

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146552.9 - phase: 0 /pseudo
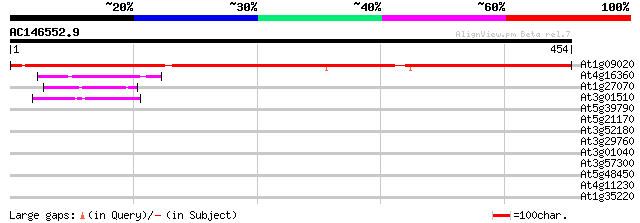
(454 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g09020 putative activator subunit of SNF1-related protein kin... 513 e-145
At4g16360 kinase like protein 53 3e-07
At1g27070 unknown protein 51 1e-06
At3g01510 unknown protein 47 2e-05
At5g39790 5'-AMP-activated protein kinase beta-1 subunit - like 41 0.001
At5g21170 AKIN beta1 41 0.001
At3g52180 unknown protein 37 0.029
At3g29760 unknown protein 31 1.2
At3g01040 unknown protein 31 1.2
At3g57300 helicase-like protein 30 2.7
At5g48450 putative protein 29 4.7
At4g11230 respiratory burst oxidase homolog F - like protein 28 8.0
At1g35220 hypothetical protein 28 8.0
>At1g09020 putative activator subunit of SNF1-related protein kinase
SNF4
Length = 487
Score = 513 bits (1320), Expect = e-145
Identities = 280/470 (59%), Positives = 333/470 (70%), Gaps = 30/470 (6%)
Query: 1 MFSPSMDSARDVGGVVAAGTVLIPVRFVWPYGGRTVYLSGSFTRWSELLQMSPVEGCPTV 60
MF ++DS+R G A+G +L P RFVWPYGGR V+LSGSFTRW+E + MSP+EGCPTV
Sbjct: 1 MFGSTLDSSR--GNSAASGQLLTPTRFVWPYGGRRVFLSGSFTRWTEHVPMSPLEGCPTV 58
Query: 61 FQVIHNLAPGYHQYKFFVDGEWRHDEHTPHITGDYGIVNTVLLATDPFVPV-LPPDIVSG 119
FQVI NL PGYHQYKFFVDGEWRHDEH P ++G+ G+VNT+ + VP P+ +
Sbjct: 59 FQVICNLTPGYHQYKFFVDGEWRHDEHQPFVSGNGGVVNTIFITGPDMVPAGFSPETLGR 118
Query: 120 SNMDVDNETFQRVVRLTDGTLSEVMPRISDVDVQTSRQRISTYLSMRTAYELLPESGKVV 179
SNMDVD+ V T E +PR+S VD++ SR RIS LS RTAYELLPESGKV+
Sbjct: 119 SNMDVDD-----VFLRTADPSQEAVPRMSGVDLELSRHRISVLLSTRTAYELLPESGKVI 173
Query: 180 TLDVDLPVKQAFHILHEQGIPMAPLWDFCKGQFVGVLSVLDFILILRELGNHGSNLTEEE 239
LDV+LPVKQAFHIL+EQGIP+APLWDF KGQFVGVL LDFILILRELG HGSNLTEEE
Sbjct: 174 ALDVNLPVKQAFHILYEQGIPLAPLWDFGKGQFVGVLGPLDFILILRELGTHGSNLTEEE 233
Query: 240 LETHTISAWKEGKWTL----------FSRRFIHAGPSDNLKDVALKILQNGISTVPIIHS 289
LETHTI+AWKEGK + + R + GP DNLKDVALKILQN ++ VP+I+S
Sbjct: 234 LETHTIAAWKEGKAHISRQYDGSGRPYPRPLVQVGPYDNLKDVALKILQNKVAAVPVIYS 293
Query: 290 SSADGSFPQLLHLASLSGILRCILGVVLVHCPYF--NFQFVQFLWAHGCP-KLGRQIAGL 346
S DGS+PQLLHLASLSGIL+CI C YF + + L C LG + +
Sbjct: 294 SLQDGSYPQLLHLASLSGILKCI-------CRYFRHSSSSLPILQQPICSIPLGTWVPRI 346
Query: 347 *QH*DQMLLLLQP*I-Y*FKFNIFFAAQVSSIPIVDESDSLLDIYCRSDITALAKDRAYT 405
+ + L L+P + A+VSSIP+VD++DSL+DIY RSDITALAKD+AY
Sbjct: 347 GESSSKPLATLRPHASLGSALALLVQAEVSSIPVVDDNDSLIDIYSRSDITALAKDKAYA 406
Query: 406 HINLDEMTVHQALQLSQDAFNP-NESRSQRCQMCLRTDSLHKVMERLANP 454
I+LD+MTVHQALQL QDA P QRC MCLR+DSL KVMERLANP
Sbjct: 407 QIHLDDMTVHQALQLGQDASPPYGIFNGQRCHMCLRSDSLVKVMERLANP 456
>At4g16360 kinase like protein
Length = 259
Score = 53.1 bits (126), Expect = 3e-07
Identities = 31/101 (30%), Positives = 46/101 (44%), Gaps = 7/101 (6%)
Query: 23 IPVRFVWPYGGRTVYLSGSFTRWSELLQMSPVEGCPTVFQVIHNLAPGYHQYKFFVDGEW 82
IP W +GG+ + + GS+ W S ++ F ++ L G ++Y+F VDG+W
Sbjct: 72 IPTMITWCHGGKEIAVEGSWDNWKT---RSRLQRSGKDFTIMKVLPSGVYEYRFIVDGQW 128
Query: 83 RHDEHTPHITGDYGIVNTVLLATDPFVPVLPPDIVSGSNMD 123
RH P D G +L D +P DI S S D
Sbjct: 129 RHAPELPLARDDAGNTFNILDLQD----YVPEDIQSISGFD 165
>At1g27070 unknown protein
Length = 532
Score = 51.2 bits (121), Expect = 1e-06
Identities = 29/76 (38%), Positives = 39/76 (51%), Gaps = 3/76 (3%)
Query: 28 VWPYGGRTVYLSGSFTRWSELLQMSPVEGCPTVFQVIHNLAPGYHQYKFFVDGEWRHDEH 87
VWP V L+GSF WS +M E VF + L PG ++ KF VDG+W+ D
Sbjct: 458 VWPNSASEVLLTGSFDGWSTQRKMKKAEN--GVFSLSLKLYPGKYEIKFIVDGQWKVDPL 515
Query: 88 TPHITGDYGIVNTVLL 103
P +T G N +L+
Sbjct: 516 RPIVTSG-GYENNLLI 530
>At3g01510 unknown protein
Length = 716
Score = 47.4 bits (111), Expect = 2e-05
Identities = 30/90 (33%), Positives = 45/90 (49%), Gaps = 4/90 (4%)
Query: 19 GTVLIPVRFVWP-YGGRTVYLSGSFT-RWSELLQMSPVEGCPTVFQVIHNLAPGYHQYKF 76
GT V FVW + G V L G FT W E ++ + +G P F+ L G + YK+
Sbjct: 610 GTPTHSVTFVWNGHEGEEVLLVGDFTGNWKEPIKATH-KGGPR-FETEVRLTQGKYYYKY 667
Query: 77 FVDGEWRHDEHTPHITGDYGIVNTVLLATD 106
++G+WRH +P D G N +++ D
Sbjct: 668 IINGDWRHSATSPTERDDRGNTNNIIVVGD 697
>At5g39790 5'-AMP-activated protein kinase beta-1 subunit - like
Length = 264
Score = 41.2 bits (95), Expect = 0.001
Identities = 27/80 (33%), Positives = 38/80 (46%), Gaps = 2/80 (2%)
Query: 25 VRFVWPYGGRTVYLSGSFTRWSELLQMSP-VEGCPTVFQVIHNLAPGYHQYKFFVDGEWR 83
V W +V + GSF WS+ +SP T F L PG ++ KF VDGEW+
Sbjct: 184 VHVFWIGMAESVQVMGSFDGWSQREDLSPEYSALFTKFSTTLFLRPGRYEMKFLVDGEWQ 243
Query: 84 HDEHTPHITGDYGIVNTVLL 103
P +G+ + N VL+
Sbjct: 244 ISPEFP-TSGEGLMENNVLV 262
>At5g21170 AKIN beta1
Length = 283
Score = 41.2 bits (95), Expect = 0.001
Identities = 30/97 (30%), Positives = 42/97 (42%), Gaps = 8/97 (8%)
Query: 23 IPVRFVWPYGGRTVYLSGSFTRWS--ELLQMSPVEGCPTVFQVIHNLAPGYHQYKFFVDG 80
IP W GG V + GS+ W + LQ S + ++ L G + YK VDG
Sbjct: 100 IPTIITWNQGGNDVAVEGSWDNWRSRKKLQKSGKD-----HSILFVLPSGIYHYKVIVDG 154
Query: 81 EWRHDEHTPHITGDYGIVNTVLLATDPFVPVLPPDIV 117
E ++ P + + G V + L FVP P IV
Sbjct: 155 ESKYIPDLPFVADEVGNVCNI-LDVHNFVPENPESIV 190
>At3g52180 unknown protein
Length = 379
Score = 36.6 bits (83), Expect = 0.029
Identities = 20/61 (32%), Positives = 32/61 (51%), Gaps = 3/61 (4%)
Query: 32 GGRTVYLSGSFTRWSELLQMSPVEGCPTVFQVIHNLAP-GYHQYKFFVDGEWRHDEHTPH 90
G V +SG W + + ++ +G T F ++ P G +YK+ +DGEW H+E P
Sbjct: 265 GFSRVEISGLDIGWGQRIPLTLDKG--TGFWILKRELPEGQFEYKYIIDGEWTHNEAEPF 322
Query: 91 I 91
I
Sbjct: 323 I 323
>At3g29760 unknown protein
Length = 533
Score = 31.2 bits (69), Expect = 1.2
Identities = 18/48 (37%), Positives = 25/48 (51%), Gaps = 5/48 (10%)
Query: 72 HQYKFFVDGE----W-RHDEHTPHITGDYGIVNTVLLATDPFVPVLPP 114
++YK+ V + W +HD P GDY NTVLL P+ +L P
Sbjct: 402 NRYKYVVFKDLNRLWEKHDPRLPWKMGDYNETNTVLLDDSPYKALLNP 449
>At3g01040 unknown protein
Length = 510
Score = 31.2 bits (69), Expect = 1.2
Identities = 26/97 (26%), Positives = 41/97 (41%), Gaps = 24/97 (24%)
Query: 135 LTDGTLSEVMPRISDVDVQTSRQRISTYLSMRTAY--ELLPESGKVVTLDVDLPVKQAFH 192
+ LSE PR +Q+ + + L+ Y EL P KVV LD D+ +++
Sbjct: 279 IAGANLSETTPRTFASKLQSRSPKYISLLNHLRIYLPELFPNLDKVVFLDDDIVIQK--- 335
Query: 193 ILHEQGIPMAPLWDF------------CKGQFVGVLS 217
++PLWD C+G+ V V+S
Sbjct: 336 -------DLSPLWDIDLNGKVNGAVETCRGEDVWVMS 365
>At3g57300 helicase-like protein
Length = 1507
Score = 30.0 bits (66), Expect = 2.7
Identities = 16/46 (34%), Positives = 30/46 (64%), Gaps = 2/46 (4%)
Query: 188 KQAFHILHEQGIPMAPLWDFCKGQFVG--VLSVLDFILILRELGNH 231
+QAF+ + I +A L+D +GQF VL++++ ++ LR++ NH
Sbjct: 839 QQAFYQAIKNKISLAELFDSNRGQFTDKKVLNLMNIVIQLRKVCNH 884
>At5g48450 putative protein
Length = 544
Score = 29.3 bits (64), Expect = 4.7
Identities = 12/38 (31%), Positives = 23/38 (59%)
Query: 13 GGVVAAGTVLIPVRFVWPYGGRTVYLSGSFTRWSELLQ 50
GG++ +IPV F P G T+++S +T+ ++L+
Sbjct: 141 GGIIVNNRAIIPVPFALPDGDVTLFISDWYTKSHKVLR 178
>At4g11230 respiratory burst oxidase homolog F - like protein
Length = 942
Score = 28.5 bits (62), Expect = 8.0
Identities = 27/98 (27%), Positives = 47/98 (47%), Gaps = 10/98 (10%)
Query: 156 RQRISTYLSMRTAYELL---PESGKVVTLDVDLPVKQAFHIL---HEQGIPMAPLWDFCK 209
RQR ++ A+ ++ PE+GK L D+P +++F L G P W +
Sbjct: 677 RQRGDWTEGIKKAFSVVCHAPEAGKSGLLRADVPNQRSFPELLIDGPYGAPAQDHWKYDV 736
Query: 210 GQFVGV-LSVLDFILILRELGNHGSNLTEEELETHTIS 246
VG+ + F+ ILR+L N N+ +++ + IS
Sbjct: 737 VLLVGLGIGATPFVSILRDLLN---NIIKQQEQAECIS 771
>At1g35220 hypothetical protein
Length = 1028
Score = 28.5 bits (62), Expect = 8.0
Identities = 14/34 (41%), Positives = 21/34 (61%), Gaps = 4/34 (11%)
Query: 172 LPESGKVVTLDVDLPVKQAF----HILHEQGIPM 201
L SG++VTLD+ LP+K + H E G+P+
Sbjct: 734 LDSSGRIVTLDIPLPLKNSDGSIPHFGDELGLPL 767
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.328 0.142 0.446
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,124,848
Number of Sequences: 26719
Number of extensions: 431309
Number of successful extensions: 1220
Number of sequences better than 10.0: 13
Number of HSP's better than 10.0 without gapping: 8
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 1203
Number of HSP's gapped (non-prelim): 14
length of query: 454
length of database: 11,318,596
effective HSP length: 103
effective length of query: 351
effective length of database: 8,566,539
effective search space: 3006855189
effective search space used: 3006855189
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.8 bits)
S2: 62 (28.5 bits)
Medicago: description of AC146552.9


