMN 353
CCG GR G C P + C +RN Y FWDAFHP+E A+ II ++ + S+ Y
Sbjct: 296 AGCCGIGRNAGQITCLPGQRPCRDRNAYVFWDAFHPTEAANVIIARRSYNAQSASDAY-- 353
Query: 354 PMNLS 358
PM++S
Sbjct: 354 PMDIS 358
>At5g08460 GDSL-motif lipase/acylhydrolase-like protein
Length = 385
Score = 229 bits (583), Expect = 2e-60
Identities = 122/332 (36%), Positives = 187/332 (55%), Gaps = 4/332 (1%)
Query: 31 AFFVFGDSVADNGNNNFLTTTARADAPPYGIDFPTHEPTGRFSNGLNIPDLTSERLGLEP 90
A FVFGDS+ DNGNNN L + AR++ PYGIDF ++PTGRFSNG I D E LGL
Sbjct: 49 AMFVFGDSLVDNGNNNHLNSLARSNYLPYGIDFAGNQPTGRFSNGKTIVDFIGELLGLPE 108
Query: 91 SLPYLSPLLVGEKLLVGANFASAGVGILNDTGFQFLQIIHIGKQLDLFNQYQQKLSAQIG 150
++ + G +L G N+ASA GIL +TG + +G+Q++ F + ++S +
Sbjct: 109 IPAFMDTVDGGVDILHGVNYASAAGGILEETGRHLGERFSMGRQVENFEKTLMEISRSMR 168
Query: 151 AEGAKQLVNKAIVLIMLGGNDFVNNYYLVPFSARSRQFSLPNYVTYLISEYKKILQRLYD 210
E K+ + K++V++ LG ND++NNY S + ++ L+S + L LY
Sbjct: 169 KESVKEYMAKSLVVVSLGNNDYINNYLKPRLFLSSSIYDPTSFADLLLSNFTTHLLELYG 228
Query: 211 LGARRVLVTGTGPMGCAPAELALKSR-NGDCDAELMRAASLYNPQLVQMITQL---NREI 266
G R+ ++ G GP+GC P +LA ++ G+C + A L+N +LV ++ +L N+
Sbjct: 229 KGFRKFVIAGVGPLGCIPDQLAAQAALPGECVEAVNEMAELFNNRLVSLVDRLNSDNKTA 288
Query: 267 GDDVFIAVNAHKMHMDFITNPKAFGFVTAKDACCGQGRFNGIGLCTPISKLCPNRNLYAF 326
+ +F+ N + +D +TNP +GF CCG GR G C P++ C R+ + F
Sbjct: 289 SEAIFVYGNTYGAAVDILTNPFNYGFEVTDRGCCGVGRNRGEITCLPLAVPCAFRDRHVF 348
Query: 327 WDAFHPSEKASRIIVQQMFIGSNLYMNPMNLS 358
WDAFHP++ + II + F GS P+NLS
Sbjct: 349 WDAFHPTQAFNLIIALRAFNGSKSDCYPINLS 380
>At1g71691 unknown protein
Length = 384
Score = 226 bits (577), Expect = 1e-59
Identities = 129/335 (38%), Positives = 187/335 (55%), Gaps = 6/335 (1%)
Query: 31 AFFVFGDSVADNGNNNFLTTTARADAPPYGIDFPTHEPTGRFSNGLNIPDLTSERLGLEP 90
A FVFGDS+ DNGNNN + + A+A+ PYGIDF PTGRF NGL + D ++ LGL P
Sbjct: 55 ALFVFGDSLIDNGNNNNIPSFAKANYFPYGIDF-NGGPTGRFCNGLTMVDGIAQLLGL-P 112
Query: 91 SLPYLSPLLVGEKLLVGANFASAGVGILNDTGFQFLQIIHIGKQLDLFNQ-YQQKLSAQI 149
+P S G+++L G N+ASA GIL DTG F+ I +Q+ F Q S
Sbjct: 113 LIPAYSEA-TGDQVLRGVNYASAAAGILPDTGGNFVGRIPFDQQIHNFETTLDQVASKSG 171
Query: 150 GAEGAKQLVNKAIVLIMLGGNDFVNNYYLVPFSARSRQFSLPNYVTYLISEYKKILQRLY 209
GA V +++ I +G ND++NNY + F R+ Q++ + L+ Y L RLY
Sbjct: 172 GAVAIADSVTRSLFFIGMGSNDYLNNYLMPNFPTRN-QYNSQQFGDLLVQHYTDQLTRLY 230
Query: 210 DLGARRVLVTGTGPMGCAPAELALKSRNGDCDAELMRAASLYNPQLVQMITQLNREIGDD 269
+LG R+ +V G G MGC P+ LA + +G C E+ + +N + MI+ LN+ + D
Sbjct: 231 NLGGRKFVVAGLGRMGCIPSILA-QGNDGKCSEEVNQLVLPFNTNVKTMISNLNQNLPDA 289
Query: 270 VFIAVNAHKMHMDFITNPKAFGFVTAKDACCGQGRFNGIGLCTPISKLCPNRNLYAFWDA 329
FI ++ M D + N A+G T CCG G+ G C P CPNR+ Y FWDA
Sbjct: 290 KFIYLDIAHMFEDIVANQAAYGLTTMDKGCCGIGKNRGQITCLPFETPCPNRDQYVFWDA 349
Query: 330 FHPSEKASRIIVQQMFIGSNLYMNPMNLSTVLAMD 364
FHP+EK + I+ ++ F G P+N+ + +++
Sbjct: 350 FHPTEKVNLIMAKKAFAGDRTVAYPINIQQLASLN 384
>At1g71250 putative GDSL-motif lipase/acylhydrolase
Length = 374
Score = 225 bits (574), Expect = 3e-59
Identities = 116/328 (35%), Positives = 178/328 (53%), Gaps = 2/328 (0%)
Query: 31 AFFVFGDSVADNGNNNFLTTTARADAPPYGIDFPTHEPTGRFSNGLNIPDLTSERLGLEP 90
A FV GDS+ D GNNNFL T ARA+ PYGID ++PTGRFSNGL DL + L +
Sbjct: 41 AMFVLGDSLVDAGNNNFLQTVARANFLPYGIDM-NYQPTGRFSNGLTFIDLLARLLEIPS 99
Query: 91 SLPYLSPLLVGEKLLVGANFASAGVGILNDTGFQFLQIIHIGKQLDLFNQYQQKLSAQIG 150
P+ P G ++L G N+ASA GIL+ +G+ + + +Q+ +L +
Sbjct: 100 PPPFADPTTSGNRILQGVNYASAAAGILDVSGYNYGGRFSLNQQMVNLETTLSQLRTMMS 159
Query: 151 AEGAKQLVNKAIVLIMLGGNDFVNNYYLVPFSARSRQFSLPNYVTYLISEYKKILQRLYD 210
+ + +++V+++ G ND++NNY + S +F P++ L+S+Y + L LY
Sbjct: 160 PQNFTDYLARSLVVLVFGSNDYINNYLMPNLYDSSIRFRPPDFANLLLSQYARQLLTLYS 219
Query: 211 LGARRVLVTGTGPMGCAPAELALKSRNGD-CDAELMRAASLYNPQLVQMITQLNREIGDD 269
LG R++ + G P+GC P + A D C + + +N L ++ QLN+
Sbjct: 220 LGLRKIFIPGVAPLGCIPNQRARGISPPDRCVDSVNQILGTFNQGLKSLVDQLNQRSPGA 279
Query: 270 VFIAVNAHKMHMDFITNPKAFGFVTAKDACCGQGRFNGIGLCTPISKLCPNRNLYAFWDA 329
+++ N + D + NP A+GF ACCG GR G C P+ CPNRN Y FWDA
Sbjct: 280 IYVYGNTYSAIGDILNNPAAYGFSVVDRACCGIGRNQGQITCLPLQTPCPNRNQYVFWDA 339
Query: 330 FHPSEKASRIIVQQMFIGSNLYMNPMNL 357
FHP++ A+ I+ ++ F G P+N+
Sbjct: 340 FHPTQTANSILARRAFYGPPSDAYPVNV 367
>At4g18965 unknown protein
Length = 361
Score = 223 bits (567), Expect = 2e-58
Identities = 127/342 (37%), Positives = 189/342 (55%), Gaps = 9/342 (2%)
Query: 9 CSYILMINLFVGFDLAYAQP-KRAFFVFGDSVADNGNNNFLTTTARADAPPYGIDFPTHE 67
C ++ + + + ++A P +F+FGDS+ D+GNNN LT+ ARA+ PYGIDF +
Sbjct: 5 CVMMMAMAIAMAMNIAMGDPIAPCYFIFGDSLVDSGNNNRLTSLARANYFPYGIDFQ-YG 63
Query: 68 PTGRFSNGLNIPDLTSERLGLEPSLPYLSPLLV--GEKLLVGANFASAGVGILNDTGFQF 125
PTGRFSNG D+ +E LG + Y++P GE +L G N+ASA GI +TG Q
Sbjct: 64 PTGRFSNGKTTVDVITELLGFDD---YITPYSEARGEDILRGVNYASAAAGIREETGRQL 120
Query: 126 -LQIIHIGKQLDLFNQYQQKLSAQIGAEGAKQLVNKAIVLIMLGGNDFVNNYYLVPFSAR 184
+I G+ + N Q ++ A ++K I I LG ND++NNY++ + +
Sbjct: 121 GARITFAGQVANHVNTVSQVVNILGDENEAANYLSKCIYSIGLGSNDYLNNYFMPVYYST 180
Query: 185 SRQFSLPNYVTYLISEYKKILQRLYDLGARRVLVTGTGPMGCAPAELALKSRNG-DCDAE 243
Q+S Y LI+ Y + L+ +Y+ GAR+ + G G +GC+P ELA SR+G CD
Sbjct: 181 GSQYSPDAYANDLINRYTEQLRIMYNNGARKFALVGIGAIGCSPNELAQNSRDGVTCDER 240
Query: 244 LMRAASLYNPQLVQMITQLNREIGDDVFIAVNAHKMHMDFITNPKAFGFVTAKDACCGQG 303
+ A ++N +LV ++ N+ F +NA+ + D + NP +GF CCG G
Sbjct: 241 INSANRIFNSKLVSLVDHFNQNTPGAKFTYINAYGIFQDMVANPSRYGFRVTNAGCCGVG 300
Query: 304 RFNGIGLCTPISKLCPNRNLYAFWDAFHPSEKASRIIVQQMF 345
R NG C P C NR+ Y FWDAFHP E A+ +I + F
Sbjct: 301 RNNGQITCLPGQAPCLNRDEYVFWDAFHPGEAANVVIGSRSF 342
>At1g74460 putative lipase/acylhydrolase
Length = 366
Score = 220 bits (560), Expect = 1e-57
Identities = 123/339 (36%), Positives = 192/339 (56%), Gaps = 10/339 (2%)
Query: 5 LMLCCSYILMINLFV-GFDLAYAQPKRAFFVFGDSVADNGNN-NFLTTTARADAPPYGID 62
+ C ++L I L + G+D Q F+FGDS++D GNN N + A A+ P YGID
Sbjct: 1 MKFCAIFVLFIVLAINGYDCKIVQ-----FIFGDSLSDVGNNKNLPRSLATANLPFYGID 55
Query: 63 FPTHEPTGRFSNGLNIPDLTSERLGLEPSLPYLSPLLVGEKLLV-GANFASAGVGILNDT 121
F P GRF+NG + D+ +++GL + +L P + + +L G N+AS G GILN+T
Sbjct: 56 FGNGLPNGRFTNGRTVSDIIGDKIGLPRPVAFLDPSMNEDVILENGVNYASGGGGILNET 115
Query: 122 GFQFLQIIHIGKQLDLFNQYQQKLSAQIGAEGAKQLVNKAIVLIMLGGNDFVNNYYLVPF 181
G F+Q + KQ++LF Q + A+IG + A + A ++ LG NDF+NNY L+P
Sbjct: 116 GGYFIQRFSLWKQIELFQGTQDVVVAKIGKKEADKFFQDARYVVALGSNDFINNY-LMPV 174
Query: 182 SARSRQFSLPNYVTYLISEYKKILQRLYDLGARRVLVTGTGPMGCAPAELALKSRNGDCD 241
+ S +++ +V YL+ + L+ L+ LGAR+++V G GPMGC P + AL S +G+C
Sbjct: 175 YSDSWKYNDQTFVDYLMETLESQLKVLHSLGARKLMVFGLGPMGCIPLQRAL-SLDGNCQ 233
Query: 242 AELMRAASLYNPQLVQMITQLNREIGDDVFIAVNAHKMHMDFITNPKAFGFVTAKDACCG 301
+ A +N M+ L ++ + + A+ + D ITNPK +GF + CC
Sbjct: 234 NKASNLAKRFNKAATTMLLDLETKLPNASYRFGEAYDLVNDVITNPKKYGFDNSDSPCCS 293
Query: 302 QGRFNGIGLCTPISKLCPNRNLYAFWDAFHPSEKASRII 340
R C P S LC +R+ Y FWD +HP++KA+ ++
Sbjct: 294 FYRIRPALTCIPASTLCKDRSKYVFWDEYHPTDKANELV 332
>At1g33811 unknown protein
Length = 370
Score = 220 bits (560), Expect = 1e-57
Identities = 127/362 (35%), Positives = 203/362 (55%), Gaps = 18/362 (4%)
Query: 11 YILMINL---FVGFDLAYAQPKR-----AFFVFGDSVADNGNNNFLTTTARADAPPYGID 62
++L+I+L GF +QP++ F+FGDS+ DNGNNN L + ARA+ PYGID
Sbjct: 6 FVLLISLNLVLFGFKTTVSQPQQQAQVPCLFIFGDSLVDNGNNNRLLSLARANYRPYGID 65
Query: 63 FPTHEPTGRFSNGLNIPDLTSERLGLEPSLPYLSPLLVGEKLLVGANFASAGVGILNDTG 122
FP TGRF+NG D ++ LG +P S + G+ +L GANFAS GI ++TG
Sbjct: 66 FP-QGTTGRFTNGRTYVDALAQILGFRNYIPPYSRIR-GQAILRGANFASGAAGIRDETG 123
Query: 123 FQFLQIIHIGKQLDLFNQYQQKLSAQIGAEG--AKQLVNKAIVLIMLGGNDFVNNYYLVP 180
+ +Q++L+ Q++ + ++ +++ I +G ND++NNY++
Sbjct: 124 DNLGAHTSMNQQVELYTTAVQQMLRYFRGDTNELQRYLSRCIFYSGMGSNDYLNNYFMPD 183
Query: 181 FSARSRQFSLPNYVTYLISEYKKILQRLYDLGARRVLVTGTGPMGCAPAELA-LKSRN-- 237
F + S ++ + LI Y + L RLY GAR+V+VTG G +GC P +LA +RN
Sbjct: 184 FYSTSTNYNDKTFAESLIKNYTQQLTRLYQFGARKVIVTGVGQIGCIPYQLARYNNRNNS 243
Query: 238 -GDCDAELMRAASLYNPQLVQMITQLNR-EIGDDVFIAVNAHKMHMDFITNPKAFGFVTA 295
G C+ ++ A ++N Q+ +++ +LN+ ++ F+ ++++K D N A+GF
Sbjct: 244 TGRCNEKINNAIVVFNTQVKKLVDRLNKGQLKGAKFVYLDSYKSTYDLAVNGAAYGFEVV 303
Query: 296 KDACCGQGRFNGIGLCTPISKLCPNRNLYAFWDAFHPSEKASRIIVQQMFIGSNLYMNPM 355
CCG GR NG C P+ CP+R Y FWDAFHP+E A+ ++ + F S Y P+
Sbjct: 304 DKGCCGVGRNNGQITCLPLQTPCPDRTKYLFWDAFHPTETANILLAKSNFY-SRAYTYPI 362
Query: 356 NL 357
N+
Sbjct: 363 NI 364
>At1g75890 family II lipase EXL2 (EXL2)
Length = 379
Score = 213 bits (543), Expect = 1e-55
Identities = 124/363 (34%), Positives = 188/363 (51%), Gaps = 22/363 (6%)
Query: 3 SSLMLCCSYILMINLFVGFDLAYAQPKR----AFFVFGDSVADNGNNN-FLTTTARADAP 57
SS C + L++ + QP A VFGDS+ D GNN+ +TT AR + P
Sbjct: 15 SSSPFWCVFFLVLLCKTSTNALVKQPPNETTPAIIVFGDSIVDAGNNDDIMTTLARCNYP 74
Query: 58 PYGIDFPTHEPTGRFSNGLNIPDLTSERLGLEPSLP-YLSPLLVGEKLLVGANFASAGVG 116
PYGIDF PTGRF NG D + + G++PS+P Y +P L E LL G FAS G G
Sbjct: 75 PYGIDFDGGIPTGRFCNGKVATDFIAGKFGIKPSIPAYRNPNLKPEDLLTGVTFASGGAG 134
Query: 117 ILNDT-----------GFQFLQ-IIHIGKQLDLFNQYQQKLSAQIGAEGAKQLVNKAIVL 164
+ T FL+ I + +QL LF +Y +K+ +G E K ++ ++ +
Sbjct: 135 YVPFTTQLSTYLFIYKPLLFLKGGIALSQQLKLFEEYVEKMKKMVGEERTKLIIKNSLFM 194
Query: 165 IMLGGNDFVNNYYLVPFSARSRQFSLPNYVTYLISEYKKILQRLYDLGARRVLVTGTGPM 224
++ G ND N Y+ +P + +Q+ + ++ T + + Q+L++ GARR+ V G P+
Sbjct: 195 VICGSNDITNTYFGLP--SVQQQYDVASFTTLMADNARSFAQKLHEYGARRIQVFGAPPV 252
Query: 225 GCAPAELALK-SRNGDCDAELMRAASLYNPQLVQMITQLNREIGDDVFIAVNAHKMHMDF 283
GC P++ L +C A LYN +L + L+R +GD I V+ + +D
Sbjct: 253 GCVPSQRTLAGGPTRNCVVRFNDATKLYNVKLAANLGSLSRTLGDKTIIYVDIYDSLLDI 312
Query: 284 ITNPKAFGFVTAKDACCGQGRFNGIGLCTPI-SKLCPNRNLYAFWDAFHPSEKASRIIVQ 342
I +P+ +GF CCG G LC + +CPNR+ Y FWD+FHP+EK RI+
Sbjct: 313 ILDPRQYGFKVVDKGCCGTGLIEVALLCNNFAADVCPNRDEYVFWDSFHPTEKTYRIMAT 372
Query: 343 QMF 345
+ F
Sbjct: 373 KYF 375
>At2g30220 putative GDSL-motif lipase/hydrolase
Length = 358
Score = 213 bits (542), Expect = 1e-55
Identities = 113/314 (35%), Positives = 173/314 (54%), Gaps = 6/314 (1%)
Query: 31 AFFVFGDSVADNGNNNFLTTTA-RADAPPYGIDFPTHEPTGRFSNGLNIPDLTSERLGLE 89
A +FGDS AD GNNN+ + +A+ PYG+D P HE GRFSNG I D+ S +L ++
Sbjct: 33 AILIFGDSTADTGNNNYYSQAVFKANHLPYGVDLPGHEANGRFSNGKLISDVISTKLNIK 92
Query: 90 PSLP-YLSPLLVGEKLLVGANFASAGVGILNDTGFQFLQIIHIGKQLDLFNQYQQKLSAQ 148
+P +L P + + ++ G FASAG G ++T + I + +Q +F Y +L
Sbjct: 93 EFVPPFLQPNISDQDIVTGVCFASAGAGYDDETSLSS-KAIPVSQQPSMFKNYIARLKGI 151
Query: 149 IGAEGAKQLVNKAIVLIMLGGNDFVNNYYLVPFSARSRQFSLPNYVTYLISEYKKILQRL 208
+G + A +++N A+V+I G NDF+ N+Y +P R ++ Y +++ ++ L
Sbjct: 152 VGDKKAMEIINNALVVISAGPNDFILNFYDIPIR-RLEYPTIYGYQDFVLKRLDGFVREL 210
Query: 209 YDLGARRVLVTGTGPMGCAPAELALKSRN--GDCDAELMRAASLYNPQLVQMITQLNREI 266
Y LG R +LV G PMGC P +L K R G C + + + LYN +LV+ + ++ +
Sbjct: 211 YSLGCRNILVGGLPPMGCLPIQLTAKLRTILGICVEQENKDSILYNQKLVKKLPEIQASL 270
Query: 267 GDDVFIAVNAHKMHMDFITNPKAFGFVTAKDACCGQGRFNGIGLCTPISKLCPNRNLYAF 326
F+ N + MD I NP +GF K CCG G LCT +SK CPN + + F
Sbjct: 271 PGSKFLYANVYDPVMDMIRNPSKYGFKETKKGCCGTGYLETSFLCTSLSKTCPNHSDHLF 330
Query: 327 WDAFHPSEKASRII 340
WD+ HPSE A + +
Sbjct: 331 WDSIHPSEAAYKYL 344
>At1g75880 family II extracellular lipase 1 like protein (EXL1)
Length = 374
Score = 212 bits (539), Expect = 3e-55
Identities = 126/353 (35%), Positives = 192/353 (53%), Gaps = 12/353 (3%)
Query: 1 MASSLMLC-CSYILMINLFVGFDLAYAQ-PKR----AFFVFGDSVADNGNNNFLTTTARA 54
++SSL+L C ++L++ A + PK A VFGDS+ D GNN+ + T AR
Sbjct: 16 LSSSLILFWCIFVLVLLSTTSTTNALVKIPKNTTVPAVIVFGDSIVDAGNNDDMITEARC 75
Query: 55 DAPPYGIDFPTHEPTGRFSNGLNIPDLTSERLGLEPSLP-YLSPLLVGEKLLVGANFASA 113
D PYGIDF TGRFSNG D+ +E LG++P++P Y +P L E+LL G FAS
Sbjct: 76 DYAPYGIDFDGGVATGRFSNGKVPGDIVAEELGIKPNIPAYRNPNLKPEELLTGVTFASG 135
Query: 114 GVGILNDTGFQFLQIIHIGKQLDLFNQYQQKLSAQIGAEGAKQLVNKAIVLIMLGGNDFV 173
G G + T + I + +QL F +Y +KL +G + K ++ ++ +++ G ND
Sbjct: 136 GAGYVPLT-TKIAGGIPLPQQLIYFEEYIEKLKQMVGEKRTKFIIKNSLFVVICGSNDIA 194
Query: 174 NNYYLVPFSARSRQFSLPNYVTYLISEYKKILQRLYDLGARRVLVTGTGPMGCAPAELAL 233
N+++ +P +++ ++ + + Q LY GARR+LV G P+GC P++ +
Sbjct: 195 NDFFTLP--PVRLHYTVASFTALMADNARSFAQTLYGYGARRILVFGAPPIGCVPSQRTV 252
Query: 234 K-SRNGDCDAELMRAASLYNPQLVQMITQLNREIGDDVFIAVNAHKMHMDFITNPKAFGF 292
DC A AA L+N +L I L+R + D I ++ + +D I NP +GF
Sbjct: 253 AGGPTRDCVARFNDAAKLFNTKLSANIDVLSRTLQDPTIIYIDIYSPLLDLILNPHQYGF 312
Query: 293 VTAKDACCGQGRFNGIGLCTP-ISKLCPNRNLYAFWDAFHPSEKASRIIVQQM 344
A CCG G LC + +CP R+ Y FWD+FHP+EKA RIIV ++
Sbjct: 313 KVANKGCCGTGLIEVTALCNNYTASVCPIRSDYVFWDSFHPTEKAYRIIVAKL 365
>At2g04570 putative GDSL-motif lipase/hydrolase
Length = 350
Score = 211 bits (537), Expect = 5e-55
Identities = 119/337 (35%), Positives = 178/337 (52%), Gaps = 6/337 (1%)
Query: 11 YILMINLFVGFDLAYAQPKRAFFVFGDSVADNGNNNFLTTTARADAPPYGIDFPTHEPTG 70
+ ++ + + + +A A VFGDS D GNNN++ T AR++ PYG DF +PTG
Sbjct: 8 FTILFLIAMSSTVTFAGKIPAIIVFGDSSVDAGNNNYIPTVARSNFEPYGRDFVGGKPTG 67
Query: 71 RFSNGLNIPDLTSERLGLEPSLP-YLSPLLVGEKLLVGANFASAGVGILNDTGFQFLQII 129
RF NG D SE LGL+P +P YL P G FASA G N T L ++
Sbjct: 68 RFCNGKIATDFMSEALGLKPIIPAYLDPSYNISDFATGVTFASAATGYDNATS-DVLSVL 126
Query: 130 HIGKQLDLFNQYQQKLSAQIGAEGAKQLVNKAIVLIMLGGNDFVNNYYLVPFSARSRQFS 189
+ KQL+ + +YQ KL A G + + + ++ LI +G NDF+ NY+ P RS Q+S
Sbjct: 127 PLWKQLEYYKEYQTKLKAYQGKDRGTETIESSLYLISIGTNDFLENYFAFP--GRSSQYS 184
Query: 190 LPNYVTYLISEYKKILQRLYDLGARRVLVTGTGPMGCAPAELALK-SRNGDCDAELMRAA 248
+ Y +L K+ +++L+ LGAR++ + G PMGC P E A G+C A
Sbjct: 185 VSLYQDFLAGIAKEFVKKLHGLGARKISLGGLPPMGCMPLERATNIGTGGECVGRYNDIA 244
Query: 249 SLYNPQLVQMITQLNREIGDDVFIAVNAHKMHMDFITNPKAFGFVTAKDACCGQGRFN-G 307
+N +L +M+ +L++E+ + N ++ M I NP +FGF ACC G F G
Sbjct: 245 VQFNSKLDKMVEKLSKELPGSNLVFSNPYEPFMRIIKNPSSFGFEVVGAACCATGMFEMG 304
Query: 308 IGLCTPISKLCPNRNLYAFWDAFHPSEKASRIIVQQM 344
G C N + Y FWD+FHP++K + I+ +
Sbjct: 305 YGCQRNNPFTCTNADKYVFWDSFHPTQKTNHIMANAL 341
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.324 0.140 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,888,878
Number of Sequences: 26719
Number of extensions: 335798
Number of successful extensions: 1216
Number of sequences better than 10.0: 115
Number of HSP's better than 10.0 without gapping: 103
Number of HSP's successfully gapped in prelim test: 12
Number of HSP's that attempted gapping in prelim test: 724
Number of HSP's gapped (non-prelim): 122
length of query: 367
length of database: 11,318,596
effective HSP length: 101
effective length of query: 266
effective length of database: 8,619,977
effective search space: 2292913882
effective search space used: 2292913882
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 61 (28.1 bits)
Medicago: description of AC146559.11


