

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146555.15 - phase: 0 /pseudo
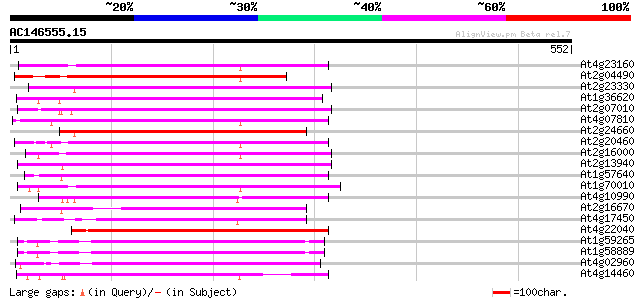
(552 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g23160 putative protein 223 2e-58
At2g04490 putative retroelement pol polyprotein 219 2e-57
At2g23330 putative retroelement pol polyprotein 216 3e-56
At1g36620 hypothetical protein 215 6e-56
At2g07010 putative retroelement pol polyprotein 214 1e-55
At4g07810 putative polyprotein 213 3e-55
At2g24660 putative retroelement pol polyprotein 209 2e-54
At2g20460 putative retroelement pol polyprotein 209 2e-54
At2g16000 putative retroelement pol polyprotein 207 2e-53
At2g13940 putative retroelement pol polyprotein 206 3e-53
At1g57640 206 4e-53
At1g70010 hypothetical protein 205 5e-53
At4g10990 putative retrotransposon polyprotein 203 2e-52
At2g16670 putative retroelement pol polyprotein 200 1e-51
At4g17450 retrotransposon like protein 200 2e-51
At4g22040 LTR retrotransposon like protein 194 1e-49
At1g59265 polyprotein, putative 194 1e-49
At1g58889 polyprotein, putative 192 4e-49
At4g02960 putative polyprotein of LTR transposon 191 7e-49
At4g14460 retrovirus-related like polyprotein 185 5e-47
>At4g23160 putative protein
Length = 1240
Score = 223 bits (569), Expect = 2e-58
Identities = 122/311 (39%), Positives = 187/311 (59%), Gaps = 12/311 (3%)
Query: 9 TTSPSPPITKTSPSPPFIPPP-IRITTRNKITPTYM*DYICNIPTTSTTNNSHVQYPISN 67
T+S S I ++ +P P + + R P Y+ DY C+ + T ++ IS
Sbjct: 9 TSSSSIDIMPSANIQNDVPEPSVHTSHRRTRKPAYLQDYYCHSVASLTIHD------ISQ 62
Query: 68 FLSHTYLSKSHSIFAMSLVSYTEPKSYDEAIKHDCWKQVMQNELTTLDQTGTWKIVDLPP 127
FLS+ +S + F + + EP +Y+EA + W M +E+ ++ T TW+I LPP
Sbjct: 63 FLSYEKVSPLYHSFLVCIAKAKEPSTYNEAKEFLVWCGAMDDEIGAMETTHTWEICTLPP 122
Query: 128 SSKPIGCIWVSKVKHNVDGSIERYKVCLVAKGYNQIEGLDYFDTFSLVAKVTTIRLVIAL 187
+ KPIGC WV K+K+N DG+IERYK LVAKGY Q EG+D+ +TFS V K+T+++L++A+
Sbjct: 123 NKKPIGCKWVYKIKYNSDGTIERYKARLVAKGYTQQEGIDFIETFSPVCKLTSVKLILAI 182
Query: 188 ASINY*FLHQLDVNNAFLHGDLHEDVYMAIPPGVSTSK-----PNQVCKLSKSLYGLKPA 242
++I LHQLD++NAFL+GDL E++YM +PPG + + PN VC L KS+YGLK A
Sbjct: 183 SAIYNFTLHQLDISNAFLNGDLDEEIYMKLPPGYAARQGDSLPPNAVCYLKKSIYGLKQA 242
Query: 243 SRKWYEKLTCLPITNGYQQATSNASLFTKKNLDSFIMLLVYVYDITLAGDFLSEITFIKN 302
SR+W+ K + I G+ Q+ S+ + F K F+ +LVYV DI + + + + +K+
Sbjct: 243 SRQWFLKFSVTLIGFGFVQSHSDHTYFLKITATLFLCVLVYVDDIIICSNNDAAVDELKS 302
Query: 303 ALNQASKSKIL 313
L K + L
Sbjct: 303 QLKSCFKLRDL 313
>At2g04490 putative retroelement pol polyprotein
Length = 1015
Score = 219 bits (559), Expect = 2e-57
Identities = 117/273 (42%), Positives = 168/273 (60%), Gaps = 22/273 (8%)
Query: 5 THIPTTSPSPPITKTSPSPPFIPPPIRITTRNKITPTYM*DYICNIPTTSTTNNSHVQYP 64
+H + S P T TS S R P ++ DY CN+ + VQYP
Sbjct: 561 SHEDSAPKSVPTTSTSRSK-----------RESKQPAHLKDYFCNL------SRKGVQYP 603
Query: 65 ISNFLSHTYLSKSHSIFAMSLVSYTEPKSYDEAIKHDCWKQVMQNELTTLDQTGTWKIVD 124
+S+++S+ LS + + S+ ++EP S+ +A K D W + M EL L+ T TW+I
Sbjct: 604 LSDYMSYDQLSTPYRAYICSVTKFSEPSSFFQAKKSDDWIKAMNAELQALEGTATWEICS 663
Query: 125 LPPSSKPIGCIWVSKVKHNVDGSIERYKVCLVAKGYNQIEGLDYFDTFSLVAKVTTIRLV 184
LP + K IGC WV KVK NVDG++ERYK LVAKGY Q EG+D+ DTFS VAK+TT++ +
Sbjct: 664 LPSNKKAIGCKWVYKVKLNVDGTLERYKARLVAKGYTQQEGVDFEDTFSPVAKMTTVKTL 723
Query: 185 IALASINY*FLHQLDVNNAFLHGDLHEDVYMAIPPGVSTSK-----PNQVCKLSKSLYGL 239
+A+A+ LHQLD++NAFL+ DL+E++YM + PG + + PN VCKL KSLYGL
Sbjct: 724 LAVAAAKKWSLHQLDISNAFLNRDLYEEIYMNLAPGYTPKEGEEIPPNAVCKLKKSLYGL 783
Query: 240 KPASRKWYEKLTCLPITNGYQQATSNASLFTKK 272
K SR+W+ K ++ G+QQ+ ++ +LF KK
Sbjct: 784 KQDSRQWFLKFRSTLLSLGFQQSHADHTLFVKK 816
>At2g23330 putative retroelement pol polyprotein
Length = 1496
Score = 216 bits (550), Expect = 3e-56
Identities = 120/322 (37%), Positives = 183/322 (56%), Gaps = 24/322 (7%)
Query: 19 TSPSPPFIPPPIRITTRNKITPTYM*DYICNIPTTSTTNNSHVQ---------------- 62
++PS P +P + R K + D++ N + T + ++
Sbjct: 891 STPSSPGLPELLGKGCREKKKSVLLKDFVTNTTSKKKTASHNIHSPSQVLPSGLPTSLSA 950
Query: 63 --------YPISNFLSHTYLSKSHSIFAMSLVSYTEPKSYDEAIKHDCWKQVMQNELTTL 114
YP+S+FL+++ S +H F +++ EPK + +AI W + M E+ L
Sbjct: 951 DSVSGKTLYPLSDFLTNSGYSANHIAFMAAILDSNEPKHFKDAILIKEWCEAMSKEIDAL 1010
Query: 115 DQTGTWKIVDLPPSSKPIGCIWVSKVKHNVDGSIERYKVCLVAKGYNQIEGLDYFDTFSL 174
+ TW I DLP K I WV K+K+N DG++ER+K LV G +Q EG+D+ +TF+
Sbjct: 1011 EANHTWDITDLPHGKKAISSKWVYKLKYNSDGTLERHKARLVVMGNHQKEGVDFKETFAP 1070
Query: 175 VAKVTTIRLVIALASINY*FLHQLDVNNAFLHGDLHEDVYMAIPPGVSTSKPNQVCKLSK 234
VAK+TT+R ++A+A+ +HQ+DV+NAFLHGDL E+VYM +PPG S P++VC+L K
Sbjct: 1071 VAKLTTVRTILAVAAAKDWEVHQMDVHNAFLHGDLEEEVYMRLPPGFKCSDPSKVCRLRK 1130
Query: 235 SLYGLKPASRKWYEKLTCLPITNGYQQATSNASLFTKKNLDSFIMLLVYVYDITLAGDFL 294
SLYGLK A R W+ KL+ G+ Q+ + SLF+ KN D+ I +LVYV D+ +AG+ L
Sbjct: 1131 SLYGLKQAPRCWFSKLSTALRNIGFTQSYEDYSLFSLKNGDTIIHVLVYVDDLIVAGNNL 1190
Query: 295 SEITFIKNALNQASKSKILVSL 316
I K+ L++ K L L
Sbjct: 1191 DAIDRFKSQLHKCFHMKDLGKL 1212
>At1g36620 hypothetical protein
Length = 1152
Score = 215 bits (547), Expect = 6e-56
Identities = 119/317 (37%), Positives = 177/317 (55%), Gaps = 16/317 (5%)
Query: 7 IPTTSPSPPITKTSPSPPFIP-----------PPIRITTRNKITPTYM*DYI-----CNI 50
+P T+P P I + SPP P PP+R R + + DY C
Sbjct: 835 VPATNPLPAIIDVNDSPPSSPIITATPAAASPPPLRRGLRQRQENVRLKDYQTYSAQCES 894
Query: 51 PTTSTTNNSHVQYPISNFLSHTYLSKSHSIFAMSLVSYTEPKSYDEAIKHDCWKQVMQNE 110
T + N YP++N++S S S+ F ++ P++Y++AI+ W+ + E
Sbjct: 895 TQTLSDNIGTCIYPMANYVSGEIFSPSNQHFLAAISMVDPPQTYNQAIREKEWRNAVFFE 954
Query: 111 LTTLDQTGTWKIVDLPPSSKPIGCIWVSKVKHNVDGSIERYKVCLVAKGYNQIEGLDYFD 170
+ L+ GTW I LP K IG WV ++K+N +G++ERYK LVA G +Q EG+D+
Sbjct: 955 VDALEDQGTWDITKLPQGVKAIGSKWVFRIKYNSNGTVERYKARLVALGNHQKEGIDFTK 1014
Query: 171 TFSLVAKVTTIRLVIALASINY*FLHQLDVNNAFLHGDLHEDVYMAIPPGVSTSKPNQVC 230
TF+ V K+ T+RL++ +A+ LHQ+DV+NAFLHGDL ED+YM PPG T+ P+ VC
Sbjct: 1015 TFAPVVKMQTVRLLLDVAAAKDWELHQMDVHNAFLHGDLKEDIYMKPPPGFKTTDPSLVC 1074
Query: 231 KLSKSLYGLKPASRKWYEKLTCLPITNGYQQATSNASLFTKKNLDSFIMLLVYVYDITLA 290
KL KS+YGLK A R W+EKL+ + G+ Q+ + SLFT + ++VYV D+ +
Sbjct: 1075 KLKKSIYGLKQAPRCWFEKLSTSLLKFGFTQSKKDYSLFTSIRGSKVLHVIVYVDDVVIC 1134
Query: 291 GDFLSEITFIKNALNQA 307
G + E K AL +
Sbjct: 1135 GKAVRENNTSKLALGSS 1151
>At2g07010 putative retroelement pol polyprotein
Length = 1413
Score = 214 bits (544), Expect = 1e-55
Identities = 125/324 (38%), Positives = 180/324 (54%), Gaps = 17/324 (5%)
Query: 8 PTTSPSPPITKTSPSPPFIPPPIRITTRNKITPTYM*DYI-----CN-------IPTTST 55
P+T+ SPP T+P PP R R P + DYI C P+TS
Sbjct: 886 PSTNVSPPQQDTTPIIENTPP--RQGKRQVQQPARLKDYILYNASCTPNTPHVLSPSTSQ 943
Query: 56 TNNS---HVQYPISNFLSHTYLSKSHSIFAMSLVSYTEPKSYDEAIKHDCWKQVMQNELT 112
+++S ++QYP+++++S S H +F ++ + EPK + E +K W M E+
Sbjct: 944 SSSSIQGNLQYPLTDYISDECFSAGHKVFLAAITANDEPKHFKEDVKVKVWNDAMYKEVD 1003
Query: 113 TLDQTGTWKIVDLPPSSKPIGCIWVSKVKHNVDGSIERYKVCLVAKGYNQIEGLDYFDTF 172
L+ TW IVDLP IG WV K K N DG++ERYK LV +G NQIEG DY +TF
Sbjct: 1004 ALEVNKTWDIVDLPTGKVAIGSQWVYKTKFNADGTVERYKARLVVQGNNQIEGEDYTETF 1063
Query: 173 SLVAKVTTIRLVIALASINY*FLHQLDVNNAFLHGDLHEDVYMAIPPGVSTSKPNQVCKL 232
+ V K+TT+R ++ L + N ++Q+DV+NAFLHGDL E+VYM +PPG S P++VC+L
Sbjct: 1064 APVVKMTTVRTLLRLVAANQWEVYQMDVHNAFLHGDLEEEVYMKLPPGFRHSHPDKVCRL 1123
Query: 233 SKSLYGLKPASRKWYEKLTCLPITNGYQQATSNASLFTKKNLDSFIMLLVYVYDITLAGD 292
KSLYGLK A R W++KL+ G+ Q + S F+ + +LVYV D+ + G+
Sbjct: 1124 RKSLYGLKQAPRCWFKKLSDALKRFGFIQGYEDYSFFSYSCKGIELRVLVYVDDLIICGN 1183
Query: 293 FLSEITFIKNALNQASKSKILVSL 316
+ K L + K L L
Sbjct: 1184 DEYMVQKFKEYLGRCFSMKDLGKL 1207
>At4g07810 putative polyprotein
Length = 1366
Score = 213 bits (541), Expect = 3e-55
Identities = 124/319 (38%), Positives = 181/319 (55%), Gaps = 11/319 (3%)
Query: 3 AHTHIPTTSPSPPITKTSPSPPFIPPPIRITTRNKIT--PTYM*DYICNIPTTSTTNNSH 60
AHTH T P++ T + F K T P+Y+ Y C+ +++ H
Sbjct: 801 AHTH---TRSLAPLSTTVTNDQFGNDMDNTLMPRKETRAPSYLSQYHCSNVLKEPSSSLH 857
Query: 61 -VQYPISNFLSHTYLSKSHSIFAMSLVSYTEPKSYDEAIKHDCWKQVMQNELTTLDQTGT 119
+ +S+ LS+ LS + +F ++++ EP ++ EA W M EL L T T
Sbjct: 858 GTAHSLSSHLSYDKLSNEYRLFCFAIIAEKEPTTFKEAALLQKWLDAMNVELDALVSTST 917
Query: 120 WKIVDLPPSSKPIGCIWVSKVKHNVDGSIERYKVCLVAKGYNQIEGLDYFDTFSLVAKVT 179
+I L + IGC WV K+K+ DG+IERYK LVA GY Q EG+DY DTFS +AK+T
Sbjct: 918 REICSLHDGKRAIGCKWVFKIKYKSDGTIERYKARLVANGYTQQEGVDYIDTFSPIAKLT 977
Query: 180 TIRLVIALASINY*FLHQLDVNNAFLHGDLHEDVYMAIPPGVSTSK-----PNQVCKLSK 234
++RL++ALA+I+ + Q+DV NAFLHGD E++YM +P G + K VC+L K
Sbjct: 978 SVRLILALAAIHNWSISQMDVTNAFLHGDFEEEIYMQLPQGYTPRKGELLPKRPVCRLVK 1037
Query: 235 SLYGLKPASRKWYEKLTCLPITNGYQQATSNASLFTKKNLDSFIMLLVYVYDITLAGDFL 294
SLYGLK ASR+W+ K + + I NG+ Q+ + +LF + D+F+ LLVYV DI L +
Sbjct: 1038 SLYGLKQASRQWFHKFSGVLIQNGFMQSLFDPTLFVRVREDTFLALLVYVDDIMLVSNKD 1097
Query: 295 SEITFIKNALNQASKSKIL 313
S + +K L + K K L
Sbjct: 1098 SAVIEVKQILAKEFKLKDL 1116
>At2g24660 putative retroelement pol polyprotein
Length = 1156
Score = 209 bits (533), Expect = 2e-54
Identities = 107/248 (43%), Positives = 154/248 (61%), Gaps = 5/248 (2%)
Query: 50 IPTTSTTNNSHVQ-----YPISNFLSHTYLSKSHSIFAMSLVSYTEPKSYDEAIKHDCWK 104
+P +S+ ++S VQ YP+S+++S S H F ++ + EPK + EA++ W
Sbjct: 593 LPDSSSQSSSMVQGTSSLYPLSDYVSDDCFSAGHKAFLAAITANDEPKHFKEAVRIKVWN 652
Query: 105 QVMQNELTTLDQTGTWKIVDLPPSSKPIGCIWVSKVKHNVDGSIERYKVCLVAKGYNQIE 164
M E+ L+ TW IVDLPP IG WV K K+N DGSIERYK LV +G Q+E
Sbjct: 653 DAMFKEVDALEINKTWDIVDLPPGKVAIGSQWVYKTKYNADGSIERYKARLVVQGNKQVE 712
Query: 165 GLDYFDTFSLVAKVTTIRLVIALASINY*FLHQLDVNNAFLHGDLHEDVYMAIPPGVSTS 224
G DY +TF+ V K+TT+R ++ L + N ++Q+DVNNAFLHGDL E+VYM +PPG S
Sbjct: 713 GEDYNETFAPVVKMTTVRTLLRLVAANQWEVYQMDVNNAFLHGDLDEEVYMKLPPGFRHS 772
Query: 225 KPNQVCKLSKSLYGLKPASRKWYEKLTCLPITNGYQQATSNASLFTKKNLDSFIMLLVYV 284
P++VC+L KSLYGLK A R W++KL+ + G+ Q + S F+ + +LVYV
Sbjct: 773 HPDKVCRLRKSLYGLKQAPRCWFKKLSDALLRFGFVQGHEDYSFFSYTRNGIELRVLVYV 832
Query: 285 YDITLAGD 292
D+ + G+
Sbjct: 833 DDLLICGN 840
>At2g20460 putative retroelement pol polyprotein
Length = 1461
Score = 209 bits (533), Expect = 2e-54
Identities = 122/316 (38%), Positives = 183/316 (57%), Gaps = 15/316 (4%)
Query: 5 THIPTTSPSPPITKTSPSPPFIPPPIRITTRNKIT--PTYM*DYICNIPTTSTTNNSHVQ 62
T + S IT PSP I P +I+ R +IT P ++ DY C N
Sbjct: 867 TPMDPLSSGNSITSHLPSPQ-ISPSTQISKR-RITKFPAHLQDYHCYFV------NKDDS 918
Query: 63 YPISNFLSHTYLSKSHSIFAMSLVSYTEPKSYDEAIKHDCWKQVMQNELTTLDQTGTWKI 122
+PIS+ LS++ +S SH ++ ++ P+SY EA W + E+ +++T TW+I
Sbjct: 919 HPISSSLSYSQISPSHMLYINNISKIPIPQSYHEAKDSKEWCGAIDQEIGAMERTDTWEI 978
Query: 123 VDLPPSSKPIGCIWVSKVKHNVDGSIERYKVCLVAKGYNQIEGLDYFDTFSLVAKVTTIR 182
LPP K +GC WV VK + DGS+ER+K +VAKGY Q EGLDY +TFS VAK+ T++
Sbjct: 979 TSLPPGKKAVGCKWVFTVKFHADGSLERFKARIVAKGYTQKEGLDYTETFSPVAKMATVK 1038
Query: 183 LVIALASINY*FLHQLDVNNAFLHGDLHEDVYMAIPPGVSTSK-----PNQVCKLSKSLY 237
L++ +++ +L+QLD++NAFL+GDL E +YM +P G + K PN VC+L KS+Y
Sbjct: 1039 LLLKVSASKKWYLNQLDISNAFLNGDLEETIYMKLPDGYADIKGTSLPPNVVCRLKKSIY 1098
Query: 238 GLKPASRKWYEKLTCLPITNGYQQATSNASLFTKKNLDSFIMLLVYVYDITLAGDFLSEI 297
GLK ASR+W+ K + + G+++ + +LF + FI+LLVYV DI +A
Sbjct: 1099 GLKQASRQWFLKFSNSLLALGFEKQHGDHTLFVRCIGSEFIVLLVYVDDIVIASTTEQAA 1158
Query: 298 TFIKNALNQASKSKIL 313
+ AL + K + L
Sbjct: 1159 QSLTEALKASFKLREL 1174
>At2g16000 putative retroelement pol polyprotein
Length = 1454
Score = 207 bits (526), Expect = 2e-53
Identities = 118/312 (37%), Positives = 182/312 (57%), Gaps = 17/312 (5%)
Query: 16 ITKTSPSPPFIP------PPIRITTRNKITPTYM*DYICNIPTTSTTNNSHVQYPISNFL 69
I+ T+ SP +P PP + R + P ++ DY CN T S +YPIS+ +
Sbjct: 865 ISDTTHSPSSLPSQISDLPPQISSQRVRKPPAHLNDYHCN------TMQSDHKYPISSTI 918
Query: 70 SHTYLSKSHSIFAMSLVSYTEPKSYDEAIKHDCWKQVMQNELTTLDQTGTWKIVDLPPSS 129
S++ +S SH + ++ P +Y EA W + + E+ +++T TW+I LP
Sbjct: 919 SYSKISPSHMCYINNITKIPIPTNYAEAQDTKEWCEAVDAEIGAMEKTNTWEITTLPKGK 978
Query: 130 KPIGCIWVSKVKHNVDGSIERYKVCLVAKGYNQIEGLDYFDTFSLVAKVTTIRLVIALAS 189
K +GC WV +K DG++ERYK LVAKGY Q EGLDY DTFS VAK+TTI+L++ +++
Sbjct: 979 KAVGCKWVFTLKFLADGNLERYKARLVAKGYTQKEGLDYTDTFSPVAKMTTIKLLLKVSA 1038
Query: 190 INY*FLHQLDVNNAFLHGDLHEDVYMAIPPGVSTSK-----PNQVCKLSKSLYGLKPASR 244
FL QLDV+NAFL+G+L E+++M IP G + K N V +L +S+YGLK ASR
Sbjct: 1039 SKKWFLKQLDVSNAFLNGELEEEIFMKIPEGYAERKGIVLPSNVVLRLKRSIYGLKQASR 1098
Query: 245 KWYEKLTCLPITNGYQQATSNASLFTKKNLDSFIMLLVYVYDITLAGDFLSEITFIKNAL 304
+W++K + ++ G+++ + +LF K F+++LVYV DI +A + + L
Sbjct: 1099 QWFKKFSSSLLSLGFKKTHGDHTLFLKMYDGEFVIVLVYVDDIVIASTSEAAAAQLTEEL 1158
Query: 305 NQASKSKILVSL 316
+Q K + L L
Sbjct: 1159 DQRFKLRDLGDL 1170
>At2g13940 putative retroelement pol polyprotein
Length = 1501
Score = 206 bits (524), Expect = 3e-53
Identities = 114/323 (35%), Positives = 177/323 (54%), Gaps = 14/323 (4%)
Query: 8 PTTSPSPPITKTSPSPPFIPPPIRITTRNKITPTYM*DYICNI--------------PTT 53
P T +P + + P PP R + R P + DY+ P+
Sbjct: 895 PNTPTTPIVVPVASPIPVSPPKQRKSKRATHPPPKLNDYVLYNAMYTPSSIHALPADPSQ 954
Query: 54 STTNNSHVQYPISNFLSHTYLSKSHSIFAMSLVSYTEPKSYDEAIKHDCWKQVMQNELTT 113
S+T +P+++++S S SH + ++ EPK + EA++ W M E+
Sbjct: 955 SSTVPGKSLFPLTDYVSDAAFSSSHRAYLAAITDNVEPKHFKEAVQIKVWNDAMFTEVDA 1014
Query: 114 LDQTGTWKIVDLPPSSKPIGCIWVSKVKHNVDGSIERYKVCLVAKGYNQIEGLDYFDTFS 173
L+ TW IVDLPP IG WV K K+N DG++ERYK LV +G Q+EG DY +TF+
Sbjct: 1015 LEINKTWDIVDLPPGKVAIGSQWVFKTKYNSDGTVERYKARLVVQGNKQVEGEDYKETFA 1074
Query: 174 LVAKVTTIRLVIALASINY*FLHQLDVNNAFLHGDLHEDVYMAIPPGVSTSKPNQVCKLS 233
V ++TT+R ++ + N ++Q+DV+NAFLHGDL E+VYM +PPG S P++VC+L
Sbjct: 1075 PVVRMTTVRTLLRNVAANQWEVYQMDVHNAFLHGDLEEEVYMKLPPGFRHSHPDKVCRLR 1134
Query: 234 KSLYGLKPASRKWYEKLTCLPITNGYQQATSNASLFTKKNLDSFIMLLVYVYDITLAGDF 293
KSLYGLK A R W++KL+ + G+ Q+ + SLF+ + + +L+YV D+ + G+
Sbjct: 1135 KSLYGLKQAPRCWFKKLSDSLLRFGFVQSYEDYSLFSYTRNNIELRVLIYVDDLLICGND 1194
Query: 294 LSEITFIKNALNQASKSKILVSL 316
+ K+ L++ K L L
Sbjct: 1195 GYMLQKFKDYLSRCFSMKDLGKL 1217
>At1g57640
Length = 1444
Score = 206 bits (523), Expect = 4e-53
Identities = 112/302 (37%), Positives = 176/302 (58%), Gaps = 7/302 (2%)
Query: 15 PITKTSPSPPFIPPPIRITTRNKITPTYM*DYICN---IPTTSTTNNSHVQYPISNFLSH 71
P++ T+P+P +R ++R P + +++ N + + S +S YPI ++
Sbjct: 860 PLSSTTPAPI----QLRRSSRQTQKPMKLKNFVTNTVSVESISPEASSSSLYPIEKYVDC 915
Query: 72 TYLSKSHSIFAMSLVSYTEPKSYDEAIKHDCWKQVMQNELTTLDQTGTWKIVDLPPSSKP 131
+ SH F ++ + EP +Y+EA+ W++ M E+ +L T+ IV+LPP +
Sbjct: 916 HRFTSSHKAFLAAVTAGMEPTTYNEAMVDKAWREAMSAEIESLRVNQTFSIVNLPPGKRA 975
Query: 132 IGCIWVSKVKHNVDGSIERYKVCLVAKGYNQIEGLDYFDTFSLVAKVTTIRLVIALASIN 191
+G WV K+K+ DG+IERYK LV G Q EG+DY +TF+ VAK++T+RL + +A+
Sbjct: 976 LGNKWVYKIKYRSDGAIERYKARLVVLGNCQKEGVDYDETFAPVAKMSTVRLFLGVAAAR 1035
Query: 192 Y*FLHQLDVNNAFLHGDLHEDVYMAIPPGVSTSKPNQVCKLSKSLYGLKPASRKWYEKLT 251
+HQ+DV+NAFLHGDL E+VYM +P G P++VC+L KSLYGLK A R W+ KL+
Sbjct: 1036 DWHVHQMDVHNAFLHGDLKEEVYMKLPQGFQCDDPSKVCRLHKSLYGLKQAPRCWFSKLS 1095
Query: 252 CLPITNGYQQATSNASLFTKKNLDSFIMLLVYVYDITLAGDFLSEITFIKNALNQASKSK 311
G+ Q+ S+ SLF+ N F+ +LVYV D+ ++G + K+ L K
Sbjct: 1096 SALKQYGFTQSLSDYSLFSYNNDGIFVHVLVYVDDLIISGSCPDAVAQFKSYLESCFHMK 1155
Query: 312 IL 313
L
Sbjct: 1156 DL 1157
>At1g70010 hypothetical protein
Length = 1315
Score = 205 bits (522), Expect = 5e-53
Identities = 120/343 (34%), Positives = 188/343 (53%), Gaps = 31/343 (9%)
Query: 8 PTTSPSPPITK----------TSPSPPFIP----------PPIRITTRNKITPTYM*DYI 47
P +P+PP+ + +S S +P P ++ + R P Y+ DY
Sbjct: 704 PDLNPTPPMQRQSSDHVNPSDSSSSVEILPSANPTNNVPEPSVQTSHRKAKKPAYLQDYY 763
Query: 48 CNIPTTSTTNNSHVQYPISNFLSHTYLSKSHSIFAMSLVSYTEPKSYDEAIKHDCWKQVM 107
C+ +ST + I FLS+ ++ + F L EP +Y EA K W+ M
Sbjct: 764 CHSVVSSTPHE------IRKFLSYDRINDPYLTFLACLDKTKEPSNYTEAEKLQVWRDAM 817
Query: 108 QNELTTLDQTGTWKIVDLPPSSKPIGCIWVSKVKHNVDGSIERYKVCLVAKGYNQIEGLD 167
E L+ T TW++ LP + IGC W+ K+K+N DGS+ERYK LVA+GY Q EG+D
Sbjct: 818 GAEFDFLEGTHTWEVCSLPADKRCIGCRWIFKIKYNSDGSVERYKARLVAQGYTQKEGID 877
Query: 168 YFDTFSLVAKVTTIRLVIALASINY*FLHQLDVNNAFLHGDLHEDVYMAIPPGVSTSK-- 225
Y +TFS VAK+ +++L++ +A+ L QLD++NAFL+GDL E++YM +P G ++ +
Sbjct: 878 YNETFSPVAKLNSVKLLLGVAARFKLSLTQLDISNAFLNGDLDEEIYMRLPQGYASRQGD 937
Query: 226 ---PNQVCKLSKSLYGLKPASRKWYEKLTCLPITNGYQQATSNASLFTKKNLDSFIMLLV 282
PN VC+L KSLYGLK ASR+WY K + + G+ Q+ + + F K + F+ +LV
Sbjct: 938 SLPPNAVCRLKKSLYGLKQASRQWYLKFSSTLLGLGFIQSYCDHTCFLKISDGIFLCVLV 997
Query: 283 YVYDITLAGDFLSEITFIKNALNQASKSKILVSLNIS*ALKFV 325
Y+ DI +A + + + +K+ + K + L L L+ V
Sbjct: 998 YIDDIIIASNNDAAVDILKSQMKSFFKLRDLGELKYFLGLEIV 1040
>At4g10990 putative retrotransposon polyprotein
Length = 1203
Score = 203 bits (516), Expect = 2e-52
Identities = 119/310 (38%), Positives = 179/310 (57%), Gaps = 27/310 (8%)
Query: 29 PIRITTRNKITPTYM*DYICNI--------PTTST---TNNSHVQ-------YPISNFLS 70
PI RN P Y+ +Y CN PTTST T +S + YP+S +S
Sbjct: 445 PIARPKRNAKAPAYLSEYHCNSVPFLSSLSPTTSTSIETPSSSIPPKKITTPYPMSTAIS 504
Query: 71 HTYLSKSHSIFAMSLVSYTEPKSYDEAIKHDCWKQVMQNELTTLDQTGTWKIVDLPPSSK 130
+ L+ + + TEPK++ +A+K + W + EL L+Q TW + L
Sbjct: 505 YDKLTPLFHSYICAYNVETEPKAFTQAMKSEKWTRAANEELHALEQNKTWIVESLTEGKN 564
Query: 131 PIGCIWVSKVKHNVDGSIERYKVCLVAKGYNQIEGLDYFDTFSLVAKVTTIRLVIALASI 190
+GC WV +K+N DGSIERYK LVA+G+ Q EG+DY +TFS VAK +++L++ LA+
Sbjct: 565 VVGCKWVFTIKYNPDGSIERYKARLVAQGFTQQEGIDYMETFSPVAKFGSVKLLLGLAAA 624
Query: 191 NY*FLHQLDVNNAFLHGDLHEDVYMAIPPGVS-------TSKPNQVCKLSKSLYGLKPAS 243
L Q+DV+NAFLHG+L E++YM++P G + SKP VC+L KSLYGLK AS
Sbjct: 625 TGWSLTQMDVSNAFLHGELDEEIYMSLPQGYTPPTGISLPSKP--VCRLLKSLYGLKQAS 682
Query: 244 RKWYEKLTCLPITNGYQQATSNASLFTKKNLDSFIMLLVYVYDITLAGDFLSEITFIKNA 303
R+WY++L+ + + + Q+ ++ ++F K + S I++LVYV D+ +A + S + +K
Sbjct: 683 RQWYKRLSSVFLGANFIQSPADNTMFVKVSCTSIIVVLVYVDDLMIASNDSSAVENLKEL 742
Query: 304 LNQASKSKIL 313
L K K L
Sbjct: 743 LRSEFKIKDL 752
>At2g16670 putative retroelement pol polyprotein
Length = 1333
Score = 200 bits (509), Expect = 1e-51
Identities = 113/284 (39%), Positives = 157/284 (54%), Gaps = 29/284 (10%)
Query: 11 SPSPPITKTSPSPPFIPPPIRITTRNKITPTYM*DYICN--IPTTSTTNNSHVQYPISNF 68
S + P T + +PP +PPP R +TRNK P + D++ N + S + + + Y +
Sbjct: 769 SRAKPTTAQAVAPPAVPPPRRQSTRNKAPPVTLKDFVVNTTVCQESPSKLNSILYQLQKR 828
Query: 69 LSHTYLSKSHSIFAMSLVSYTEPKSYDEAIKHDCWKQVMQNELTTLDQTGTWKIVDLPPS 128
S SH+ + + ++ TW I DLPP
Sbjct: 829 DDTRRFSASHTTYV---------------------------AIDAQEENHTWTIEDLPPG 861
Query: 129 SKPIGCIWVSKVKHNVDGSIERYKVCLVAKGYNQIEGLDYFDTFSLVAKVTTIRLVIALA 188
+ IG WV KVKHN DGS+ERYK LVA G Q EG DY +TF+ VAK+ T+RL + +A
Sbjct: 862 KRAIGSQWVYKVKHNSDGSVERYKARLVALGNKQKEGEDYGETFAPVAKMATVRLFLDVA 921
Query: 189 SINY*FLHQLDVNNAFLHGDLHEDVYMAIPPGVSTSKPNQVCKLSKSLYGLKPASRKWYE 248
+HQ+DV+NAFLHGDL E+VYM +PPG S PN+VC+L K+LYGLK A R W+E
Sbjct: 922 VKRNWEIHQMDVHNAFLHGDLREEVYMKLPPGFEASHPNKVCRLRKALYGLKQAPRCWFE 981
Query: 249 KLTCLPITNGYQQATSNASLFTKKNLDSFIMLLVYVYDITLAGD 292
KLT G+QQ+ ++ SLFT I +L+YV D+ + G+
Sbjct: 982 KLTTALKRYGFQQSLADYSLFTLVKGSVRIKILIYVDDLIITGN 1025
>At4g17450 retrotransposon like protein
Length = 1433
Score = 200 bits (508), Expect = 2e-51
Identities = 114/293 (38%), Positives = 166/293 (55%), Gaps = 31/293 (10%)
Query: 5 THIPTTSPSPPITKTSPSPPFIPPPIRITTRNKITPTYM*DYICNIPTTSTTNNSHVQYP 64
T I T P ++ + PF P ++ R K P ++ D+ C TT P
Sbjct: 857 TSIIDTHPHQDVSSSKALVPFDP----LSKRQKKPPKHLQDFHCYNNTTE---------P 903
Query: 65 ISNFLSHTYLSKSHSIFAMSLVSYTEPKSYDEAIKHDCWKQVMQNELTTLDQTGTWKIVD 124
F+++ + + P+ Y EA W M+ E+ + +T TW +V
Sbjct: 904 FHAFINN-------------ITNAVIPQRYSEAKDFKAWCDAMKEEIGAMVRTNTWSVVS 950
Query: 125 LPPSSKPIGCIWVSKVKHNVDGSIERYKVCLVAKGYNQIEGLDYFDTFSLVAKVTTIRLV 184
LPP+ K IGC WV +KHN DGSIERYK LVAKGY Q EGLDY +TFS VAK+T++R++
Sbjct: 951 LPPNKKAIGCKWVFTIKHNADGSIERYKARLVAKGYTQEEGLDYEETFSPVAKLTSVRMM 1010
Query: 185 IALASINY*FLHQLDVNNAFLHGDLHEDVYMAIPPGVS-----TSKPNQVCKLSKSLYGL 239
+ LA+ +HQLD++NAFL+GDL E++YM IPPG + P+ +C+L KS+YGL
Sbjct: 1011 LLLAAKMKWSVHQLDISNAFLNGDLDEEIYMKIPPGYADLVGEALPPHAICRLHKSIYGL 1070
Query: 240 KPASRKWYEKLTCLPITNGYQQATSNASLFTKKNLDSFIMLLVYVYDITLAGD 292
K ASR+WY KL+ G+Q++ ++ +LF K + +LVYV DI + +
Sbjct: 1071 KQASRQWYLKLSNTLKGMGFQKSNADHTLFIKYANGVLMGVLVYVDDIMIVSN 1123
>At4g22040 LTR retrotransposon like protein
Length = 1109
Score = 194 bits (493), Expect = 1e-49
Identities = 102/252 (40%), Positives = 153/252 (60%), Gaps = 1/252 (0%)
Query: 62 QYPISNFLSHTYLSKSHSIFAMSLVSYTEPKSYDEAIKHDCWKQVMQNELTTLDQTGTWK 121
++P S + + S SH F ++ + EP +Y+EA+ W++ M E+ +L T+
Sbjct: 572 EFPYSKMSCNRFTS-SHKAFLAAVTAGMEPTTYNEAMVDKAWREAMSAEIESLRVNQTFS 630
Query: 122 IVDLPPSSKPIGCIWVSKVKHNVDGSIERYKVCLVAKGYNQIEGLDYFDTFSLVAKVTTI 181
IV+LPP + +G WV K+K+ DG+IERYK LV G Q EG+DY +TF+ VAK++T+
Sbjct: 631 IVNLPPGKRALGNKWVYKIKYRSDGAIERYKARLVVLGNCQKEGVDYDETFAPVAKMSTV 690
Query: 182 RLVIALASINY*FLHQLDVNNAFLHGDLHEDVYMAIPPGVSTSKPNQVCKLSKSLYGLKP 241
RL + +A+ +HQ+DV+NAFLHGDL E+VYM +P G P++VC+L KSLYGLK
Sbjct: 691 RLFLGVAAARDWHVHQMDVHNAFLHGDLKEEVYMKLPQGFQCDDPSKVCRLHKSLYGLKQ 750
Query: 242 ASRKWYEKLTCLPITNGYQQATSNASLFTKKNLDSFIMLLVYVYDITLAGDFLSEITFIK 301
A R W+ KL+ G+ Q+ S+ SLF+ N F+ +LVYV D+ ++G + K
Sbjct: 751 APRCWFSKLSSALKQYGFTQSLSDYSLFSYNNDGVFVHVLVYVDDLIISGSCPDAVAQFK 810
Query: 302 NALNQASKSKIL 313
+ L K L
Sbjct: 811 SYLESCFHMKDL 822
>At1g59265 polyprotein, putative
Length = 1466
Score = 194 bits (492), Expect = 1e-49
Identities = 123/309 (39%), Positives = 171/309 (54%), Gaps = 30/309 (9%)
Query: 8 PTTSPSPPITKTSPSPPFI----PPPI-RITTRNKITPTYM*DYICNIPTTSTTNNSHVQ 62
PTTS S + TSP+PP I PPP+ +I N P N + T + +
Sbjct: 888 PTTSASS--SSTSPTPPSILIHPPPPLAQIVNNNNQAP-------LNTHSMGTRAKAGII 938
Query: 63 YPISNFLSHTYLSKSHSIFAMSLVSYTEPKSYDEAIKHDCWKQVMQNELTTLDQTGTWKI 122
P + A+SL + +EP++ +A+K + W+ M +E+ TW +
Sbjct: 939 KPNPKY-----------SLAVSLAAESEPRTAIQALKDERWRNAMGSEINAQIGNHTWDL 987
Query: 123 VDLPPSSKPI-GCIWVSKVKHNVDGSIERYKVCLVAKGYNQIEGLDYFDTFSLVAKVTTI 181
V PPS I GC W+ K+N DGS+ RYK LVAKGYNQ GLDY +TFS V K T+I
Sbjct: 988 VPPPPSHVTIVGCRWIFTKKYNSDGSLNRYKARLVAKGYNQRPGLDYAETFSPVIKSTSI 1047
Query: 182 RLVIALASINY*FLHQLDVNNAFLHGDLHEDVYMAIPPG-VSTSKPNQVCKLSKSLYGLK 240
R+V+ +A + QLDVNNAFL G L +DVYM+ PPG + +PN VCKL K+LYGLK
Sbjct: 1048 RIVLGVAVDRSWPIRQLDVNNAFLQGTLTDDVYMSQPPGFIDKDRPNYVCKLRKALYGLK 1107
Query: 241 PASRKWYEKLTCLPITNGYQQATSNASLFTKKNLDSFIMLLVYVYDITLAGDFLSEITFI 300
A R WY +L +T G+ + S+ SLF + S + +LVYV DI + G ++ T +
Sbjct: 1108 QAPRAWYVELRNYLLTIGFVNSVSDTSLFVLQRGKSIVYMLVYVDDILITG---NDPTLL 1164
Query: 301 KNALNQASK 309
N L+ S+
Sbjct: 1165 HNTLDNLSQ 1173
>At1g58889 polyprotein, putative
Length = 1466
Score = 192 bits (488), Expect = 4e-49
Identities = 122/309 (39%), Positives = 170/309 (54%), Gaps = 30/309 (9%)
Query: 8 PTTSPSPPITKTSPSPPFI----PPPI-RITTRNKITPTYM*DYICNIPTTSTTNNSHVQ 62
PTTS S + TSP+PP I PPP+ +I N P N + T + +
Sbjct: 888 PTTSASS--SSTSPTPPSILIHPPPPLAQIVNNNNQAP-------LNTHSMGTRAKAGII 938
Query: 63 YPISNFLSHTYLSKSHSIFAMSLVSYTEPKSYDEAIKHDCWKQVMQNELTTLDQTGTWKI 122
P + A+SL + +EP++ +A+K + W+ M +E+ TW +
Sbjct: 939 KPNPKY-----------SLAVSLAAESEPRTAIQALKDERWRNAMGSEINAQIGNHTWDL 987
Query: 123 VDLPPSSKPI-GCIWVSKVKHNVDGSIERYKVCLVAKGYNQIEGLDYFDTFSLVAKVTTI 181
V PPS I GC W+ K+N DGS+ RYK VAKGYNQ GLDY +TFS V K T+I
Sbjct: 988 VPPPPSHVTIVGCRWIFTKKYNSDGSLNRYKARFVAKGYNQRPGLDYAETFSPVIKSTSI 1047
Query: 182 RLVIALASINY*FLHQLDVNNAFLHGDLHEDVYMAIPPG-VSTSKPNQVCKLSKSLYGLK 240
R+V+ +A + QLDVNNAFL G L +DVYM+ PPG + +PN VCKL K+LYGLK
Sbjct: 1048 RIVLGVAVDRSWPIRQLDVNNAFLQGTLTDDVYMSQPPGFIDKDRPNYVCKLRKALYGLK 1107
Query: 241 PASRKWYEKLTCLPITNGYQQATSNASLFTKKNLDSFIMLLVYVYDITLAGDFLSEITFI 300
A R WY +L +T G+ + S+ SLF + S + +LVYV DI + G ++ T +
Sbjct: 1108 QAPRAWYVELRNYLLTIGFVNSVSDTSLFVLQRGKSIVYMLVYVDDILITG---NDPTLL 1164
Query: 301 KNALNQASK 309
N L+ S+
Sbjct: 1165 HNTLDNLSQ 1173
>At4g02960 putative polyprotein of LTR transposon
Length = 1456
Score = 191 bits (486), Expect = 7e-49
Identities = 122/311 (39%), Positives = 167/311 (53%), Gaps = 29/311 (9%)
Query: 6 HIPT--------TSPSPPITKTSPSPPFIPPPIRITTRNKITPTYM*DYICNIPTTSTTN 57
HIPT SPS T T P PP +P P I N P N + +T
Sbjct: 865 HIPTPSTSISEPNSPSSSSTSTPPLPPVLPAPPIIQV-NAQAPV-------NTHSMATRA 916
Query: 58 NSHVQYPISNFLSHTYLSKSHSIFAMSLVSYTEPKSYDEAIKHDCWKQVMQNELTTLDQT 117
++ P + +A SL + +EP++ +A+K D W+Q M +E+
Sbjct: 917 KDGIRKPNQKYS-----------YATSLAANSEPRTAIQAMKDDRWRQAMGSEINAQIGN 965
Query: 118 GTWKIVDLPPSSKPI-GCIWVSKVKHNVDGSIERYKVCLVAKGYNQIEGLDYFDTFSLVA 176
TW +V PP S I GC W+ K N DGS+ RYK LVAKGYNQ GLDY +TFS V
Sbjct: 966 HTWDLVPPPPPSVTIVGCRWIFTKKFNSDGSLNRYKARLVAKGYNQRPGLDYAETFSPVI 1025
Query: 177 KVTTIRLVIALASINY*FLHQLDVNNAFLHGDLHEDVYMAIPPG-VSTSKPNQVCKLSKS 235
K T+IR+V+ +A + QLDVNNAFL G L ++VYM+ PPG V +P+ VC+L K+
Sbjct: 1026 KSTSIRIVLGVAVDRSWPIRQLDVNNAFLQGTLTDEVYMSQPPGFVDKDRPDYVCRLRKA 1085
Query: 236 LYGLKPASRKWYEKLTCLPITNGYQQATSNASLFTKKNLDSFIMLLVYVYDITLAGDFLS 295
+YGLK A R WY +L +T G+ + S+ SLF + S I +LVYV DI + G+
Sbjct: 1086 IYGLKQAPRAWYVELRTYLLTVGFVNSISDTSLFVLQRGRSIIYMLVYVDDILITGNDTV 1145
Query: 296 EITFIKNALNQ 306
+ +AL+Q
Sbjct: 1146 LLKHTLDALSQ 1156
>At4g14460 retrovirus-related like polyprotein
Length = 1489
Score = 185 bits (470), Expect = 5e-47
Identities = 115/352 (32%), Positives = 179/352 (50%), Gaps = 72/352 (20%)
Query: 7 IPTTSPSPPI-----TKTSPSPPFIPP--------------PIRITTRNKITPTYM*DYI 47
IPTT+ S + +S P IPP PI + R P+Y+ +Y
Sbjct: 899 IPTTTDSRTADNHASSSSSALPSIIPPSSNTETQDIDSNAVPITRSKRTTRAPSYLSEYH 958
Query: 48 CNI--------PT-------------TSTTNNSHVQYPISNFLSHTYLSKSHSIFAMSLV 86
C++ PT T+++ YPIS +S+ + + +
Sbjct: 959 CSLVPSISTLPPTDSSIPIHPLPEIFTASSPKKTTPYPISTVVSYDKYTPLCQSYIFAYN 1018
Query: 87 SYTEPKSYDEAIKHDCWKQVMQNELTTLDQTGTWKIVDLPPSSKPIGCIWVSKVKHNVDG 146
+ TEPK++ +A+K + W +V EL ++ TW + LPP +GC WV +K+N DG
Sbjct: 1019 TETEPKTFSQAMKSEKWIRVAVEELQAMELNKTWSVESLPPDKNVVGCKWVFTIKYNPDG 1078
Query: 147 SIERYKVCLVAKGYNQIEGLDYFDTFSLVAKVTTIRLVIALASINY*FLHQLDVNNAFLH 206
++ERYK LVA+G+ Q EG+D+ DTFS VAK+T+ ++++ LA+I L Q+DV++AFLH
Sbjct: 1079 TVERYKARLVAQGFTQQEGIDFLDTFSPVAKLTSAKMMLGLAAITGWTLTQMDVSDAFLH 1138
Query: 207 GDLHEDVYMAIPPGVSTS-----KPNQVCKLSKSLYGLKPASRKWYEKLTCLPITNGYQQ 261
GDL E+++M++P G + PN VC+L KS+YGLK ASR+WY++
Sbjct: 1139 GDLDEEIFMSLPQGYTPPAGTILPPNPVCRLLKSIYGLKQASRQWYKR------------ 1186
Query: 262 ATSNASLFTKKNLDSFIMLLVYVYDITLAGDFLSEITFIKNALNQASKSKIL 313
F+ LVY+ DI +A + +E+ +K L K K L
Sbjct: 1187 ---------------FVAALVYIDDIMIASNNDAEVENLKALLRSEFKIKDL 1223
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.336 0.146 0.457
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,183,098
Number of Sequences: 26719
Number of extensions: 458741
Number of successful extensions: 5115
Number of sequences better than 10.0: 124
Number of HSP's better than 10.0 without gapping: 108
Number of HSP's successfully gapped in prelim test: 18
Number of HSP's that attempted gapping in prelim test: 3476
Number of HSP's gapped (non-prelim): 1314
length of query: 552
length of database: 11,318,596
effective HSP length: 104
effective length of query: 448
effective length of database: 8,539,820
effective search space: 3825839360
effective search space used: 3825839360
T: 11
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.7 bits)
S2: 63 (28.9 bits)
Medicago: description of AC146555.15


