

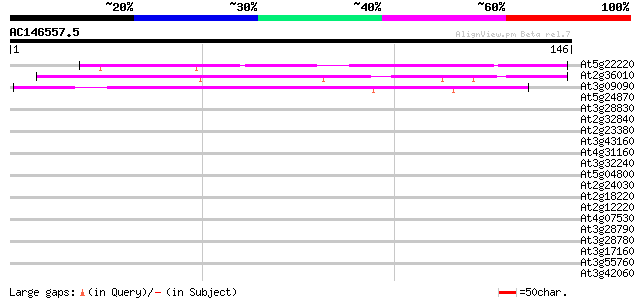
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146557.5 - phase: 0
(146 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g22220 E2F transcription factor-1 E2F1 94 2e-20
At2g36010 E2F transcription factor-3 E2F3 83 6e-17
At3g09090 defective in exine formation (DEX1) 39 7e-04
At5g24870 RING finger-like protein 35 0.018
At3g28830 hypothetical protein 32 0.092
At2g32840 unknown protein 32 0.092
At2g23380 curly leaf protein (polycomb-group) 32 0.092
At3g43160 putative protein 32 0.12
At4g31160 unknown protein 31 0.27
At3g32240 hypothetical protein 30 0.59
At5g04800 40S ribosomal protein S17 29 0.78
At2g24030 hypothetical protein 29 1.0
At2g18220 unknown protein 29 1.0
At2g12220 En/Spm-like transposon protein 29 1.0
At4g07530 putative myosin-like protein 28 1.7
At3g28790 unknown protein 28 1.7
At3g28780 histone-H4-like protein 28 1.7
At3g17160 unknown protein 28 1.7
At3g55760 unknown protein 28 2.3
At3g42060 putative protein 28 2.3
>At5g22220 E2F transcription factor-1 E2F1
Length = 469
Score = 94.4 bits (233), Expect = 2e-20
Identities = 60/134 (44%), Positives = 77/134 (56%), Gaps = 17/134 (12%)
Query: 19 QRQL--PFSSMKPPFLAAADYHRFAPDHRRN-----QDLETEAIVVKTPQLKRKSEAADF 71
+RQL SSMKPP +A +YHRF R + ++AIV+K+ LKRK++ +
Sbjct: 13 KRQLHPSLSSMKPPLVAPGEYHRFDAAETRGGGAVADQVVSDAIVIKST-LKRKTDLVNQ 71
Query: 72 EADSGDRMTPGSTAAANSSVQTPVSGKAGKGGKSSRMTKCNRSGTQTPGSNIGSPSGNNL 131
+ + T +QTPVSGK GK K+SR K N+SGT GSN GSP GNN
Sbjct: 72 IVEVNELNT--------GVLQTPVSGKGGKAKKTSRSAKSNKSGTLASGSNAGSP-GNNF 122
Query: 132 TPAGPCRYDSSLGI 145
AG CRYDSSLG+
Sbjct: 123 AQAGTCRYDSSLGL 136
>At2g36010 E2F transcription factor-3 E2F3
Length = 483
Score = 82.8 bits (203), Expect = 6e-17
Identities = 61/154 (39%), Positives = 79/154 (50%), Gaps = 23/154 (14%)
Query: 8 PATAAPDQIMQQRQLPFSSMKPPFLAAADYHRFAPDHRRNQ------------DLETEAI 55
P T+ P +R L F+S KPPF + DYHRF P N D E +A+
Sbjct: 28 PVTSTPVIPPIRRHLAFASTKPPFHPSDDYHRFNPSSLSNNNDRSFVHGCGVVDREEDAV 87
Query: 56 VVKTPQLKRKSEAADFEADSGDRMT-PGSTAAANSSVQTPVSGKAGKGGKSSRMTKC--N 112
VV++P KRK+ A S + T G T +S QTP KGG+ + +K N
Sbjct: 88 VVRSPSRKRKATMDMVVAPSNNGFTSSGFTNIPSSPCQTP-----RKGGRVNIKSKAKGN 142
Query: 113 RSGTQTP-GSNIGSPSGNNLTPAGPCRYDSSLGI 145
+S QTP +N GSP LTP+G CRYDSSLG+
Sbjct: 143 KSTPQTPISTNAGSPI--TLTPSGSCRYDSSLGL 174
>At3g09090 defective in exine formation (DEX1)
Length = 896
Score = 39.3 bits (90), Expect = 7e-04
Identities = 38/138 (27%), Positives = 58/138 (41%), Gaps = 12/138 (8%)
Query: 2 SGTRTPPATAAPDQIMQQRQLPFSSMKPPFLAAADYHRFAPDHRRNQDLETEAIVVKTPQ 61
S T+T T P+ + SM A Y D +R ++ +TEAIV TP+
Sbjct: 199 STTQTNATTTTPNVTV--------SMTKEVHGANSYVSTQEDQKRPENNQTEAIVKPTPE 250
Query: 62 LKRKSEAADFEADSGDRMTPGSTAAANSSVQT-PVSGKAGKGGKSSRMTKCNRS---GTQ 117
L S A + + T GS N +V T V G K+ + K N S ++
Sbjct: 251 LHNSSMDAGANNLAANATTAGSRENLNRNVTTNEVDQSKISGDKNETVIKLNTSTGNSSE 310
Query: 118 TPGSNIGSPSGNNLTPAG 135
T G++ S + +T +G
Sbjct: 311 TLGTSGNSSTAETVTKSG 328
>At5g24870 RING finger-like protein
Length = 520
Score = 34.7 bits (78), Expect = 0.018
Identities = 28/100 (28%), Positives = 45/100 (45%), Gaps = 8/100 (8%)
Query: 44 HRRNQDLETEAIVVKTPQLKR----KSEAADFEADSGDRMTPGSTAAANSSVQTPVSGKA 99
H+R + +A V + Q R K+ D SG + P ++ S + K
Sbjct: 165 HQRPDLVSRDARVSNSEQNARASVNKNGLRDLRNKSGSDVLPSNSTPTRKS---NIFRKK 221
Query: 100 GKGGKSSRMTKCNRS-GTQTPGSNIGSPSGNNLTPAGPCR 138
G+SS ++ N++ G+ G NI SP GN +T + P R
Sbjct: 222 TSDGESSSSSRGNKTEGSVVGGKNISSPQGNGITMSEPRR 261
>At3g28830 hypothetical protein
Length = 536
Score = 32.3 bits (72), Expect = 0.092
Identities = 22/81 (27%), Positives = 36/81 (44%), Gaps = 4/81 (4%)
Query: 63 KRKSEAADFEADSGDRMTPGSTAAANSSVQTPVSGKAGKGGKSSRMTKCNRSGTQTPGSN 122
K K ++ + SG T ++ S+ K GG ++ +K + G+ T G
Sbjct: 218 KSKESSSSGSSASGSVATKSKESSGGSAA---TKSKESSGGSAATKSKESSGGSATTGKT 274
Query: 123 IGSPSGN-NLTPAGPCRYDSS 142
GSPSG+ +P+G SS
Sbjct: 275 SGSPSGSPKASPSGSVSGKSS 295
>At2g32840 unknown protein
Length = 337
Score = 32.3 bits (72), Expect = 0.092
Identities = 32/134 (23%), Positives = 50/134 (36%), Gaps = 21/134 (15%)
Query: 6 TPPATAAPDQIMQQRQLPFSSMKP-PFLAAADYHRFAPDHRRN--QDLETEAIVVKTPQL 62
TP TA+ + Q + S +P P A+ Y AP HR + Q++ T + ++
Sbjct: 22 TPIVTASESPVTQPNTVITPSSQPQPQTPASSYRAIAPLHRHHPHQNIYTNPLPIRRSNS 81
Query: 63 KRKSEAADFEADSGDRMTPGSTAAANSSVQTPVSGKAGKGGKSSRMTKCNRSGTQTPGSN 122
S D + P G +G+G T+ R + +
Sbjct: 82 VTNSPHQPPHPDPSSLIYP--------------FGSSGRGFP----TRPVRQNSNSVADP 123
Query: 123 IGSPSGNNLTPAGP 136
+GSPS TP GP
Sbjct: 124 VGSPSPGGYTPRGP 137
>At2g23380 curly leaf protein (polycomb-group)
Length = 902
Score = 32.3 bits (72), Expect = 0.092
Identities = 21/78 (26%), Positives = 38/78 (47%), Gaps = 3/78 (3%)
Query: 45 RRNQDLETEAIVVKTPQLKRKSEAADFEADSGDRMTPGSTAAANSSVQTPVSGKAGKGGK 104
+RN++ E + KT + ++K+EA+D ++ + +P ++ T S K K G
Sbjct: 412 KRNKNRVAERVPRKTQKRQKKTEASDSDSIASGSCSPSDAKHKDNEDATSSSQKHVKSGN 471
Query: 105 SSRMTKCNRSGTQTPGSN 122
S K ++GT SN
Sbjct: 472 SG---KSRKNGTPAEVSN 486
>At3g43160 putative protein
Length = 295
Score = 32.0 bits (71), Expect = 0.12
Identities = 19/53 (35%), Positives = 27/53 (50%), Gaps = 1/53 (1%)
Query: 77 DRMTPGSTAAANSSVQTPVSGKAGKGGKSSRMTKCNRS-GTQTPGSNIGSPSG 128
D + ST NSS +T S K G+S + + +S GT GS++GS G
Sbjct: 116 DYVLESSTGTENSSTRTAESSTRTKNGRSKSIVESGQSMGTPLLGSDLGSDRG 168
>At4g31160 unknown protein
Length = 1846
Score = 30.8 bits (68), Expect = 0.27
Identities = 31/103 (30%), Positives = 42/103 (40%), Gaps = 16/103 (15%)
Query: 33 AAADYHRFAPDHRRNQDLETEA---IVVKTPQLKRKSEAADFEA-----DSGDRMTPGST 84
A AD A + DL+ +A + P+ ++ SE D D G+R ST
Sbjct: 1237 ALADTSETAAELVLKNDLDADAQFKTPISFPRKRKLSELRDSSVPGKRIDLGERRN--ST 1294
Query: 85 AAANSSVQTPVSGKAGKGGKSSRMTKCNRSGTQTPGSNIGSPS 127
A S +QTP S SSR+ G TP S + PS
Sbjct: 1295 FADGSGLQTPASALDANQSGSSRL------GQMTPASQLRLPS 1331
>At3g32240 hypothetical protein
Length = 487
Score = 29.6 bits (65), Expect = 0.59
Identities = 16/61 (26%), Positives = 27/61 (44%)
Query: 76 GDRMTPGSTAAANSSVQTPVSGKAGKGGKSSRMTKCNRSGTQTPGSNIGSPSGNNLTPAG 135
GD +PG+T+A + Q G + T T +P + GSP+ + +P+
Sbjct: 22 GDATSPGNTSAVQDNRQAHPRHSRRHSGSPAMSTGSPAQSTGSPAQSTGSPALFSGSPSS 81
Query: 136 P 136
P
Sbjct: 82 P 82
>At5g04800 40S ribosomal protein S17
Length = 141
Score = 29.3 bits (64), Expect = 0.78
Identities = 17/65 (26%), Positives = 28/65 (42%), Gaps = 2/65 (3%)
Query: 43 DHRRNQDLETEAIVVKTPQLKRKSEAADFEADSGDRMTPGSTAAANSSVQTPVSGKAGKG 102
+ R D + +KT ++K E + A G TPG +A P++ A G
Sbjct: 77 ERERRMDFVPDESAIKTDEIKVDKETLEMLASLGMSDTPGISAVEPQQAMAPIA--AFGG 134
Query: 103 GKSSR 107
G++ R
Sbjct: 135 GRAPR 139
>At2g24030 hypothetical protein
Length = 440
Score = 28.9 bits (63), Expect = 1.0
Identities = 22/72 (30%), Positives = 36/72 (49%), Gaps = 14/72 (19%)
Query: 43 DHRRNQDLET---EAIVVKTPQLKRKSEAADFEADSGDRMTPGSTAAANSSVQTPVSGKA 99
+ R+ +D++T E + VK P+ K +SEA ++G+ + S + P SGK
Sbjct: 230 EKRKAEDIQTGLNEDLQVKRPKAK-ESEAKAMSLETGEIV----------SSKVPCSGKL 278
Query: 100 GKGGKSSRMTKC 111
G G K+ T C
Sbjct: 279 GSGKKAEVGTYC 290
>At2g18220 unknown protein
Length = 779
Score = 28.9 bits (63), Expect = 1.0
Identities = 26/100 (26%), Positives = 40/100 (40%), Gaps = 12/100 (12%)
Query: 28 KPPFLAAADYHRFAPDHRRNQDLETEAIVVKTPQLKRKSEAADFEADSGDRMTPGSTA-- 85
K P L A+ R R +E++ IV + + K+ + + D DRM G+ A
Sbjct: 609 KTPLLQYAEIIRQRAQQRNESLVESDVIVGENSAVFGKNAPSSDDEDDEDRMEKGAAAFN 668
Query: 86 -----AANSSVQTPVSGKAGK-----GGKSSRMTKCNRSG 115
++S + P K K GGKS K + G
Sbjct: 669 SSWLPGSDSKEKEPEEEKTKKKKRKRGGKSKTEKKQDEQG 708
>At2g12220 En/Spm-like transposon protein
Length = 781
Score = 28.9 bits (63), Expect = 1.0
Identities = 19/55 (34%), Positives = 28/55 (50%), Gaps = 5/55 (9%)
Query: 81 PGSTAAANSSVQTPVSG-KAGKGGKS----SRMTKCNRSGTQTPGSNIGSPSGNN 130
P ++ SS+Q S K+ G KS S +K + SG++TP N +PS N
Sbjct: 628 PSASGNQKSSLQPSGSASKSSAGAKSAASKSTSSKSSASGSETPLENANAPSSTN 682
>At4g07530 putative myosin-like protein
Length = 786
Score = 28.1 bits (61), Expect = 1.7
Identities = 22/83 (26%), Positives = 36/83 (42%), Gaps = 2/83 (2%)
Query: 47 NQDLETEAIVVKTPQLKRKSEAADFEADSGDRMTPGSTAAANSSVQTPVSGKAGKGGKSS 106
N + + VV++ + + A + + G+ T G + A +S SG++ GG+ S
Sbjct: 407 NNQVSGSSAVVRSDGVPSSPKRARTQTERGEDGTYGCYSGAAASADRRSSGRSHGGGRGS 466
Query: 107 R--MTKCNRSGTQTPGSNIGSPS 127
R NRS SN G S
Sbjct: 467 RGGGRSSNRSQEVRQTSNSGGGS 489
>At3g28790 unknown protein
Length = 608
Score = 28.1 bits (61), Expect = 1.7
Identities = 17/57 (29%), Positives = 28/57 (48%), Gaps = 1/57 (1%)
Query: 80 TPGSTAAANSSVQTPVSGKAG-KGGKSSRMTKCNRSGTQTPGSNIGSPSGNNLTPAG 135
TP + + + TP +GK KG +S+ M K + S +++ + GS S T G
Sbjct: 302 TPSTPTPSTPAPSTPAAGKTSEKGSESASMKKESNSKSESESAASGSVSKTKETNKG 358
>At3g28780 histone-H4-like protein
Length = 614
Score = 28.1 bits (61), Expect = 1.7
Identities = 17/48 (35%), Positives = 24/48 (49%), Gaps = 1/48 (2%)
Query: 82 GSTAAANSSVQTPVSGKAGKGGKSSRMTKCNRSGTQTPGSNIGSPSGN 129
GSTAA +S ++ G A GG S + G+ T + G PSG+
Sbjct: 509 GSTAAGGTS-ESASGGSATAGGASGGTYTDSTGGSPTGSPSAGGPSGS 555
Score = 26.9 bits (58), Expect = 3.9
Identities = 16/43 (37%), Positives = 22/43 (50%), Gaps = 2/43 (4%)
Query: 102 GGKSSRMTKCNRSGTQTPGSNIGSPSGNNLTPAGPCRYDSSLG 144
GG SS + SG+ PGS+ GSPS + +P Y + G
Sbjct: 128 GGGSSGLGSSGDSGS--PGSDSGSPSADTGSPTDGGSYGDTTG 168
>At3g17160 unknown protein
Length = 133
Score = 28.1 bits (61), Expect = 1.7
Identities = 22/88 (25%), Positives = 39/88 (44%), Gaps = 6/88 (6%)
Query: 48 QDLETEAIVVKTPQLKRKSEAADFE-ADSGDRMTPGSTAAANSSVQTPVSGKAGKGGKSS 106
+DL TE +V +++ + +A+DF+ ++G G V +S AGK S
Sbjct: 49 EDLGTEYLVRPVGEVEDEDDASDFDPEENGLDEEEGDEEIEEDDVDEDISLSAGKSEPLS 108
Query: 107 RMTKCNRSGTQTPGSNIGSPSGNNLTPA 134
+ R S G +G+N+ P+
Sbjct: 109 K-----RKRVAKDHSEQGDVTGDNIRPS 131
>At3g55760 unknown protein
Length = 578
Score = 27.7 bits (60), Expect = 2.3
Identities = 18/68 (26%), Positives = 30/68 (43%), Gaps = 4/68 (5%)
Query: 69 ADFEADSGDRMTPGSTAAANSSV-QTPVSGKAGKGGKSSRMTKCNRSGTQTPGSNIGS-- 125
A + G P S V +TP + K G ++ + + SGT+TPG + S
Sbjct: 144 ASNNSGDGKEGFPNEDNTVTSEVTETPKNAKLGMWSRTVYVPRSETSGTETPGPDFWSWT 203
Query: 126 -PSGNNLT 132
P G+ ++
Sbjct: 204 PPQGSEIS 211
>At3g42060 putative protein
Length = 712
Score = 27.7 bits (60), Expect = 2.3
Identities = 18/58 (31%), Positives = 24/58 (41%), Gaps = 3/58 (5%)
Query: 72 EADSGDRMTPGSTAAANSSVQTPVSGKA---GKGGKSSRMTKCNRSGTQTPGSNIGSP 126
E + DR T + A A V +G GG SR +R +T SN+G P
Sbjct: 422 EVPNKDRGTEVAAARAGKKVLKETTGSVPFPSVGGSRSRKRDSSRRSPETGPSNVGKP 479
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.309 0.125 0.359
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,488,801
Number of Sequences: 26719
Number of extensions: 140427
Number of successful extensions: 425
Number of sequences better than 10.0: 51
Number of HSP's better than 10.0 without gapping: 27
Number of HSP's successfully gapped in prelim test: 24
Number of HSP's that attempted gapping in prelim test: 383
Number of HSP's gapped (non-prelim): 60
length of query: 146
length of database: 11,318,596
effective HSP length: 90
effective length of query: 56
effective length of database: 8,913,886
effective search space: 499177616
effective search space used: 499177616
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 55 (25.8 bits)
Medicago: description of AC146557.5


