

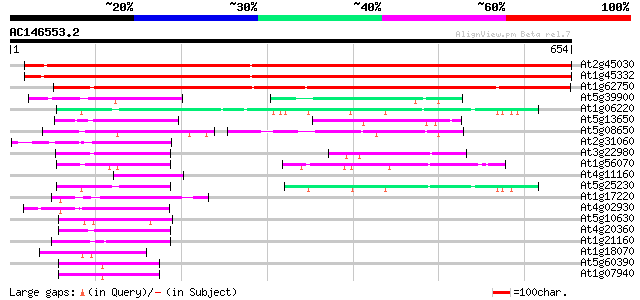
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146553.2 + phase: 0 /pseudo
(654 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g45030 putative mitochondrial translation elongation factor G 1028 0.0
At1g45332 mitochondrial elongation factor, putative 1024 0.0
At1g62750 elongation factor G, putative 487 e-138
At5g39900 GTP-binding protein-like 92 1e-18
At1g06220 elongation factor like protein 92 1e-18
At5g13650 GTP-binding protein typA (tyrosine phosphorylated prot... 91 2e-18
At5g08650 GTP-binding protein LepA homolog 91 3e-18
At2g31060 putative GTP-binding protein 87 2e-17
At3g22980 eukaryotic translation elongation factor 2, putative 79 8e-15
At1g56070 elongation factor like protein 74 2e-13
At4g11160 translation initiation factor IF-2 like protein 59 1e-08
At5g25230 translation Elongation Factor 2 - like protein 58 2e-08
At1g17220 putative translation initiation factor IF2 57 3e-08
At4g02930 mitochondrial elongation factor Tu 56 7e-08
At5g10630 putative protein 55 9e-08
At4g20360 translation elongation factor EF-Tu precursor, chlorop... 53 5e-07
At1g21160 transcription factor, putative 48 2e-05
At1g18070 unknown protein 46 7e-05
At5g60390 translation elongation factor eEF-1 alpha chain (gene A4) 43 5e-04
At1g07940 elongation factor 1-alpha 43 5e-04
>At2g45030 putative mitochondrial translation elongation factor G
Length = 754
Score = 1028 bits (2659), Expect = 0.0
Identities = 509/638 (79%), Positives = 576/638 (89%), Gaps = 4/638 (0%)
Query: 18 SLIAGTFH-IRHFSAGNVARATAATIDKDPWWKESMEKVRNIGISAHIDSGKTTLTEWIL 76
+L+ G FH IRHFSAG ARA +K+PWWKESM+K+RNIGISAHIDSGKTTLTE +L
Sbjct: 29 ALLTGDFHLIRHFSAGTAARAVKD--EKEPWWKESMDKLRNIGISAHIDSGKTTLTERVL 86
Query: 77 FYTGKIHLMYEVRSKDGMGPKMDFKPLEIIMGITIKSAATYCNWKGSKITIIDTPGHVDF 136
FYTG+IH ++EVR +DG+G KMD LE GITI+SAATYC WK K+ IIDTPGHVDF
Sbjct: 87 FYTGRIHEIHEVRGRDGVGAKMDSMDLEREKGITIQSAATYCTWKDYKVNIIDTPGHVDF 146
Query: 137 TIEVERALRVLDGAVLVLCSVGGVQCQSITVDRQMRRYQVPRIAFINKLDRPGADPWKVI 196
TIEVERALRVLDGA+LVLCSVGGVQ QSITVDRQMRRY+VPR+AFINKLDR GADPWKV+
Sbjct: 147 TIEVERALRVLDGAILVLCSVGGVQSQSITVDRQMRRYEVPRVAFINKLDRMGADPWKVL 206
Query: 197 TQARSKLRHHCAALQVPIGLESDFKGVVDLVKLKAYCFDGQYGQNVVVGEVPADMEALVA 256
QAR+KLRHH AA+QVPIGLE +F+G++DL+ +KAY F G G+NVV G++PADME LV
Sbjct: 207 NQARAKLRHHSAAVQVPIGLEENFQGLIDLIHVKAYFFHGSSGENVVAGDIPADMEGLVG 266
Query: 257 EKRRELIETVSEVDDVLAEAFLSDDENISAADLEGAIRRATIARKFIPVFMGSAVKNTGV 316
+KRRELIETVSEVDDVLAE FL+D E +SAA+LE AIRRATIA+KF+PVFMGSA KN GV
Sbjct: 267 DKRRELIETVSEVDDVLAEKFLND-EPVSAAELEEAIRRATIAQKFVPVFMGSAFKNKGV 325
Query: 317 QPLLDGVVSYLPCPIEVSNYALDQSKNEEKVQLTGSPDGPLVALAFKLEQTKFGQLTYLR 376
QPLLDGVVS+LP P EV+NYALDQ+ NEE+V LTGSPDGPLVALAFKLE+ +FGQLTYLR
Sbjct: 326 QPLLDGVVSFLPSPNEVNNYALDQNNNEERVTLTGSPDGPLVALAFKLEEGRFGQLTYLR 385
Query: 377 VYEGVIRKGDFIVNVSTGKKIKVPRLVQMHSNEMNDIEEAHAGQIVAVFGVDCASSDTFT 436
VYEGVI+KGDFI+NV+TGK+IKVPRLV+MHSN+M DI+EAHAGQIVAVFG++CAS DTFT
Sbjct: 386 VYEGVIKKGDFIINVNTGKRIKVPRLVRMHSNDMEDIQEAHAGQIVAVFGIECASGDTFT 445
Query: 437 DGSVKYTMTSMNVPEPVMSLAVQPVSKDSGGKFSKALNRFQREDPTFRVSLDPESGQTII 496
DGSVKYTMTSMNVPEPVMSLAVQPVSKDSGG+FSKALNRFQ+EDPTFRV LDPESGQTII
Sbjct: 446 DGSVKYTMTSMNVPEPVMSLAVQPVSKDSGGQFSKALNRFQKEDPTFRVGLDPESGQTII 505
Query: 497 SGMGELHLDIYVKRIKMEYGVDATVGKPRVNFRETVTQRADFDYLHKKQSGGQGQYGRVI 556
SGMGELHLDIYV+R++ EY VDATVGKPRVNFRET+TQRA+FDYLHKKQSGG GQYGRV
Sbjct: 506 SGMGELHLDIYVERMRREYKVDATVGKPRVNFRETITQRAEFDYLHKKQSGGAGQYGRVT 565
Query: 557 GYIEPLPAGSGTKFEFDNMLVGQAIPSNFFPAIEKGFKEAANSGALIGHPVQNLRVVLTD 616
GY+EPLP GS KFEF+NM+VGQAIPS F PAIEKGFKEAANSG+LIGHPV+NLR+VLTD
Sbjct: 566 GYVEPLPPGSKEKFEFENMIVGQAIPSGFIPAIEKGFKEAANSGSLIGHPVENLRIVLTD 625
Query: 617 GAAHDVDSSELAFKLASIYAFRECYTASRPVILEPVML 654
GA+H VDSSELAFK+A+IYAFR CYTA+RPVILEPVML
Sbjct: 626 GASHAVDSSELAFKMAAIYAFRLCYTAARPVILEPVML 663
>At1g45332 mitochondrial elongation factor, putative
Length = 754
Score = 1024 bits (2648), Expect = 0.0
Identities = 508/638 (79%), Positives = 575/638 (89%), Gaps = 4/638 (0%)
Query: 18 SLIAGTFH-IRHFSAGNVARATAATIDKDPWWKESMEKVRNIGISAHIDSGKTTLTEWIL 76
+L+ G F IRHFSAG AR A +K+PWWKESM+K+RNIGISAHIDSGKTTLTE +L
Sbjct: 29 ALLTGDFQLIRHFSAGTAARV--AKDEKEPWWKESMDKLRNIGISAHIDSGKTTLTERVL 86
Query: 77 FYTGKIHLMYEVRSKDGMGPKMDFKPLEIIMGITIKSAATYCNWKGSKITIIDTPGHVDF 136
FYTG+IH ++EVR +DG+G KMD LE GITI+SAATYC WK K+ IIDTPGHVDF
Sbjct: 87 FYTGRIHEIHEVRGRDGVGAKMDSMDLEREKGITIQSAATYCTWKDYKVNIIDTPGHVDF 146
Query: 137 TIEVERALRVLDGAVLVLCSVGGVQCQSITVDRQMRRYQVPRIAFINKLDRPGADPWKVI 196
TIEVERALRVLDGA+LVLCSVGGVQ QSITVDRQMRRY+VPR+AFINKLDR GADPWKV+
Sbjct: 147 TIEVERALRVLDGAILVLCSVGGVQSQSITVDRQMRRYEVPRVAFINKLDRMGADPWKVL 206
Query: 197 TQARSKLRHHCAALQVPIGLESDFKGVVDLVKLKAYCFDGQYGQNVVVGEVPADMEALVA 256
QAR+KLRHH AA+QVPIGLE +F+G++DL+ +KAY F G G+NVV G++PADME LVA
Sbjct: 207 NQARAKLRHHSAAVQVPIGLEENFQGLIDLIHVKAYFFHGSSGENVVAGDIPADMEGLVA 266
Query: 257 EKRRELIETVSEVDDVLAEAFLSDDENISAADLEGAIRRATIARKFIPVFMGSAVKNTGV 316
EKRRELIETVSEVDDVLAE FL+D E +SA++LE AIRRATIA+ F+PVFMGSA KN GV
Sbjct: 267 EKRRELIETVSEVDDVLAEKFLND-EPVSASELEEAIRRATIAQTFVPVFMGSAFKNKGV 325
Query: 317 QPLLDGVVSYLPCPIEVSNYALDQSKNEEKVQLTGSPDGPLVALAFKLEQTKFGQLTYLR 376
QPLLDGVVS+LP P EV+NYALDQ+ NEE+V LTGSPDGPLVALAFKLE+ +FGQLTYLR
Sbjct: 326 QPLLDGVVSFLPSPNEVNNYALDQNNNEERVTLTGSPDGPLVALAFKLEEGRFGQLTYLR 385
Query: 377 VYEGVIRKGDFIVNVSTGKKIKVPRLVQMHSNEMNDIEEAHAGQIVAVFGVDCASSDTFT 436
VYEGVI+KGDFI+NV+TGK+IKVPRLV+MHSN+M DI+EAHAGQIVAVFG++CAS DTFT
Sbjct: 386 VYEGVIKKGDFIINVNTGKRIKVPRLVRMHSNDMEDIQEAHAGQIVAVFGIECASGDTFT 445
Query: 437 DGSVKYTMTSMNVPEPVMSLAVQPVSKDSGGKFSKALNRFQREDPTFRVSLDPESGQTII 496
DGSVKYTMTSMNVPEPVMSLAVQPVSKDSGG+FSKALNRFQ+EDPTFRV LDPESGQTII
Sbjct: 446 DGSVKYTMTSMNVPEPVMSLAVQPVSKDSGGQFSKALNRFQKEDPTFRVGLDPESGQTII 505
Query: 497 SGMGELHLDIYVKRIKMEYGVDATVGKPRVNFRETVTQRADFDYLHKKQSGGQGQYGRVI 556
SGMGELHLDIYV+R++ EY VDATVGKPRVNFRET+TQRA+FDYLHKKQSGG GQYGRV
Sbjct: 506 SGMGELHLDIYVERMRREYKVDATVGKPRVNFRETITQRAEFDYLHKKQSGGAGQYGRVT 565
Query: 557 GYIEPLPAGSGTKFEFDNMLVGQAIPSNFFPAIEKGFKEAANSGALIGHPVQNLRVVLTD 616
GY+EPLP GS KFEF+NM+VGQAIPS F PAIEKGFKEAANSG+LIGHPV+NLR+VLTD
Sbjct: 566 GYVEPLPPGSKEKFEFENMIVGQAIPSGFIPAIEKGFKEAANSGSLIGHPVENLRIVLTD 625
Query: 617 GAAHDVDSSELAFKLASIYAFRECYTASRPVILEPVML 654
GA+H VDSSELAFK+A+IYAFR CYTA+RPVILEPVML
Sbjct: 626 GASHAVDSSELAFKMAAIYAFRLCYTAARPVILEPVML 663
>At1g62750 elongation factor G, putative
Length = 783
Score = 487 bits (1254), Expect = e-138
Identities = 272/607 (44%), Positives = 380/607 (61%), Gaps = 12/607 (1%)
Query: 52 MEKVRNIGISAHIDSGKTTLTEWILFYTGKIHLMYEVRSKDGMGPKMDFKPLEIIMGITI 111
++ RNIGI AHID+GKTT TE IL+YTG+ + + EV MD+ E GITI
Sbjct: 93 LKDYRNIGIMAHIDAGKTTTTERILYYTGRNYKIGEVHEGTAT---MDWMEQEQERGITI 149
Query: 112 KSAATYCNWKGSKITIIDTPGHVDFTIEVERALRVLDGAVLVLCSVGGVQCQSITVDRQM 171
SAAT W +I IIDTPGHVDFT+EVERALRVLDGA+ + SV GV+ QS TV RQ
Sbjct: 150 TSAATTTFWDKHRINIIDTPGHVDFTLEVERALRVLDGAICLFDSVAGVEPQSETVWRQA 209
Query: 172 RRYQVPRIAFINKLDRPGADPWKVITQARSKLRHHCAALQVPIGLESDFKGVVDLVKLKA 231
+Y VPRI F+NK+DR GA+ ++ + L LQ+PIG E FKGVVDLV++KA
Sbjct: 210 DKYGVPRICFVNKMDRLGANFFRTRDMIVTNLGAKPLVLQIPIGAEDVFKGVVDLVRMKA 269
Query: 232 YCFDG-QYGQNVVVGEVPADMEALVAEKRRELIETVSEVDDVLAEAFLSDDENISAADLE 290
+ G + G ++P D+E L E R ++E + ++DD + E +L E A ++
Sbjct: 270 IVWSGEELGAKFSYEDIPEDLEDLAQEYRAAMMELIVDLDDEVMENYLEGVEP-DEATVK 328
Query: 291 GAIRRATIARKFIPVFMGSAVKNTGVQPLLDGVVSYLPCPIEVSNYALDQSKNEEKVQLT 350
+R+ TI KF+P+ GSA KN GVQPLLD VV YLP P+EV +N E + +
Sbjct: 329 RLVRKGTITGKFVPILCGSAFKNKGVQPLLDAVVDYLPSPVEVPPMNGTDPENPE-ITII 387
Query: 351 GSPDG--PLVALAFKLEQTKF-GQLTYLRVYEGVIRKGDFIVNVSTGKKIKVPRLVQMHS 407
PD P LAFK+ F G LT++RVY G I G +++N + GKK ++ RL++MH+
Sbjct: 388 RKPDDDEPFAGLAFKIMSDPFVGSLTFVRVYSGKISAGSYVLNANKGKKERIGRLLEMHA 447
Query: 408 NEMNDIEEAHAGQIVAVFGV-DCASSDTFTDGSVKYTMTSMNVPEPVMSLAVQPVSKDSG 466
N D++ A G I+A+ G+ D + +T +D + M+ P+PV+ +A++P +K
Sbjct: 448 NSREDVKVALTGDIIALAGLKDTITGETLSDPENPVVLERMDFPDPVIKVAIEPKTKADI 507
Query: 467 GKFSKALNRFQREDPTFRVSLDPESGQTIISGMGELHLDIYVKRIKMEYGVDATVGKPRV 526
K + L + +EDP+F S D E QT+I GMGELHL+I V R+K E+ V+A VG P+V
Sbjct: 508 DKMATGLIKLAQEDPSFHFSRDEEMNQTVIEGMGELHLEIIVDRLKREFKVEANVGAPQV 567
Query: 527 NFRETVTQRADFDYLHKKQSGGQGQYGRVIGYIEPLPAGSGTKFEFDNMLVGQAIPSNFF 586
N+RE++++ A+ Y HKKQSGGQGQ+ + EPL AGSG +EF + + G A+P +
Sbjct: 568 NYRESISKIAEVKYTHKKQSGGQGQFADITVRFEPLEAGSG--YEFKSEIKGGAVPREYI 625
Query: 587 PAIEKGFKEAANSGALIGHPVQNLRVVLTDGAAHDVDSSELAFKLASIYAFRECYTASRP 646
P + KG +E ++G L G PV ++R L DG+ HDVDSS LAF+LA+ AFRE + P
Sbjct: 626 PGVMKGLEECMSTGVLAGFPVVDVRACLVDGSYHDVDSSVLAFQLAARGAFREGMRKAGP 685
Query: 647 VILEPVM 653
+LEP+M
Sbjct: 686 RMLEPIM 692
>At5g39900 GTP-binding protein-like
Length = 661
Score = 91.7 bits (226), Expect = 1e-18
Identities = 64/189 (33%), Positives = 93/189 (48%), Gaps = 20/189 (10%)
Query: 22 GTFHIRHFSAGNVARATAATIDKDPWWKESMEKVRNIGISAHIDSGKTTLTEWILFYTGK 81
G + FS+ + + TID K EK+RN I AHID GK+TL + ++ TG
Sbjct: 36 GLYQAYGFSSDSRQSSKEPTIDLT---KFPSEKIRNFSIIAHIDHGKSTLADRLMELTGT 92
Query: 82 IHLMYEVRSKDGMGPKMDFKPLEIIMGITIKSAATYCNWK---------GSKITIIDTPG 132
I K G G L+ GIT+K+ ++ G + +IDTPG
Sbjct: 93 I--------KKGHGQPQYLDKLQRERGITVKAQTATMFYENKVEDQEASGYLLNLIDTPG 144
Query: 133 HVDFTIEVERALRVLDGAVLVLCSVGGVQCQSITVDRQMRRYQVPRIAFINKLDRPGADP 192
HVDF+ EV R+L GA+LV+ + GVQ Q++ + + INK+D+P ADP
Sbjct: 145 HVDFSYEVSRSLSACQGALLVVDAAQGVQAQTVANFYLAFEANLTIVPVINKIDQPTADP 204
Query: 193 WKVITQARS 201
+V Q +S
Sbjct: 205 ERVKAQLKS 213
Score = 42.4 bits (98), Expect = 8e-04
Identities = 53/236 (22%), Positives = 95/236 (39%), Gaps = 34/236 (14%)
Query: 305 VFMGSAVKNTGVQPLLDGVVSYLPCPIEVSNYALDQSKNEEKVQLTGSPDGPLVALAFKL 364
V + SA G++ +L V+ +P P +S + PL L F
Sbjct: 222 VLLVSAKTGLGLEHVLPAVIERIPPPPGIS-------------------ESPLRMLLFDS 262
Query: 365 EQTKF-GQLTYLRVYEGVIRKGDFIVNVSTGKKIKVPRLVQMHSNEMNDIEEAHAGQI-V 422
++ G + Y+ V +G++ KGD + ++G+ +V + MH E+ GQ+
Sbjct: 263 FFNEYKGVICYVSVVDGMLSKGDKVSFAASGQSYEVLDVGIMHP-ELTSTGMLLTGQVGY 321
Query: 423 AVFGVDCASSDTFTDGSVKYTMTSMNVP--EPVMSLAVQPVSKDSGGKFSK---ALNRFQ 477
V G+ D + T +P +PV + V G F A+ +
Sbjct: 322 IVTGMRTTKEARIGDTIYRTKTTVEPLPGFKPVRHMVFSGVYPADGSDFEALGHAMEKLT 381
Query: 478 REDPTFRVSLDPESGQTIISG-----MGELHLDIYVKRIKMEYGVDATVGKPRVNF 528
D + VS+ E+ + G +G LH+D++ +R++ EYG P V +
Sbjct: 382 CNDAS--VSVAKETSTALGMGFRCGFLGLLHMDVFHQRLEQEYGTQVISTIPTVPY 435
>At1g06220 elongation factor like protein
Length = 987
Score = 91.7 bits (226), Expect = 1e-18
Identities = 152/686 (22%), Positives = 258/686 (37%), Gaps = 140/686 (20%)
Query: 55 VRNIGISAHIDSGKTTLTEWILFYTGKI---------HLMYEVRSKDGMGPKMDFKPLEI 105
VRN+ + H+ GKT + ++ T + H+ Y D + K + +
Sbjct: 138 VRNVALVGHLQHGKTVFMDMLVEQTHHMSTFNAKNEKHMKYTDTRVDEQERNISIKAVPM 197
Query: 106 IMGITIKSAATY-CNWKGSKITIIDTPGHVDFTIEVERALRVLDGAVLVLCSVGGVQCQS 164
+ + + +Y CN I+DTPGHV+F+ E+ +LR+ DGAVL++ + GV +
Sbjct: 198 SLVLEDSRSKSYLCN-------IMDTPGHVNFSDEMTASLRLADGAVLIVDAAEGVMVNT 250
Query: 165 ITVDRQMRRYQVPRIAFINKLDRPGADPWKVITQARSKLRHHCAALQVPIGLESDFKGVV 224
R + +P + INK+DR + A KLRH + I S G +
Sbjct: 251 ERAIRHAIQDHLPIVVVINKVDRLITELKLPPRDAYYKLRHTIEVINNHISAASTTAGDL 310
Query: 225 DLVKLKA--YCF-DGQYGQNVVVGEVPADMEALVAEKRRELIETVSEV-DDVLAEAFLSD 280
L+ A CF G G + + A M A + ++ + S + DV + SD
Sbjct: 311 PLIDPAAGNVCFASGTAGWSFTLQSF-AKMYAKLHGVAMDVDKFASRLWGDVY---YHSD 366
Query: 281 DENISAADLEGAIRRATIARKFIPVF--------------------MGSAVKNT----GV 316
+ G RA + P++ +G + N+ V
Sbjct: 367 TRVFKRSPPVGGGERAFVQFILEPLYKIYSQVIGEHKKSVETTLAELGVTLSNSAYKLNV 426
Query: 317 QPLL---------------DGVVSYLPCPIEVSNYALDQSKNEEK------VQLTGSPDG 355
+PLL D +V ++P P E + +D S K + P G
Sbjct: 427 RPLLRLACSSVFGSASGFTDMLVKHIPSPREAAARKVDHSYTGTKDSPIYESMVECDPSG 486
Query: 356 PLVALAFKLEQTKFGQL--TYLRVYEGVIRKGDFIVNVSTGKK---------IKVPRLVQ 404
PL+ KL + + RVY G ++ G + + G +V +L
Sbjct: 487 PLMVNVTKLYPKSDTSVFDVFGRVYSGRLQTGQSVRVLGEGYSPEDEEDMTIKEVTKLWI 546
Query: 405 MHSNEMNDIEEAHAGQIVAVFGVDCASSDTFT------DGSVKYTMTSMNVPEPVMSLAV 458
+ + A G V + GVD + T T D V PV+ A
Sbjct: 547 YQARYRIPVSSAPPGSWVLIEGVDASIMKTATLCNASYDEDVYIFRALQFNTLPVVKTAT 606
Query: 459 QPVSKDSGGKFSKALNRFQREDPTFRVSLDPESGQTIISGMGELHLDIYVKRIKMEYG-V 517
+P++ K + L + + P ++ ESG+ I G GEL+LD +K ++ Y V
Sbjct: 607 EPLNPSELPKMVEGLRKISKSYP-LAITKVEESGEHTILGTGELYLDSIMKDLRELYSEV 665
Query: 518 DATVGKPRVNFRETVTQRADFDYLHKKQSGGQGQYGRVIGYIEPLPAGSG---------- 567
+ V P V+F ETV + + K + + ++ EPL G
Sbjct: 666 EVKVADPVVSFCETVVESSSM----KCFAETPNKKNKITMIAEPLDRGLAEDIENGVVSI 721
Query: 568 ------------TKFEFD-----------------NMLVGQAIPS----NFFPAIE---- 590
TK+++D N+L+ +P+ N A++
Sbjct: 722 DWNRKQLGDFFRTKYDWDLLAARSIWAFGPDKQGPNILLDDTLPTEVDRNLMMAVKDSIV 781
Query: 591 KGFKEAANSGALIGHPVQNLRVVLTD 616
+GF+ A G L P++N++ + D
Sbjct: 782 QGFQWGAREGPLCDEPIRNVKFKIVD 807
>At5g13650 GTP-binding protein typA (tyrosine phosphorylated protein
A)
Length = 609
Score = 91.3 bits (225), Expect = 2e-18
Identities = 59/144 (40%), Positives = 81/144 (55%), Gaps = 5/144 (3%)
Query: 53 EKVRNIGISAHIDSGKTTLTEWILFYTGKIHLMYEVRSKDGMGPKMDFKPLEIIMGITIK 112
+ VRNI I AH+D GKTTL + +L K+ +V + MD LE GITI
Sbjct: 14 DNVRNIAIVAHVDHGKTTLVDSML-RQAKVFRDNQVMQER----IMDSNDLERERGITIL 68
Query: 113 SAATYCNWKGSKITIIDTPGHVDFTIEVERALRVLDGAVLVLCSVGGVQCQSITVDRQMR 172
S T +K +K+ IIDTPGH DF EVER L ++DG +LV+ SV G Q+ V ++
Sbjct: 69 SKNTSITYKNTKVNIIDTPGHSDFGGEVERVLNMVDGVLLVVDSVEGPMPQTRFVLKKAL 128
Query: 173 RYQVPRIAFINKLDRPGADPWKVI 196
+ + +NK+DRP A P V+
Sbjct: 129 EFGHAVVVVVNKIDRPSARPEFVV 152
Score = 50.4 bits (119), Expect = 3e-06
Identities = 47/187 (25%), Positives = 85/187 (45%), Gaps = 15/187 (8%)
Query: 354 DGPLVALAFKLEQTKF-GQLTYLRVYEGVIRKGDFIVNVSTGKKIKVPRLVQMHSNEMN- 411
DG L LA +E + G++ R++ GV+RKG + ++ + R+ ++ E
Sbjct: 213 DGALQMLATNIEYDEHKGRIAIGRLHAGVLRKGMDVRVCTSEDSCRFARVSELFVYEKFY 272
Query: 412 --DIEEAHAGQIVAVFGVDCAS-SDTFTDGSVKYTMTSMNVPEPVMSLAVQPVSKDSGGK 468
+ AG I AV G+D +T D + ++ V EP + ++ + G+
Sbjct: 273 RVPTDSVEAGDICAVCGIDNIQIGETIADKVHGKPLPTIKVEEPTVKMSFSVNTSPFSGR 332
Query: 469 FSKALNRFQREDPTFR-----VSLDPESGQT----IISGMGELHLDIYVKRIKMEYGVDA 519
K + D R +++ E G+T I+SG G LH+ I ++ ++ E G +
Sbjct: 333 EGKYVTSRNLRDRLNRELERNLAMKVEDGETADTFIVSGRGTLHITILIENMRRE-GYEF 391
Query: 520 TVGKPRV 526
VG P+V
Sbjct: 392 MVGPPKV 398
>At5g08650 GTP-binding protein LepA homolog
Length = 675
Score = 90.5 bits (223), Expect = 3e-18
Identities = 68/218 (31%), Positives = 104/218 (47%), Gaps = 24/218 (11%)
Query: 39 AATIDKDPWWKESMEKVRNIGISAHIDSGKTTLTEWILFYTGKIHLMYEVRSKDGMGPKM 98
AA +D K + +RN I AHID GK+TL + +L TG V+++D +
Sbjct: 64 AARSGQDRLLKVPISNIRNFSIIAHIDHGKSTLADKLLQVTGT------VQNRDMKEQFL 117
Query: 99 DFKPLEIIMGITIKSAATYCNWKGSK----ITIIDTPGHVDFTIEVERALRVLDGAVLVL 154
D LE GITIK A + + +IDTPGHVDF+ EV R+L +GA+LV+
Sbjct: 118 DNMDLERERGITIKLQAARMRYVYEDTPFCLNLIDTPGHVDFSYEVSRSLAACEGALLVV 177
Query: 155 CSVGGVQCQSITVDRQMRRYQVPRIAFINKLDRPGADPWKVITQARSKLRHHCA-----A 209
+ GV+ Q++ + I +NK+D PGA+P KV+ + + C+ +
Sbjct: 178 DASQGVEAQTLANVYLALENNLEIIPVLNKIDLPGAEPEKVLREIEEVIGLDCSKAIFCS 237
Query: 210 LQVPIGLESDFKGVVDLV---------KLKAYCFDGQY 238
+ IG+ +V + L+A FD Y
Sbjct: 238 AKEGIGITEILDAIVQRIPAPLDTAGKPLRALIFDSYY 275
Score = 65.1 bits (157), Expect = 1e-10
Identities = 67/291 (23%), Positives = 118/291 (40%), Gaps = 50/291 (17%)
Query: 254 LVAEKRRELIETVSEVD--DVLAEAFLSDDENISAADLEGAIRRATIARKFIPVFMGSAV 311
L E E+I ++++D E L + E + D AI SA
Sbjct: 193 LALENNLEIIPVLNKIDLPGAEPEKVLREIEEVIGLDCSKAI-------------FCSAK 239
Query: 312 KNTGVQPLLDGVVSYLPCPIEVSNYALDQSKNEEKVQLTGSPDGPLVALAFKLEQTKF-G 370
+ G+ +LD +V +P P++ + PL AL F + G
Sbjct: 240 EGIGITEILDAIVQRIPAPLDTAGK-------------------PLRALIFDSYYDPYRG 280
Query: 371 QLTYLRVYEGVIRKGDFIVNVSTGKKIKVPRLVQMHSNEMNDIEEAHAGQIVAVFG---- 426
+ Y RV +G ++KGD I +++GK + + N++ ++E +AG++ +
Sbjct: 281 VIVYFRVIDGKVKKGDRIFFMASGKDYFADEVGVLSPNQI-QVDELYAGEVGYIAASVRS 339
Query: 427 -VDCASSDTFTDGSVKYTMTSMNVPE--PVMSLAVQPVSKDSGGKFSKALNRFQREDPTF 483
D DT T S K + E P++ + PV D AL + Q D
Sbjct: 340 VADARVGDTITHYSRKAESSLPGYEEATPMVFCGLFPVDADQFPDLRDALEKLQLNDAAL 399
Query: 484 RVSLDPESGQTIISG-----MGELHLDIYVKRIKMEYGVDATVGKPRVNFR 529
+ +PE+ + G +G LH++I +R++ EY ++ P V +R
Sbjct: 400 K--FEPETSSAMGFGFRCGFLGLLHMEIVQERLEREYNLNLITTAPSVVYR 448
>At2g31060 putative GTP-binding protein
Length = 664
Score = 87.4 bits (215), Expect = 2e-17
Identities = 63/188 (33%), Positives = 95/188 (50%), Gaps = 32/188 (17%)
Query: 3 RFGYALCSSTLTTEGSLIAGTFHIRHFSAGNVARATAATIDK--DPWWKESMEKVRNIGI 60
+FGY+L S +R FSA + A A + DP ++RN+ +
Sbjct: 26 KFGYSLSS---------------LRSFSAATASTAAAGAPNSSLDP------NRLRNVAV 64
Query: 61 SAHIDSGKTTLTEWILFYTGKIHLMYEVRSKDGMGPKMDFKPLEIIMGITIKSAATYCNW 120
AH+D GKTTL + +L G + +E MD LE GITI S T W
Sbjct: 65 IAHVDHGKTTLMDRLLRQCGA-DIPHE--------RAMDSINLERERGITISSKVTSIFW 115
Query: 121 KGSKITIIDTPGHVDFTIEVERALRVLDGAVLVLCSVGGVQCQSITVDRQMRRYQVPRIA 180
K +++ ++DTPGH DF EVER + +++GA+LV+ + G Q+ V + +Y + I
Sbjct: 116 KDNELNMVDTPGHADFGGEVERVVGMVEGAILVVDAGEGPLAQTKFVLAKALKYGLRPIL 175
Query: 181 FINKLDRP 188
+NK+DRP
Sbjct: 176 LLNKVDRP 183
>At3g22980 eukaryotic translation elongation factor 2, putative
Length = 1015
Score = 79.0 bits (193), Expect = 8e-15
Identities = 50/134 (37%), Positives = 76/134 (56%), Gaps = 3/134 (2%)
Query: 54 KVRNIGISAHIDSGKTTLTEWILFYTGKIHLMYEVRSKDGMGPKMDFKPLEIIMGITIKS 113
KVRNI I AH+D GKTTL + ++ +G L + K MD+ E IT+KS
Sbjct: 8 KVRNICILAHVDHGKTTLADHLIASSGGGVLHPRLAGKLRF---MDYLDEEQRRAITMKS 64
Query: 114 AATYCNWKGSKITIIDTPGHVDFTIEVERALRVLDGAVLVLCSVGGVQCQSITVDRQMRR 173
++ +K + +ID+PGH+DF EV A R+ DGA++++ +V GV Q+ V RQ
Sbjct: 65 SSISLKYKDYSLNLIDSPGHMDFCSEVSTAARLSDGALVLVDAVEGVHIQTHAVLRQAWI 124
Query: 174 YQVPRIAFINKLDR 187
++ +NK+DR
Sbjct: 125 EKLTPCLVLNKIDR 138
Score = 57.4 bits (137), Expect = 2e-08
Identities = 45/178 (25%), Positives = 83/178 (46%), Gaps = 18/178 (10%)
Query: 372 LTYLRVYEGVIRKGD--FIVNV--------STGKKIKVPRLVQMH---SNEMNDIEEAHA 418
L + R++ GV+R G F++ S+ K I+ L ++ + + E A
Sbjct: 441 LAFARIFSGVLRAGQRVFVITALYDPLKGESSHKYIQEAELHSLYLMMGQGLTPVTEVKA 500
Query: 419 GQIVAVFGVDCASSDTFTDGSVK--YTMTSMNVP-EPVMSLAVQPVSKDSGGKFSKALNR 475
G +VA+ G+ S + T S + + + SM P + +A++P K L
Sbjct: 501 GNVVAIRGLGPYISKSATLSSTRNCWPLASMEFQVSPTLRVAIEPSDPADMSALMKGLRL 560
Query: 476 FQREDPTFRVSLDPESGQTIISGMGELHLDIYVKRIKMEYG-VDATVGKPRVNFRETV 532
R DP +++ G+ +++ GE+HL+ VK +K + V+ V P V++RET+
Sbjct: 561 LNRADPFVEITVSAR-GEHVLAAAGEVHLERCVKDLKERFAKVNLEVSPPLVSYRETI 617
>At1g56070 elongation factor like protein
Length = 843
Score = 74.3 bits (181), Expect = 2e-13
Identities = 51/149 (34%), Positives = 80/149 (53%), Gaps = 21/149 (14%)
Query: 55 VRNIGISAHIDSGKTTLTEWILFYTGKIHLMYEVRSKDGMGPKMDFKPLEIIMGITIKSA 114
+RN+ + AH+D GK+TLT+ ++ G I + G D + E GITIKS
Sbjct: 19 IRNMSVIAHVDHGKSTLTDSLVAAAGII-----AQEVAGDVRMTDTRADEAERGITIKST 73
Query: 115 A----------TYCNWKGSK------ITIIDTPGHVDFTIEVERALRVLDGAVLVLCSVG 158
+ ++ G++ I +ID+PGHVDF+ EV ALR+ DGA++V+ +
Sbjct: 74 GISLYYEMTDESLKSFTGARDGNEYLINLIDSPGHVDFSSEVTAALRITDGALVVVDCIE 133
Query: 159 GVQCQSITVDRQMRRYQVPRIAFINKLDR 187
GV Q+ TV RQ ++ + +NK+DR
Sbjct: 134 GVCVQTETVLRQALGERIRPVLTVNKMDR 162
Score = 62.8 bits (151), Expect = 6e-10
Identities = 73/287 (25%), Positives = 123/287 (42%), Gaps = 37/287 (12%)
Query: 319 LLDGVVSYLPCPIEVSNYAL---------DQSKNEEKVQLTGSPDGPLVALAFKL--EQT 367
LL+ ++ +LP P Y + DQ N + P+GPL+ K+
Sbjct: 332 LLEMMIFHLPSPHTAQRYRVENLYEGPLDDQYANAIR---NCDPNGPLMLYVSKMIPASD 388
Query: 368 KFGQLTYLRVYEGVIRKGDFI----VNVSTGKKI-----KVPRLVQMHSNEMNDIEEAHA 418
K + RV+ G + G + N G+K V R V +E+
Sbjct: 389 KGRFFAFGRVFAGKVSTGMKVRIMGPNYIPGEKKDLYTKSVQRTVIWMGKRQETVEDVPC 448
Query: 419 GQIVAVFGVDCASSDTFTDGSVK----YTMTSMNVP-EPVMSLAVQPVSKDSGGKFSKAL 473
G VA+ G+D + T + K + + +M PV+ +AVQ K + L
Sbjct: 449 GNTVAMVGLDQFITKNATLTNEKEVDAHPIRAMKFSVSPVVRVAVQCKVASDLPKLVEGL 508
Query: 474 NRFQREDPTFRVSLDPESGQTIISGMGELHLDIYVKRIKMEY--GVDATVGKPRVNFRET 531
R + DP +++ ESG+ I++G GELHL+I +K ++ ++ G + P V+FRET
Sbjct: 509 KRLAKSDPMVVCTME-ESGEHIVAGAGELHLEICLKDLQDDFMGGAEIIKSDPVVSFRET 567
Query: 532 VTQRADFDYLHKKQSGGQGQYGRVIGYIEPLPAGSGTKFEFDNMLVG 578
V R+ + K + ++ R+ Y+E P G D+ +G
Sbjct: 568 VCDRSTRTVMSKSPN----KHNRL--YMEARPMEEGLAEAIDDGRIG 608
>At4g11160 translation initiation factor IF-2 like protein
Length = 743
Score = 58.5 bits (140), Expect = 1e-08
Identities = 33/81 (40%), Positives = 44/81 (53%)
Query: 122 GSKITIIDTPGHVDFTIEVERALRVLDGAVLVLCSVGGVQCQSITVDRQMRRYQVPRIAF 181
G+ IT +DTPGH F+ R V D VLV+ + GV Q++ R VP +
Sbjct: 267 GTSITFLDTPGHAAFSEMRARGAAVTDIVVLVVAADDGVMPQTLEAIAHARSANVPVVVA 326
Query: 182 INKLDRPGADPWKVITQARSK 202
INK D+PGA+P KV Q S+
Sbjct: 327 INKCDKPGANPEKVKYQLTSE 347
>At5g25230 translation Elongation Factor 2 - like protein
Length = 973
Score = 57.8 bits (138), Expect = 2e-08
Identities = 38/143 (26%), Positives = 69/143 (47%), Gaps = 17/143 (11%)
Query: 55 VRNIGISAHIDSGKTTLTEWILFYTGKI---------HLMYEVRSKDGMGPKMDFKPLEI 105
VRN+ + H+ GKT + ++ T ++ H+ Y D + K + +
Sbjct: 124 VRNVALVGHLQHGKTVFMDMLVEQTHRMSTFNAENDKHMRYTDTRVDEQERNISIKAVPM 183
Query: 106 IMGITIKSAATY-CNWKGSKITIIDTPGHVDFTIEVERALRVLDGAVLVLCSVGGVQCQS 164
+ + + +Y CN I+DTPG+V+F+ E+ +LR+ DGAV ++ + GV +
Sbjct: 184 SLVLEDSRSKSYLCN-------IMDTPGNVNFSDEMTASLRLADGAVFIVDAAQGVMVNT 236
Query: 165 ITVDRQMRRYQVPRIAFINKLDR 187
R + +P + INK+DR
Sbjct: 237 ERAIRHAIQDHLPIVVVINKVDR 259
Score = 52.0 bits (123), Expect = 1e-06
Identities = 79/367 (21%), Positives = 134/367 (35%), Gaps = 76/367 (20%)
Query: 321 DGVVSYLPCPIEVSNYALDQSKNEEK------VQLTGSPDGPLVALAFKLEQTKFGQL-- 372
D +V ++P P E + +D S K + P GPL+ KL +
Sbjct: 432 DMLVKHIPSPREAAARKVDHSYTGTKDSPIYESMVECDPSGPLMVNVTKLYPKSDTSVFD 491
Query: 373 TYLRVYEGVIRKGDFIVNVSTGKKIK---------VPRLVQMHSNEMNDIEEAHAGQIVA 423
+ RVY G ++ G + + G + V +L + + A G V
Sbjct: 492 VFGRVYSGRLQTGQSVRVLGEGYSPEDEEDMTIKEVTKLWIYQARYRIPVSSAPPGSWVL 551
Query: 424 VFGVDCASSDTFT------DGSVKYTMTSMNVPEPVMSLAVQPVSKDSGGKFSKALNRFQ 477
+ GVD + T T D V PV+ A +P++ K + L +
Sbjct: 552 IEGVDASIMKTATLCNASYDEDVYIFRALKFNTLPVVKTATEPLNPSELPKMVEGLRKIS 611
Query: 478 REDPTFRVSLDPESGQTIISGMGELHLDIYVKRIKMEYG-VDATVGKPRVNFRETVTQRA 536
+ P ++ ESG+ I G GEL+LD +K ++ Y V V P V+F ETV + +
Sbjct: 612 KSYPLAITKVE-ESGEHTILGTGELYLDSIIKDLRELYSEVQVKVADPVVSFCETVVESS 670
Query: 537 DFDYLHKKQSGGQGQYGRVIGYIEPLPAGSG----------------------TKFEFD- 573
K + + ++ EPL G TK+++D
Sbjct: 671 SM----KCFAETPNKKNKLTMIAEPLDRGLAEDIENGVVSIDWNRVQLGDFFRTKYDWDL 726
Query: 574 ----------------NMLVGQAIPS--------NFFPAIEKGFKEAANSGALIGHPVQN 609
N+L+ +P+ +I +GF+ A G L P++N
Sbjct: 727 LAARSIWAFGPDKQGTNILLDDTLPTEVDRNLMMGVKDSIVQGFQWGAREGPLCDEPIRN 786
Query: 610 LRVVLTD 616
++ + D
Sbjct: 787 VKFKIVD 793
>At1g17220 putative translation initiation factor IF2
Length = 1026
Score = 57.0 bits (136), Expect = 3e-08
Identities = 51/188 (27%), Positives = 83/188 (44%), Gaps = 34/188 (18%)
Query: 49 KESMEKVRN----IGISAHIDSGKTTLTEWILFYTGKIHLMYEVRSKDGMGPKMDFKPLE 104
+E ++K+ + I I H+D GKTTL ++I +V + + G
Sbjct: 491 EEDLDKLEDRPPVITIMGHVDHGKTTLLDYIR--------KSKVAASEAGG--------- 533
Query: 105 IIMGITIKSAATYCNWKGSKITIIDTPGHVDFTIEVERALRVLDGAVLVLCSVGGVQCQS 164
I GI + + K +DTPGH F R RV D A++V+ + G++ Q+
Sbjct: 534 ITQGIGAYKVSVPVDGKLQSCVFLDTPGHEAFGAMRARGARVTDIAIIVVAADDGIRPQT 593
Query: 165 ITVDRQMRRYQVPRIAFINKLDRPGADPWKVITQARSKLRHHCAALQVPIGL-ESDFKGV 223
+ VP + INK+D+ GA P +V+ + S IGL D+ G
Sbjct: 594 NEAIAHAKAAAVPIVIAINKIDKEGASPDRVMQELSS------------IGLMPEDWGGD 641
Query: 224 VDLVKLKA 231
V +V++ A
Sbjct: 642 VPMVQISA 649
>At4g02930 mitochondrial elongation factor Tu
Length = 454
Score = 55.8 bits (133), Expect = 7e-08
Identities = 48/176 (27%), Positives = 77/176 (43%), Gaps = 13/176 (7%)
Query: 17 GSLIAGTFHIRHFSAGNVARATAATIDKDPWWKESMEKVRN-----IGISAHIDSGKTTL 71
G+ + ++ I H G+ +++T +W+ RN +G H+D GKTTL
Sbjct: 26 GASVTSSYSISHSIGGD--DLSSSTFGTSSFWRSMATFTRNKPHVNVGTIGHVDHGKTTL 83
Query: 72 TEWILFYTGKIHLMYEVRSKDGMGPKMDFKPLEIIMGITIKSAATYCNWKGSKITIIDTP 131
T I K+ L E ++K ++D P E GITI +A +D P
Sbjct: 84 TAAIT----KV-LAEEGKAKAIAFDEIDKAPEEKKRGITIATAHVEYETAKRHYAHVDCP 138
Query: 132 GHVDFTIEVERALRVLDGAVLVLCSVGGVQCQSITVDRQMRRYQVPR-IAFINKLD 186
GH D+ + +DG +LV+ G Q+ R+ VP + F+NK+D
Sbjct: 139 GHADYVKNMITGAAQMDGGILVVSGPDGPMPQTKEHILLARQVGVPSLVCFLNKVD 194
>At5g10630 putative protein
Length = 804
Score = 55.5 bits (132), Expect = 9e-08
Identities = 49/152 (32%), Positives = 71/152 (46%), Gaps = 19/152 (12%)
Query: 57 NIGISAHIDSGKTTLTEWILFYTGKIHLM----YEVRSK-DGMGP-----KMDFKPLEII 106
N+ I H+DSGK+TL+ +L G+I YE +K G G +D E
Sbjct: 378 NLAIVGHVDSGKSTLSGRLLHLLGRISQKQMHKYEKEAKLQGKGSFAYAWALDESAEERE 437
Query: 107 MGITIKSAATYCNWKGSKITIIDTPGHVDFTIEVERALRVLDGAVLVL-CSVGGVQC--- 162
GIT+ A Y N K + ++D+PGH DF + D A+LV+ SVG +
Sbjct: 438 RGITMTVAVAYFNSKRHHVVLLDSPGHKDFVPNMIAGATQADAAILVIDASVGAFEAGFD 497
Query: 163 ----QSITVDRQMRRYQVPR-IAFINKLDRPG 189
Q+ R +R + V + I INK+D G
Sbjct: 498 NLKGQTREHARVLRGFGVEQVIVAINKMDIVG 529
>At4g20360 translation elongation factor EF-Tu precursor,
chloroplast
Length = 476
Score = 53.1 bits (126), Expect = 5e-07
Identities = 38/132 (28%), Positives = 57/132 (42%), Gaps = 6/132 (4%)
Query: 57 NIGISAHIDSGKTTLTEWILFYTGKIHLMYEVRSKDGMGPKMDFKPLEIIMGITIKSAAT 116
NIG H+D GKTTLT + I + + +D P E GITI +A
Sbjct: 81 NIGTIGHVDHGKTTLTAALTMALASIGSSVAKKYDE-----IDAAPEERARGITINTATV 135
Query: 117 YCNWKGSKITIIDTPGHVDFTIEVERALRVLDGAVLVLCSVGGVQCQSITVDRQMRRYQV 176
+ +D PGH D+ + +DGA+LV+ G Q+ ++ V
Sbjct: 136 EYETENRHYAHVDCPGHADYVKNMITGAAQMDGAILVVSGADGPMPQTKEHILLAKQVGV 195
Query: 177 P-RIAFINKLDR 187
P + F+NK D+
Sbjct: 196 PDMVVFLNKEDQ 207
>At1g21160 transcription factor, putative
Length = 1088
Score = 48.1 bits (113), Expect = 2e-05
Identities = 46/141 (32%), Positives = 64/141 (44%), Gaps = 7/141 (4%)
Query: 49 KESMEKVRN--IGISAHIDSGKTTLTEWILFYTGKIHLMYEVRSKDGMGPKMDFKPLEII 106
KE E +R+ I H+DSGKT L + I + + + G F P E I
Sbjct: 485 KEVEENLRSPICCIMGHVDSGKTKLLDCIRGTNVQEGEAGGITQQIGA----TFFPAENI 540
Query: 107 MGITIKSAATYCNWKGSKITIIDTPGHVDFTIEVERALRVLDGAVLVLCSVGGVQCQSIT 166
T K K I +IDTPGH FT R + D A+LV+ + G++ Q+I
Sbjct: 541 RERT-KELQANAKLKVPGILVIDTPGHESFTNLRSRGSNLCDLAILVVDIMRGLEPQTIE 599
Query: 167 VDRQMRRYQVPRIAFINKLDR 187
+RR V I +NK+DR
Sbjct: 600 SLNLLRRRNVKFIIALNKVDR 620
>At1g18070 unknown protein
Length = 532
Score = 45.8 bits (107), Expect = 7e-05
Identities = 34/135 (25%), Positives = 62/135 (45%), Gaps = 10/135 (7%)
Query: 35 ARATAATIDKDPWWKESMEKVRNIGISAHIDSGKTTLTEWILFYTGKIH----LMYEVRS 90
A+ AA + + + + ++ N+ H+D+GK+T+ ILF +G++ YE +
Sbjct: 81 AQEKAAKEEAEDVAEANKKRHLNVVFIGHVDAGKSTIGGQILFLSGQVDDRQIQKYEKEA 140
Query: 91 KDG------MGPKMDFKPLEIIMGITIKSAATYCNWKGSKITIIDTPGHVDFTIEVERAL 144
KD M MD E + G T++ + + ++ TI+D PGH + +
Sbjct: 141 KDKSRESWYMAYIMDTNEEERLKGKTVEVGRAHFETESTRFTILDAPGHKSYVPNMISGA 200
Query: 145 RVLDGAVLVLCSVGG 159
D VLV+ + G
Sbjct: 201 SQADIGVLVISARKG 215
>At5g60390 translation elongation factor eEF-1 alpha chain (gene A4)
Length = 449
Score = 43.1 bits (100), Expect = 5e-04
Identities = 37/128 (28%), Positives = 54/128 (41%), Gaps = 10/128 (7%)
Query: 57 NIGISAHIDSGKTTLTEWILFYTGKI--HLMYEVRSKDGMGPKMDFKPLEII-------- 106
NI + H+DSGK+T T +++ G I ++ + K FK ++
Sbjct: 9 NIVVIGHVDSGKSTTTGHLIYKLGGIDKRVIERFEKEAAEMNKRSFKYAWVLDKLKAERE 68
Query: 107 MGITIKSAATYCNWKGSKITIIDTPGHVDFTIEVERALRVLDGAVLVLCSVGGVQCQSIT 166
GITI A T+ID PGH DF + D AVL++ S G I+
Sbjct: 69 RGITIDIALWKFETTKYYCTVIDAPGHRDFIKNMITGTSQADCAVLIIDSTTGGFEAGIS 128
Query: 167 VDRQMRRY 174
D Q R +
Sbjct: 129 KDGQTREH 136
>At1g07940 elongation factor 1-alpha
Length = 449
Score = 43.1 bits (100), Expect = 5e-04
Identities = 37/128 (28%), Positives = 54/128 (41%), Gaps = 10/128 (7%)
Query: 57 NIGISAHIDSGKTTLTEWILFYTGKI--HLMYEVRSKDGMGPKMDFKPLEII-------- 106
NI + H+DSGK+T T +++ G I ++ + K FK ++
Sbjct: 9 NIVVIGHVDSGKSTTTGHLIYKLGGIDKRVIERFEKEAAEMNKRSFKYAWVLDKLKAERE 68
Query: 107 MGITIKSAATYCNWKGSKITIIDTPGHVDFTIEVERALRVLDGAVLVLCSVGGVQCQSIT 166
GITI A T+ID PGH DF + D AVL++ S G I+
Sbjct: 69 RGITIDIALWKFETTKYYCTVIDAPGHRDFIKNMITGTSQADCAVLIIDSTTGGFEAGIS 128
Query: 167 VDRQMRRY 174
D Q R +
Sbjct: 129 KDGQTREH 136
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.136 0.396
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,122,961
Number of Sequences: 26719
Number of extensions: 612907
Number of successful extensions: 1517
Number of sequences better than 10.0: 31
Number of HSP's better than 10.0 without gapping: 26
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 1439
Number of HSP's gapped (non-prelim): 46
length of query: 654
length of database: 11,318,596
effective HSP length: 106
effective length of query: 548
effective length of database: 8,486,382
effective search space: 4650537336
effective search space used: 4650537336
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 63 (28.9 bits)
Medicago: description of AC146553.2


