

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146553.13 + phase: 0
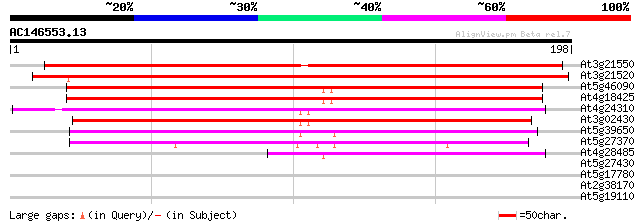
(198 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g21550 unknown protein 244 2e-65
At3g21520 unknown protein 194 3e-50
At5g46090 putative protein 162 1e-40
At4g18425 unknown protein 156 6e-39
At4g24310 unknown protein 151 2e-37
At3g02430 hypothetical protein 147 4e-36
At5g39650 unknown protein 125 1e-29
At5g27370 putative protein 116 9e-27
At4g28485 putative protein 82 2e-16
At5g27430 signal peptidase 28 3.2
At5g17780 putative protein 28 4.1
At2g38170 Ca2+ antiporter like protein 28 4.1
At5g19110 dermal glycoprotein - like 27 9.2
>At3g21550 unknown protein
Length = 184
Score = 244 bits (623), Expect = 2e-65
Identities = 115/183 (62%), Positives = 142/183 (76%), Gaps = 2/183 (1%)
Query: 13 NKGASTMTGKTFSGVGNLVKLLPTGTVFLFQYLSPVVTNNGHCSTINKYLSGILLVICGF 72
+K + +T+SGVG+L+KLLPTGTVFLFQ+L+PV+TNNGHC INKYL+G+L+VIC F
Sbjct: 2 SKTFKAIRDRTYSGVGDLIKLLPTGTVFLFQFLNPVLTNNGHCLLINKYLTGVLIVICAF 61
Query: 73 NCAFTSFTDSYTGSDGQRHYGIVTMNGLWPSPGSDSVDLSAYKLRFGDFVHAFLSVIVFA 132
+C FT FTDSY DG HYG+ T+ GLWP S SVDLS+ +LR GDFVHAF S+IVF+
Sbjct: 62 SCCFTCFTDSYRTRDGYVHYGVATVKGLWPD--SSSVDLSSKRLRVGDFVHAFFSLIVFS 119
Query: 133 VLGLLDTNVVHCFYPKFESSEKILMQVLPPVIGVVSGAVFMIFPSYRHGIGYPTSSDTND 192
V+ LLD N V+CFYP F S+ KI + VLPPVIGV+SGAVF +FPS RHGIG P+ +D
Sbjct: 120 VISLLDANTVNCFYPGFGSAGKIFLMVLPPVIGVISGAVFTVFPSRRHGIGNPSDHSEDD 179
Query: 193 TSE 195
SE
Sbjct: 180 ASE 182
>At3g21520 unknown protein
Length = 207
Score = 194 bits (492), Expect = 3e-50
Identities = 93/191 (48%), Positives = 125/191 (64%), Gaps = 2/191 (1%)
Query: 9 KTSTNKGASTM--TGKTFSGVGNLVKLLPTGTVFLFQYLSPVVTNNGHCSTINKYLSGIL 66
KT++ + M T K+ +G+ +L+KLLPTGT+F++ L+PV+TN+G CST NK +S IL
Sbjct: 10 KTNSPASSENMANTNKSLTGLESLIKLLPTGTLFIYLLLNPVLTNDGECSTGNKVMSSIL 69
Query: 67 LVICGFNCAFTSFTDSYTGSDGQRHYGIVTMNGLWPSPGSDSVDLSAYKLRFGDFVHAFL 126
+ +C F+C F+ FTDS+ G DG R +GIVT GLW SVDLS YKLR DFVHA
Sbjct: 70 VALCSFSCVFSCFTDSFKGVDGSRKFGIVTKKGLWTYAEPGSVDLSKYKLRIADFVHAGF 129
Query: 127 SVIVFAVLGLLDTNVVHCFYPKFESSEKILMQVLPPVIGVVSGAVFMIFPSYRHGIGYPT 186
+ VF L LLD N CFYP+F ++K L+ LPP +GV S +F +FPS R GIGY
Sbjct: 130 VLAVFGTLVLLDANTASCFYPRFRETQKTLVMALPPAVGVASATIFALFPSKRSGIGYAP 189
Query: 187 SSDTNDTSEKT 197
++ E+T
Sbjct: 190 IAEEVGAEEET 200
>At5g46090 putative protein
Length = 214
Score = 162 bits (409), Expect = 1e-40
Identities = 82/171 (47%), Positives = 109/171 (62%), Gaps = 3/171 (1%)
Query: 21 GKTFSGVGNLVKLLPTGTVFLFQYLSPVVTNNGHCSTINKYLSGILLVICGFNCAFTSFT 80
GKTF NL LLPTGTV FQ LSP+ TN G C +++++ +L+ ICGF+C SFT
Sbjct: 43 GKTFQTTANLANLLPTGTVLAFQILSPICTNVGRCDLTSRFMTALLVSICGFSCFILSFT 102
Query: 81 DSYTGSDGQRHYGIVTMNGLWPSPGSDSV--DLS-AYKLRFGDFVHAFLSVIVFAVLGLL 137
DSY +G YG T++G W GS ++ +LS +YKLRF DFVHA +S +VF + L
Sbjct: 103 DSYKDLNGSVCYGFATIHGFWIIDGSATLPQELSKSYKLRFIDFVHAIMSFLVFGAVVLF 162
Query: 138 DTNVVHCFYPKFESSEKILMQVLPPVIGVVSGAVFMIFPSYRHGIGYPTSS 188
D NVV+CFYP+ + L+ LP +GV VF FP+ RHGIG+P S+
Sbjct: 163 DQNVVNCFYPEPSAEVVELLTTLPVAVGVFCSMVFAKFPTTRHGIGFPLSA 213
>At4g18425 unknown protein
Length = 213
Score = 156 bits (395), Expect = 6e-39
Identities = 78/171 (45%), Positives = 108/171 (62%), Gaps = 3/171 (1%)
Query: 21 GKTFSGVGNLVKLLPTGTVFLFQYLSPVVTNNGHCSTINKYLSGILLVICGFNCAFTSFT 80
G+TF +L LLPTGTV FQ LSP+ +N G C ++K ++ L+ ICGF+C SFT
Sbjct: 42 GQTFQTTAHLANLLPTGTVLAFQLLSPIFSNGGQCDLVSKIMTSTLVAICGFSCFILSFT 101
Query: 81 DSYTGSDGQRHYGIVTMNGLWPSPGSDSV--DLS-AYKLRFGDFVHAFLSVIVFAVLGLL 137
DSY +G YG+ T++G W GS ++ +LS YKLRF DFVHAF+S+ VF + L
Sbjct: 102 DSYKDKNGTICYGLATIHGFWIIDGSTTLPQELSKRYKLRFIDFVHAFMSLFVFGAVVLF 161
Query: 138 DTNVVHCFYPKFESSEKILMQVLPPVIGVVSGAVFMIFPSYRHGIGYPTSS 188
D N V+CF+P + ++ LP +GV S +F FP+ R+GIG+P SS
Sbjct: 162 DRNAVNCFFPSPSAEALEVLTALPVGVGVFSSMLFATFPTTRNGIGFPLSS 212
>At4g24310 unknown protein
Length = 213
Score = 151 bits (381), Expect = 2e-37
Identities = 77/193 (39%), Positives = 111/193 (56%), Gaps = 7/193 (3%)
Query: 2 SEKASSSKTSTNKGASTMTGKTFSGVGNLVKLLPTGTVFLFQYLSPVVTNNGHCSTINKY 61
SE + T++ A + +T + NL LLPTGT+ F L PV T+NG C +
Sbjct: 19 SESVPQLRRQTSQHA--VMSQTLTSAANLANLLPTGTLLAFTLLIPVFTSNGSCDYPTQV 76
Query: 62 LSGILLVICGFNCAFTSFTDSYTGSDGQRHYGIVTMNGLW----PSP-GSDSVDLSAYKL 116
L+ +LL + +C +SFTDS DG +YG T G+W P P G +LS Y++
Sbjct: 77 LTIVLLTLLSISCFLSSFTDSVKAEDGNVYYGFATRKGMWVFDYPDPDGLGLPNLSKYRI 136
Query: 117 RFGDFVHAFLSVIVFAVLGLLDTNVVHCFYPKFESSEKILMQVLPPVIGVVSGAVFMIFP 176
R D++HA LSV+VF + L D N V CFYP E K ++ ++P +GV+ G +F++FP
Sbjct: 137 RIIDWIHAVLSVLVFGAVALRDKNAVSCFYPAPEQETKKVLDIVPMGVGVICGMLFLVFP 196
Query: 177 SYRHGIGYPTSSD 189
+ RHGIGYP + D
Sbjct: 197 ARRHGIGYPVTGD 209
>At3g02430 hypothetical protein
Length = 219
Score = 147 bits (371), Expect = 4e-36
Identities = 71/167 (42%), Positives = 103/167 (61%), Gaps = 5/167 (2%)
Query: 23 TFSGVGNLVKLLPTGTVFLFQYLSPVVTNNGHCSTINKYLSGILLVICGFNCAFTSFTDS 82
T + NL LLPTGT+ FQ L+PV T+NG C ++L+ +LL + +C +SFTDS
Sbjct: 44 TLTSAANLSNLLPTGTLLAFQLLTPVFTSNGVCDHATRFLTAVLLFLLAASCFVSSFTDS 103
Query: 83 YTGSDGQRHYGIVTMNGLW----PSP-GSDSVDLSAYKLRFGDFVHAFLSVIVFAVLGLL 137
DG ++G VT G+W P P G DL+ Y++RF D++HA LSV+VF + L
Sbjct: 104 VKADDGTIYFGFVTFKGMWVVDYPDPSGLGLPDLAKYRMRFVDWIHATLSVLVFGAVALR 163
Query: 138 DTNVVHCFYPKFESSEKILMQVLPPVIGVVSGAVFMIFPSYRHGIGY 184
D + CFYP E+ K ++ ++P +GV+ +FM+FP+ RHGIGY
Sbjct: 164 DKYITDCFYPSPEAETKHVLDIVPVGVGVMCSLLFMVFPARRHGIGY 210
>At5g39650 unknown protein
Length = 244
Score = 125 bits (314), Expect = 1e-29
Identities = 67/180 (37%), Positives = 98/180 (54%), Gaps = 15/180 (8%)
Query: 22 KTFSGVGNLVKLLPTGTVFLFQYLSPVVTNNGHCSTINKYLSGILLVICGFNCAFTSFTD 81
KT S LV LPTGT+ +F+ + P + +G C+ IN + +LL++C +C F FTD
Sbjct: 64 KTVSKTSMLVNFLPTGTLLMFEMVLPSIYRDGDCNGINTLMIHLLLLLCAMSCFFFHFTD 123
Query: 82 SYTGSDGQRHYGIVTMNGLW-----PSPGSDSVDLSA----------YKLRFGDFVHAFL 126
S+ SDG+ +YG VT GL P P D+ A YKL DFVHA +
Sbjct: 124 SFKASDGKIYYGFVTPRGLAVFMKPPPPEFGGGDVIAEAEIPVTDDRYKLTVNDFVHAVM 183
Query: 127 SVIVFAVLGLLDTNVVHCFYPKFESSEKILMQVLPPVIGVVSGAVFMIFPSYRHGIGYPT 186
SV+VF + D V C +P E +M+ P ++G+V A+F++FP+ R+G+G T
Sbjct: 184 SVLVFMAIAFSDRRVTGCLFPGKEKEMDQVMESFPIMVGIVCSALFLVFPTTRYGVGCMT 243
>At5g27370 putative protein
Length = 191
Score = 116 bits (290), Expect = 9e-27
Identities = 69/180 (38%), Positives = 98/180 (54%), Gaps = 18/180 (10%)
Query: 22 KTFSGVGNLVKLLPTGTVFLFQYLSPVVTNNGHCST--INKYLSGILLVICGFNCAFTSF 79
++ GN LLPTGT +F+ L P +N G C+ +NK L+ L+ C C F+SF
Sbjct: 7 RSLPSAGNFANLLPTGTALIFETLLPSFSNGGECNNKPVNKLLTITLISFCAAACFFSSF 66
Query: 80 TDSYTGSDGQRHYGIVTMNGL-----WPSPGSD-SVDLSA-----YKLRFGDFVHAFLSV 128
TDSY G DG+ +YGI T NGL +P G D L+A YKL F DFVHAF+SV
Sbjct: 67 TDSYVGQDGRIYYGIATSNGLHILNDYPDEGYDPESGLTADKRERYKLSFVDFVHAFVSV 126
Query: 129 IVFAVLGLLDTNVVHCFYPKFESSE-----KILMQVLPPVIGVVSGAVFMIFPSYRHGIG 183
IVF L + ++ C P+ + + ++++ ++ ++ F IFPS R GIG
Sbjct: 127 IVFLALAVESSDFRRCLLPEDDENSWGGHFVLMIKYFAVMVLTMASFFFAIFPSKRRGIG 186
>At4g28485 putative protein
Length = 165
Score = 81.6 bits (200), Expect = 2e-16
Identities = 40/101 (39%), Positives = 60/101 (58%), Gaps = 3/101 (2%)
Query: 92 YGIVTMNGLWPSPGSDSV---DLSAYKLRFGDFVHAFLSVIVFAVLGLLDTNVVHCFYPK 148
YG+ T +GL GS ++ + YKL+ DF+HA +S++VF + + D NV C +P
Sbjct: 65 YGLATWSGLLVMDGSITLTEEEKEKYKLKILDFIHAIMSMLVFFAVSMFDQNVTRCLFPV 124
Query: 149 FESSEKILMQVLPPVIGVVSGAVFMIFPSYRHGIGYPTSSD 189
K ++ LP VIGV+ GA F+ FP+ RHGIG P + +
Sbjct: 125 PSEETKEILTSLPFVIGVICGAFFLAFPTRRHGIGSPLTKE 165
>At5g27430 signal peptidase
Length = 167
Score = 28.1 bits (61), Expect = 3.2
Identities = 14/47 (29%), Positives = 24/47 (50%), Gaps = 1/47 (2%)
Query: 67 LVICGFNCAFTSFTDSYTGSDGQRHYGIVTMNGLWPSP-GSDSVDLS 112
+ I F CA SF+D+++ + I+ +N P G+D V L+
Sbjct: 16 VTILAFICAIASFSDNFSNQNPSAQIQILNINWFQKQPHGNDEVSLT 62
>At5g17780 putative protein
Length = 419
Score = 27.7 bits (60), Expect = 4.1
Identities = 13/26 (50%), Positives = 17/26 (65%)
Query: 113 AYKLRFGDFVHAFLSVIVFAVLGLLD 138
++ +FG +HA LS IVF L LLD
Sbjct: 2 SFPRKFGTAIHAALSFIVFFFLDLLD 27
>At2g38170 Ca2+ antiporter like protein
Length = 463
Score = 27.7 bits (60), Expect = 4.1
Identities = 13/45 (28%), Positives = 20/45 (43%)
Query: 61 YLSGILLVICGFNCAFTSFTDSYTGSDGQRHYGIVTMNGLWPSPG 105
Y+ G++L++C F A F D H G MN + + G
Sbjct: 413 YMKGLVLLLCYFIIAICFFVDKLPQKQNAIHLGHQAMNNVVTATG 457
>At5g19110 dermal glycoprotein - like
Length = 405
Score = 26.6 bits (57), Expect = 9.2
Identities = 11/28 (39%), Positives = 16/28 (56%)
Query: 142 VHCFYPKFESSEKILMQVLPPVIGVVSG 169
+H F P F SS+ + + L P+ G SG
Sbjct: 203 IHYFIPPFNSSDNPIPRTLTPIKGTDSG 230
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.136 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,538,986
Number of Sequences: 26719
Number of extensions: 193253
Number of successful extensions: 423
Number of sequences better than 10.0: 13
Number of HSP's better than 10.0 without gapping: 12
Number of HSP's successfully gapped in prelim test: 1
Number of HSP's that attempted gapping in prelim test: 402
Number of HSP's gapped (non-prelim): 13
length of query: 198
length of database: 11,318,596
effective HSP length: 94
effective length of query: 104
effective length of database: 8,807,010
effective search space: 915929040
effective search space used: 915929040
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 57 (26.6 bits)
Medicago: description of AC146553.13


