

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146557.6 - phase: 0 /pseudo
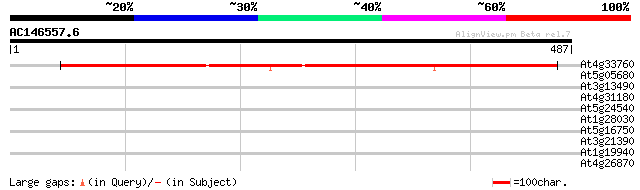
(487 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g33760 aspartate--tRNA ligase like protein 477 e-135
At5g05680 unknown protein 35 0.071
At3g13490 lysyl-tRNA synthetase like protein 33 0.27
At4g31180 aspartate--tRNA ligase - like protein 31 1.3
At5g24540 beta-glucosidase 30 3.0
At1g28030 hypothetical protein 30 3.0
At5g16750 WD40-repeat protein 30 3.9
At3g21390 unknown protein 29 5.1
At1g19940 putative protein 29 6.7
At4g26870 putative aspartate-tRNA ligase 28 8.7
>At4g33760 aspartate--tRNA ligase like protein
Length = 664
Score = 477 bits (1228), Expect = e-135
Identities = 259/472 (54%), Positives = 321/472 (67%), Gaps = 44/472 (9%)
Query: 45 KPKPLNQS-LQWISRTHHCGELSSNDVGKTVRLCGWAELHRSLGGVAFLILRDQTGIIQV 103
KP P S L+W+SRT CGELS NDVGK V LCGW LHR GG+ FL LRD TGI+QV
Sbjct: 61 KPVPSPPSVLRWVSRTELCGELSVNDVGKRVHLCGWVALHRVHGGLTFLNLRDHTGIVQV 120
Query: 104 KTRLDEFPVAHSANKNLRREYVAAIQGVVRSRSTQSINYEMKTGFIEVAANEVQELNSRN 163
+T DEFP AH ++R EYV ++G VRSR +S+N +MKTGF+EV A V+ LN
Sbjct: 121 RTLPDEFPEAHGLINDMRLEYVVLVEGTVRSRPNESVNKKMKTGFVEVVAEHVEILNPVR 180
Query: 164 AKLTLIPELNSDDAEGSPNEEIRLRYRCLDLRKQQMNSNILLRHNVVKLIRRYLEDIHGF 223
KL + +D+ + EEIRLR+RCLDLR+QQM +NI+LRHNVVKLIRRYLED HGF
Sbjct: 181 TKLPFLVT-TADENKDLIKEEIRLRFRCLDLRRQQMKNNIVLRHNVVKLIRRYLEDRHGF 239
Query: 224 VE-----------------------------------PCHKALRYISKC*W*LVLINIIK 248
+E K + +S + +
Sbjct: 240 IEIETPILSRSTPEGARDYLVPSRIQSGTFYALPQSPQLFKQMLMVSGFDKYYQIARCFR 299
Query: 249 *QDLRAVHDRQPEFTQLDMEMAFTPLEDILSLNEELIRKVFLEIKGVELPNPFPRLTYAE 308
+DLRA DRQPEFTQLDMEMAF P+ED+L LNE+LIRKVF EIKG++LP+PFPRLTYA+
Sbjct: 300 DEDLRA--DRQPEFTQLDMEMAFMPMEDMLKLNEDLIRKVFSEIKGIQLPDPFPRLTYAD 357
Query: 309 AMNRYGSDHPDTRFDLELKDVSDIFSGTSFKIFSDSLECGGVIKVLCVPSGATKDSYIL- 367
AM+RYGSD PDTRFDLELKDVS++F+ +SF++F+++LE GG+IKVLCVP GA K S
Sbjct: 358 AMDRYGSDRPDTRFDLELKDVSNVFTESSFRVFTEALESGGIIKVLCVPLGAKKYSNSAL 417
Query: 368 ---DIYNEAEKYGAKGLTTLWITKNG-IGGFSSLVSSMDSATIEELLRRCSAGPSDLILF 423
DIYNEA K GAKGL L + NG I G ++LVSS+DSA +++C A P DLILF
Sbjct: 418 KKGDIYNEAMKSGAKGLPFLKVLDNGEIEGIAALVSSLDSAGKINFVKQCGAAPGDLILF 477
Query: 424 HAGHHASVNKTLGHLRVYVAHKLGLIDHAKHSILWITDFPMFKWDDSKQRLE 475
G SVNKTL LR++VAH + LIDH+KHSILW+TDFPMF+W++ +QRLE
Sbjct: 478 GVGPVTSVNKTLDRLRLFVAHDMDLIDHSKHSILWVTDFPMFEWNEPEQRLE 529
>At5g05680 unknown protein
Length = 810
Score = 35.4 bits (80), Expect = 0.071
Identities = 33/113 (29%), Positives = 51/113 (44%), Gaps = 27/113 (23%)
Query: 313 YGSDHPDTRFDLELKDVSDIFSGTSFKIFSDSLECGGVIKVLC--VPSGAT-KDSYILDI 369
+G DH RF T F +F+D G I +LC VP G+ K +++I
Sbjct: 226 FGGDHLWDRF-------------TVFILFTD-----GSIYILCPVVPFGSVYKWESVMEI 267
Query: 370 YNEAEKYGAKGLTTLWITKNGIGGFSSLVSSMDSATIEELLRRCSAGPSDLIL 422
YN+A YG K +L ++ SSL AT +L + + G + L++
Sbjct: 268 YNDANMYGVKSSNSLAVSN------SSLAIEWLEATFPDLTEQGTRGENILVV 314
>At3g13490 lysyl-tRNA synthetase like protein
Length = 602
Score = 33.5 bits (75), Expect = 0.27
Identities = 32/94 (34%), Positives = 47/94 (49%), Gaps = 8/94 (8%)
Query: 19 SLRKTKIPLPYYEKLKPKTKKAITFKKPKPLNQSLQWISRTHHCGELSSNDVGKTVRLCG 78
S+R K+ + L+P K ++K NQ LQ I + GE S N++ V + G
Sbjct: 86 SIRLKKVEELRGQGLEPYAYK---WEKSHSANQ-LQEIYKHLANGEESDNEID-CVSIAG 140
Query: 79 WAELHRSLGGVAFLILRDQTGIIQV---KTRLDE 109
R+ G +AFL LRD +G IQ+ K RL +
Sbjct: 141 RVVARRAFGKLAFLTLRDDSGTIQLYCEKERLSD 174
>At4g31180 aspartate--tRNA ligase - like protein
Length = 558
Score = 31.2 bits (69), Expect = 1.3
Identities = 39/150 (26%), Positives = 60/150 (40%), Gaps = 29/150 (19%)
Query: 89 VAFLILRDQTGIIQV------KTRLDEFPVAHSANKNLRREYVAAIQGVVRSRSTQSINY 142
+ F++LR+ +Q KT++ V + K L RE + GVV ++
Sbjct: 126 LGFVVLRESGSTVQCVVSQSEKTKVGANMVKYL--KQLSRESFVDVIGVV------TLPK 177
Query: 143 EMKTGF---IEVAANEVQELNSRNAKLTLIPE------------LNSDDAEGSPNEEIRL 187
E TG +E+ +V +N AKL L E L + N++ RL
Sbjct: 178 EPLTGTTQQVEIQVRKVYCINKSLAKLPLSVEDAARSEADIEASLQTPSPAARVNQDTRL 237
Query: 188 RYRCLDLRKQQMNSNILLRHNVVKLIRRYL 217
YR LDLR + L++ V R L
Sbjct: 238 NYRVLDLRTPANQAIFQLQYEVEYAFREKL 267
>At5g24540 beta-glucosidase
Length = 534
Score = 30.0 bits (66), Expect = 3.0
Identities = 37/147 (25%), Positives = 54/147 (36%), Gaps = 47/147 (31%)
Query: 344 SLECGGVIKVLCVPSGATKD-SYILDIYNEAEKYGAKGLTTLWITKNGIGGFSSLVSSMD 402
SLE G + P G K +YI + YN T++IT+NG + +
Sbjct: 381 SLESDGTKILWSYPEGLRKILNYIKNKYNNP---------TIYITENGFDDYEN-----G 426
Query: 403 SATIEELLRRCSAGPSDLILFHAGHHASVNKTL--------GHLR--------------- 439
+ T EE+L + I +H H + K + G+
Sbjct: 427 TVTREEILE-----DTKRIEYHQKHLQELQKAITEDGCDVKGYFTWSLLDNFEWEHGYAV 481
Query: 440 ----VYVAHKLGLIDHAKHSILWITDF 462
YV +K GL HAKHS +W F
Sbjct: 482 RFGLYYVDYKNGLQRHAKHSAMWFKHF 508
>At1g28030 hypothetical protein
Length = 322
Score = 30.0 bits (66), Expect = 3.0
Identities = 29/107 (27%), Positives = 50/107 (46%), Gaps = 13/107 (12%)
Query: 273 PLEDILSLNEELIRKVFLEIKGVELPNPFPRLTYAEAMNRYGSDHPDTRFDL--ELKDVS 330
PLE LS ++ + +L I PR+ E M YG D+P+ DL +L
Sbjct: 73 PLETKLSTKTDVHYEGYLTI---------PRVPIQEGMGFYGIDNPNVVNDLTHKLWPQG 123
Query: 331 DIFSGTSFKIFSDSL-ECGGVIKVLCVPSGATKDSYILDIYNEAEKY 376
+IF G + + F++ L E ++ + + S + Y+ + N A K+
Sbjct: 124 NIFVGKNVQSFAEKLIELNLTVRTMTLESFGL-EKYMEEHLNAANKH 169
>At5g16750 WD40-repeat protein
Length = 876
Score = 29.6 bits (65), Expect = 3.9
Identities = 27/92 (29%), Positives = 42/92 (45%), Gaps = 15/92 (16%)
Query: 392 GGFSSLVSSMDSA---TIE---ELLRRCSAGPSDLILFHAGHHASVN----KTLGHLRVY 441
G ++V S DS+ TIE + L + P D +LF AGH + +TL +R +
Sbjct: 39 GDVINIVDSTDSSVKSTIEGESDTLTALALSPDDKLLFSAGHSRQIRVWDLETLKCIRSW 98
Query: 442 VAHK---LGLIDHAKHSILWI--TDFPMFKWD 468
H+ +G+ HA +L D + WD
Sbjct: 99 KGHEGPVMGMACHASGGLLATAGADRKVLVWD 130
>At3g21390 unknown protein
Length = 335
Score = 29.3 bits (64), Expect = 5.1
Identities = 19/65 (29%), Positives = 30/65 (45%), Gaps = 2/65 (3%)
Query: 336 TSFKIFSDSLECGGVIKVLCVPSGATKDSYILDIYNEAEKYGAKGLTTLWITKNGIGGFS 395
+SF++F L G V K++C P K + ++ KYGA+ L KN G
Sbjct: 233 SSFQLFLCGLASGTVSKLVCHPLDVVKKRFQVEGLQRHPKYGAR--VELNAYKNMFDGLG 290
Query: 396 SLVSS 400
++ S
Sbjct: 291 QILRS 295
>At1g19940 putative protein
Length = 515
Score = 28.9 bits (63), Expect = 6.7
Identities = 11/17 (64%), Positives = 13/17 (75%)
Query: 446 LGLIDHAKHSILWITDF 462
L L+DHAK S+ W TDF
Sbjct: 129 LNLLDHAKDSLKWTTDF 145
>At4g26870 putative aspartate-tRNA ligase
Length = 532
Score = 28.5 bits (62), Expect = 8.7
Identities = 42/174 (24%), Positives = 69/174 (39%), Gaps = 27/174 (15%)
Query: 70 VGKTVRLCGWAELHRSLGGVAFLILRDQTGIIQV---KTRLDEFPVAHSANKNLRREYVA 126
VG V + G +R +G F+ILR+ +Q +TR+ + K L RE V
Sbjct: 84 VGSEVSIRGRLHKNRLVGTKLFVILRESGFTVQCVVEETRVGANMIKFV--KQLSRESVV 141
Query: 127 AIQGVVRSRSTQSINYEMKTGFIEVAANEVQELNSRNAKLTLIPELNSDDAEGSPN---- 182
+ GVV S + TG + V+++ + L +P + D A +
Sbjct: 142 ELIGVV------SHPKKPLTGTTQQVEIHVRKMYCLSRSLPNLPLVVEDAARSESDIEKS 195
Query: 183 -----------EEIRLRYRCLDLRKQQMNSNILLRHNVVKLIRRYLEDIHGFVE 225
++ RL R LD+R + ++ V R YL+ GF+E
Sbjct: 196 GKDGKQAARVLQDTRLNNRVLDIRTPANQAIFRIQCQVQIAFREYLQS-KGFLE 248
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.324 0.139 0.420
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,694,165
Number of Sequences: 26719
Number of extensions: 454606
Number of successful extensions: 1451
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 4
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 1441
Number of HSP's gapped (non-prelim): 13
length of query: 487
length of database: 11,318,596
effective HSP length: 103
effective length of query: 384
effective length of database: 8,566,539
effective search space: 3289550976
effective search space used: 3289550976
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 62 (28.5 bits)
Medicago: description of AC146557.6


