

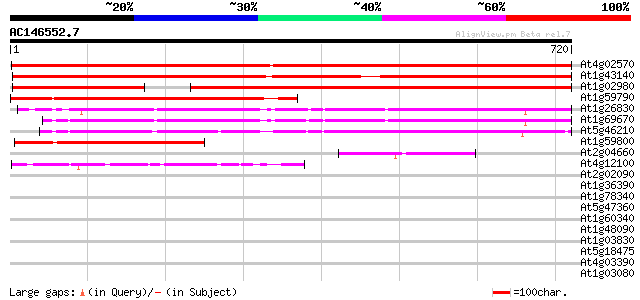
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146552.7 + phase: 0 /pseudo
(720 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g02570 putative cullin-like 1 protein 1203 0.0
At1g43140 hypothetical protein 939 0.0
At1g02980 669 0.0
At1g59790 hypothetical protein 427 e-120
At1g26830 cullin 3-like protein 387 e-108
At1g69670 putative cullin 374 e-104
At5g46210 cullin 342 4e-94
At1g59800 hypothetical protein 275 5e-74
At2g04660 unknown protein 64 2e-10
At4g12100 unknown protein (At4g12100) 59 1e-08
At2g02090 putative helicase 34 0.33
At1g36390 putative heat shock protein 33 0.56
At1g78340 GST7 like protein 33 0.73
At5g47360 putative protein 32 1.6
At1g60340 32 1.6
At1g48090 unknown protein 31 2.1
At1g03830 unknown protein 31 2.1
At5g18475 putative protein 31 2.8
At4g03390 SRF3 (LRR receptor-like protein kinase like protein) 30 3.6
At1g03080 unknown protein 30 3.6
>At4g02570 putative cullin-like 1 protein
Length = 738
Score = 1203 bits (3112), Expect = 0.0
Identities = 583/718 (81%), Positives = 657/718 (91%), Gaps = 3/718 (0%)
Query: 3 ERKTIDLEQGWDFMHRGIMKLKNILEGLPEPQFSPEDYMMLYTTIYNMCTQKPPNDYSQP 62
ERKTIDLEQGWD+M GI KLK ILEGL EP F E YMMLYTTIYNMCTQKPP+DYSQ
Sbjct: 2 ERKTIDLEQGWDYMQTGITKLKRILEGLNEPAFDSEQYMMLYTTIYNMCTQKPPHDYSQQ 61
Query: 63 LYDKYKEAFEEYIVSTVLPSLREKHDEFMLRELVKRWANHKIMVRWLSRFFHYLDRYFIA 122
LYDKY+EAFEEYI STVLP+LREKHDEFMLREL KRW+NHK+MVRWLSRFF+YLDRYFIA
Sbjct: 62 LYDKYREAFEEYINSTVLPALREKHDEFMLRELFKRWSNHKVMVRWLSRFFYYLDRYFIA 121
Query: 123 RRSLPPLNEVGLACFRDLVYKELHGKMRDAIISLIDQEREGEQIDRALLKNVLDIFVEIG 182
RRSLPPLNEVGL CFRDLVY ELH K++ A+I+L+D+EREGEQIDRALLKNVLDI+VEIG
Sbjct: 122 RRSLPPLNEVGLTCFRDLVYNELHSKVKQAVIALVDKEREGEQIDRALLKNVLDIYVEIG 181
Query: 183 MGKMDHYENDFEADMLKDTSAYYSRKASNWILEDSCPDYMLKAEECLRREKDRVAHYLHS 242
MG+M+ YE DFE+ ML+DTS+YYSRKAS+WI EDSCPDYMLK+EECL++E++RVAHYLHS
Sbjct: 182 MGQMERYEEDFESFMLQDTSSYYSRKASSWIQEDSCPDYMLKSEECLKKERERVAHYLHS 241
Query: 243 SSEPKLLEKVQNELLSVYASQLLEKEHSGCHALLRDDKCEDLSRMFRLFSKIPRGLDPVS 302
SSEPKL+EKVQ+ELL V+ASQLLEKEHSGC ALLRDDK +DLSRM+RL+ KI RGL+PV+
Sbjct: 242 SSEPKLVEKVQHELLVVFASQLLEKEHSGCRALLRDDKVDDLSRMYRLYHKILRGLEPVA 301
Query: 303 SIFKQHVTTEGMALVKHAEDAASNKKAEKRDIVGTQEQVFVRKVIELHDKYLAYVNSCFQ 362
+IFKQHVT EG ALV+ AED A+N+ A + QEQV +RKVIELHDKY+ YV CFQ
Sbjct: 302 NIFKQHVTAEGNALVQQAEDTATNQVANTASV---QEQVLIRKVIELHDKYMVYVTECFQ 358
Query: 363 NHTLFHKALKEAFEVFCNKGVAGNSSAELLATFCDNILKKGGSEKLSDEAIEETLEKVVK 422
NHTLFHKALKEAFE+FCNK VAG+SSAELLATFCDNILKKGGSEKLSDEAIE+TLEKVVK
Sbjct: 359 NHTLFHKALKEAFEIFCNKTVAGSSSAELLATFCDNILKKGGSEKLSDEAIEDTLEKVVK 418
Query: 423 LLAYISDKDLFAEFYRKKLARRLLFDKSANDDHERSILTKLKQQCGGQFTSKMEGMVTDL 482
LLAYISDKDLFAEFYRKKLARRLLFD+SANDDHERSILTKLKQQCGGQFTSKMEGMVTDL
Sbjct: 419 LLAYISDKDLFAEFYRKKLARRLLFDRSANDDHERSILTKLKQQCGGQFTSKMEGMVTDL 478
Query: 483 TLAKENQTSFEEYLSNTPNADPGIDLTVTVLTTGFWPSYKSFDLNLPAEMVKCVEVFKEF 542
TLA+ENQ SFE+YL + P A+PGIDLTVTVLTTGFWPSYKSFD+NLP+EM+KCVEVFK F
Sbjct: 479 TLARENQNSFEDYLGSNPAANPGIDLTVTVLTTGFWPSYKSFDINLPSEMIKCVEVFKGF 538
Query: 543 YSTKTKHRKLTWIYSLGTCNISGKFDPKTVELVVTTYQASALLLFNSSDRLSYSEIMTQL 602
Y TKTKHRKLTWIYSLGTC+I+GKFD K +EL+V+TYQA+ LLLFN++D+LSY+EI+ QL
Sbjct: 539 YETKTKHRKLTWIYSLGTCHINGKFDQKAIELIVSTYQAAVLLLFNTTDKLSYTEILAQL 598
Query: 603 NLLDEDVIRLLHSLSCAKYKILIKEPNTKTILPTDYFEFNAKFTDKMRRIKIPLPPVDEK 662
NL ED++RLLHSLSCAKYKIL+KEPNTKT+ D FEFN+KFTD+MRRIKIPLPPVDE+
Sbjct: 599 NLSHEDLVRLLHSLSCAKYKILLKEPNTKTVSQNDAFEFNSKFTDRMRRIKIPLPPVDER 658
Query: 663 KKVIEDVDKDRRYAIDASIVRIMKSRKVLGYQQLVMECVEQLGRMFKPDVKAI*KRIE 720
KKV+EDVDKDRRYAIDA+IVRIMKSRKVLG+QQLV ECVEQL RMFKPD+KAI KR+E
Sbjct: 659 KKVVEDVDKDRRYAIDAAIVRIMKSRKVLGHQQLVSECVEQLSRMFKPDIKAIKKRME 716
>At1g43140 hypothetical protein
Length = 711
Score = 939 bits (2428), Expect = 0.0
Identities = 469/718 (65%), Positives = 565/718 (78%), Gaps = 31/718 (4%)
Query: 4 RKTIDLEQGWDFMHRGIMKLKNILEGLPEPQFSPEDYMMLYTTIYNMCTQKPPNDYSQPL 63
++ I LE+GW M G+ KL+ ILE L EP F P Y+ LYT IY+MC Q+PPNDYSQ L
Sbjct: 2 KELILLEEGWSVMKTGVAKLQRILEDLSEPPFDPGQYINLYTIIYDMCLQQPPNDYSQEL 61
Query: 64 YDKYKEAFEEYIVSTVLPSLREKHDEFMLRELVKRWANHKIMVRWLSRFFHYLDRYFIAR 123
Y+KY+ + Y TVLPS+RE+H E+MLRELVKRWANHKI+VRWLSRF YLDR+++AR
Sbjct: 62 YNKYRGVVDHYNKETVLPSMRERHGEYMLRELVKRWANHKILVRWLSRFCFYLDRFYVAR 121
Query: 124 RSLPPLNEVGLACFRDLVYKELHGKMRDAIISLIDQEREGEQIDRALLKNVLDIFVEIGM 183
R LP LN+VG F DLVY+E+ + +D +++LI +EREGEQIDR L+KNV+D++ G+
Sbjct: 122 RGLPTLNDVGFTSFHDLVYQEIQSEAKDVLLALIHKEREGEQIDRTLVKNVIDVYCGNGV 181
Query: 184 GKMDHYENDFEADMLKDTSAYYSRKASNWILEDSCPDYMLKAEECLRREKDRVAHYLHSS 243
G+M YE DFE+ +L+DT++YYSRKAS W EDSCPDYMLKAEECL+ EK+RV +YLHS+
Sbjct: 182 GQMVIYEEDFESFLLQDTASYYSRKASRWSQEDSCPDYMLKAEECLKLEKERVTNYLHST 241
Query: 244 SEPKLLEKVQNELLSVYASQLLEKEHSGCHALLRDDKCEDLSRMFRLFSKIPRGLDPVSS 303
+EPKL+EKVQNELL V A QL+E EHSGC ALLRDDK DLSRM+RL+ IP+GL+P++
Sbjct: 242 TEPKLVEKVQNELLVVVAKQLIENEHSGCLALLRDDKMGDLSRMYRLYRLIPQGLEPIAD 301
Query: 304 IFKQHVTTEGMALVKHAEDAASNKKAEKRDIVGTQEQVFVRKVIELHDKYLAYVNSCFQN 363
+FKQHVT EG AL+K A DAA+N+ A + QV VRK IELHDKY+ YV+ CFQ
Sbjct: 302 LFKQHVTAEGNALIKQAADAATNQDA-------SASQVLVRKEIELHDKYMVYVDECFQK 354
Query: 364 HTLFHKALKEAFEVFCNKGVAGNSSAELLATFCDNILK-KGGSEKLSDEAIEETLEKVVK 422
H+LFHK LKEAFEVFCNK VAG SSAE+LAT+CDNILK +GGSEKLSDEA E TLEKVV
Sbjct: 355 HSLFHKLLKEAFEVFCNKTVAGASSAEILATYCDNILKTRGGSEKLSDEATEITLEKVVN 414
Query: 423 LLAYISDKDLFAEFYRKKLARRLLFDKSANDDHERSILTKLKQQCGGQFTSKMEGMVTDL 482
LL YISDKDLFAEFYRKK ARRLLFD+S + VTD+
Sbjct: 415 LLVYISDKDLFAEFYRKKQARRLLFDRSG-----------------------IMKEVTDI 451
Query: 483 TLAKENQTSFEEYLSNTPNADPGIDLTVTVLTTGFWPSYKSFDLNLPAEMVKCVEVFKEF 542
TLA+E QT+F +YLS GID TVTVLTTGFWPSYK+ DLNLP EMV CVE FK F
Sbjct: 452 TLARELQTNFVDYLSANMTTKLGIDFTVTVLTTGFWPSYKTTDLNLPTEMVNCVEAFKVF 511
Query: 543 YSTKTKHRKLTWIYSLGTCNISGKFDPKTVELVVTTYQASALLLFNSSDRLSYSEIMTQL 602
Y TKT R+L+WIYSLGTC+I GKF+ KT+ELVV+TYQA+ LLLFN+++RLSY+EI QL
Sbjct: 512 YGTKTNSRRLSWIYSLGTCHILGKFEKKTMELVVSTYQAAVLLLFNNAERLSYTEISEQL 571
Query: 603 NLLDEDVIRLLHSLSCAKYKILIKEPNTKTILPTDYFEFNAKFTDKMRRIKIPLPPVDEK 662
NL ED++RLLHSLSC KYKILIKEP ++TI TD FEFN+KFTDKMR+I++PLPP+DE+
Sbjct: 572 NLSHEDLVRLLHSLSCLKYKILIKEPMSRTISKTDTFEFNSKFTDKMRKIRVPLPPMDER 631
Query: 663 KKVIEDVDKDRRYAIDASIVRIMKSRKVLGYQQLVMECVEQLGRMFKPDVKAI*KRIE 720
KKV+EDVDKDRRYAIDA++VRIMKSRKVL +QQLV ECVE L +MFKPD+K I KRIE
Sbjct: 632 KKVVEDVDKDRRYAIDAALVRIMKSRKVLAHQQLVSECVEHLSKMFKPDIKMIKKRIE 689
>At1g02980
Length = 648
Score = 669 bits (1727), Expect = 0.0
Identities = 333/490 (67%), Positives = 404/490 (81%), Gaps = 3/490 (0%)
Query: 233 KDRVAHYLHSSSEPKLLE-KVQNELLSVYASQLLEKEHSGCHALLRDDKCEDLSRMFRLF 291
+D V L S ++ +L KVQNELL V A QL+E EHSGC ALLRDDK +DL+RM+RL+
Sbjct: 138 RDLVYQELQSKAKDAVLALKVQNELLVVVAKQLIENEHSGCRALLRDDKMDDLARMYRLY 197
Query: 292 SKIPRGLDPVSSIFKQHVTTEGMALVKHAEDAASNKKAEKRDIVGTQEQVFVRKVIELHD 351
IP+GLDPV+ +FKQH+T EG AL+K A +AA++K A + Q+QV +R++I+LHD
Sbjct: 198 HPIPQGLDPVADLFKQHITVEGSALIKQATEAATDKAASTSGLK-VQDQVLIRQLIDLHD 256
Query: 352 KYLAYVNSCFQNHTLFHKALKEAFEVFCNKGVAGNSSAELLATFCDNILKKGGS-EKLSD 410
K++ YV+ CFQ H+LFHKALKEAFEVFCNK VAG SSAE+LAT+CDNILK GG EKL +
Sbjct: 257 KFMVYVDECFQKHSLFHKALKEAFEVFCNKTVAGVSSAEILATYCDNILKTGGGIEKLEN 316
Query: 411 EAIEETLEKVVKLLAYISDKDLFAEFYRKKLARRLLFDKSANDDHERSILTKLKQQCGGQ 470
E +E TLEKVVKLL YISDKDLFAEF+RKK ARRLLFD++ ND HERS+LTK K+ G Q
Sbjct: 317 EDLELTLEKVVKLLVYISDKDLFAEFFRKKQARRLLFDRNGNDYHERSLLTKFKELLGAQ 376
Query: 471 FTSKMEGMVTDLTLAKENQTSFEEYLSNTPNADPGIDLTVTVLTTGFWPSYKSFDLNLPA 530
FTSKMEGM+TD+TLAKE+QT+F E+LS G+D TVTVLTTGFWPSYK+ DLNLP
Sbjct: 377 FTSKMEGMLTDMTLAKEHQTNFVEFLSVNKTKKLGMDFTVTVLTTGFWPSYKTTDLNLPI 436
Query: 531 EMVKCVEVFKEFYSTKTKHRKLTWIYSLGTCNISGKFDPKTVELVVTTYQASALLLFNSS 590
EMV CVE FK +Y TKT R+L+WIYSLGTC ++GKFD KT+E+VVTTYQA+ LLLFN++
Sbjct: 437 EMVNCVEAFKAYYGTKTNSRRLSWIYSLGTCQLAGKFDKKTIEIVVTTYQAAVLLLFNNT 496
Query: 591 DRLSYSEIMTQLNLLDEDVIRLLHSLSCAKYKILIKEPNTKTILPTDYFEFNAKFTDKMR 650
+RLSY+EI+ QLNL ED+ RLLHSLSC KYKILIKEP ++ I TD FEFN+KFTDKMR
Sbjct: 497 ERLSYTEILEQLNLGHEDLARLLHSLSCLKYKILIKEPMSRNISNTDTFEFNSKFTDKMR 556
Query: 651 RIKIPLPPVDEKKKVIEDVDKDRRYAIDASIVRIMKSRKVLGYQQLVMECVEQLGRMFKP 710
RI++PLPP+DE+KK++EDVDKDRRYAIDA++VRIMKSRKVLG+QQLV ECVE L +MFKP
Sbjct: 557 RIRVPLPPMDERKKIVEDVDKDRRYAIDAALVRIMKSRKVLGHQQLVSECVEHLSKMFKP 616
Query: 711 DVKAI*KRIE 720
D+K I KRIE
Sbjct: 617 DIKMIKKRIE 626
Score = 191 bits (484), Expect = 2e-48
Identities = 92/171 (53%), Positives = 119/171 (68%), Gaps = 1/171 (0%)
Query: 4 RKTIDLEQGWDFMHRGIMKLKNILEGLP-EPQFSPEDYMMLYTTIYNMCTQKPPNDYSQP 62
+K LE GW M G+ KL+ ILE +P EP F P M LYTT++N+CTQKPPNDYSQ
Sbjct: 3 KKDSVLEAGWSVMEAGVAKLQKILEEVPDEPPFDPVQRMQLYTTVHNLCTQKPPNDYSQQ 62
Query: 63 LYDKYKEAFEEYIVSTVLPSLREKHDEFMLRELVKRWANHKIMVRWLSRFFHYLDRYFIA 122
+YD+Y + +Y TVLP++REKH E+MLRELVKRWAN KI+VRWLS FF YLDR++
Sbjct: 63 IYDRYGGVYVDYNKQTVLPAIREKHGEYMLRELVKRWANQKILVRWLSHFFEYLDRFYTR 122
Query: 123 RRSLPPLNEVGLACFRDLVYKELHGKMRDAIISLIDQEREGEQIDRALLKN 173
R S P L+ VG FRDLVY+EL K +DA+++L Q + + L++N
Sbjct: 123 RGSHPTLSAVGFISFRDLVYQELQSKAKDAVLALKVQNELLVVVAKQLIEN 173
>At1g59790 hypothetical protein
Length = 374
Score = 427 bits (1099), Expect = e-120
Identities = 211/371 (56%), Positives = 269/371 (71%), Gaps = 21/371 (5%)
Query: 1 MSERKTIDLEQGWDFMHRGIMKLKNILEGLPEPQFSPEDYMMLYTTIYNMCTQKPPNDYS 60
MS + I E+GW + +GI KL ILEG PEP F + LYT IY+MC Q+ +DYS
Sbjct: 6 MSRPRQIKFEEGWSNIQKGITKLIRILEGEPEPTFYFSECFKLYTIIYDMCVQR--SDYS 63
Query: 61 QPLYDKYKEAFEEYIVSTVLPSLREKHDEFMLRELVKRWANHKIMVRWLSRFFHYLDRYF 120
Q LY+KY++ E+Y + TVLPSLREKHDE MLRELVKRW NHKIMV+WLS+FF Y+DR+
Sbjct: 64 QQLYEKYRKVIEDYTIQTVLPSLREKHDEDMLRELVKRWNNHKIMVKWLSKFFVYIDRHL 123
Query: 121 IARRSLP--PLNEVGLACFRDLVYKELHGKMRDAIISLIDQEREGEQIDRALLKNVLDIF 178
+ R +P L+EVGL CF DLVY E+ ++ +I+LI +EREGEQIDRAL+KNVLDI+
Sbjct: 124 VRRSKIPIPSLDEVGLTCFLDLVYCEMQSTAKEVVIALIHKEREGEQIDRALVKNVLDIY 183
Query: 179 VEIGMGKMDHYENDFEADMLKDTSAYYSRKASNWILEDSCPDYMLKAEECLRREKDRVAH 238
VE GMG ++ YE DFE+ ML+DT++YYSRKAS W EDSCPDYM+K EECL+ E++RV H
Sbjct: 184 VENGMGTLEKYEEDFESFMLQDTASYYSRKASRWTEEDSCPDYMIKVEECLKMERERVTH 243
Query: 239 YLHSSSEPKLLEKVQNELLSVYASQLLEKEHSGCHALLRDDKCEDLSRMFRLFSKIPRGL 298
YLHS +EPKL+EK+QNELL + LE EHSG ALLRDDK DLSR++RL+ IP+ L
Sbjct: 244 YLHSITEPKLVEKIQNELLVMVTKNRLENEHSGFSALLRDDKKNDLSRIYRLYLPIPKRL 303
Query: 299 DPVSSIFKQHVTTEGMALVKHAEDAASNKKAEKRDIVGTQEQVFVRKVIELHDKYLAYVN 358
V+ +FK+H+T EG AL+K A+D +N + +IELH+K++ YV
Sbjct: 304 GRVADLFKKHITEEGNALIKQADDKTTN-----------------QLLIELHNKFIVYVI 346
Query: 359 SCFQNHTLFHK 369
CFQNHTLFHK
Sbjct: 347 ECFQNHTLFHK 357
>At1g26830 cullin 3-like protein
Length = 732
Score = 387 bits (995), Expect = e-108
Identities = 247/727 (33%), Positives = 390/727 (52%), Gaps = 58/727 (7%)
Query: 10 EQGWDFMHRGIMKLKNILEGLPEPQFSPEDYMMLYTTIYNMCTQKPPNDYSQPLYDKYKE 69
++ W + R I ++ N S + LY YNM K + + LY +
Sbjct: 26 DKTWQILERAIHQIYN-------QDASGLSFEELYRNAYNMVLHK----FGEKLYTGF-- 72
Query: 70 AFEEYIVSTVLPSLREKHDEF-------MLRELVKRWANHKIMVRWLSRFFHYLDRYFIA 122
++T+ L+EK L EL K+W H + + Y+DR +I
Sbjct: 73 ------IATMTSHLKEKSKLIEAAQGGSFLEELNKKWNEHNKALEMIRDILMYMDRTYIE 126
Query: 123 RRSLPPLNEVGLACFRDLV--YKELHGKMRDAIISLIDQEREGEQIDRALLKNVLDIFVE 180
++ +GL +RD V + ++H ++ + ++ L+ +ER GE IDR L++NV+ +F++
Sbjct: 127 STKKTHVHPMGLNLWRDNVVHFTKIHTRLLNTLLDLVQKERIGEVIDRGLMRNVIKMFMD 186
Query: 181 IGMGKMDHYENDFEADMLKDTSAYYSRKASNWILEDSCPDYMLKAEECLRREKDRVAHYL 240
+G Y+ DFE L +S +Y ++ +I C DY+ K+E+ L E +RVAHYL
Sbjct: 187 LGESV---YQEDFEKPFLDASSEFYKVESQEFIESCDCGDYLKKSEKRLTEEIERVAHYL 243
Query: 241 HSSSEPKLLEKVQNELLSVYASQLLEKEHSGCHALLRDDKCEDLSRMFRLFSKIPRGLDP 300
+ SE K+ V+ E+++ + +L+ E+SG +L +DK EDL RM+ LF ++ GL
Sbjct: 244 DAKSEEKITSVVEKEMIANHMQRLVHMENSGLVNMLLNDKYEDLGRMYNLFRRVTNGLVT 303
Query: 301 VSSIFKQHVTTEGMALVKHAEDAASNKKAEKRDIVGTQEQVFVRKVIELHDKYLAYVNSC 360
V + H+ G LV E + +D V FV+++++ DKY +N+
Sbjct: 304 VRDVMTSHLREMGKQLVTDPE--------KSKDPV-----EFVQRLLDERDKYDKIINTA 350
Query: 361 FQNHTLFHKALKEAFEVFCNKGVAGNSSAELLATFCDNILKKGGSEKLSDEAIEETLEKV 420
F N F AL +FE F N S E ++ F D+ L+K G + ++D +E L+KV
Sbjct: 351 FGNDKTFQNALNSSFEYFINLNA---RSPEFISLFVDDKLRK-GLKGITDVDVEVILDKV 406
Query: 421 VKLLAYISDKDLFAEFYRKKLARRLLFDKSANDDHERSILTKLKQQCGGQFTSKMEGMVT 480
+ L Y+ +KD+F ++Y++ LA+RLL K+ +DD ERS++ KLK +CG QFTSK+EGM T
Sbjct: 407 MMLFRYLQEKDVFEKYYKQHLAKRLLSGKTVSDDAERSLIVKLKTECGYQFTSKLEGMFT 466
Query: 481 DLTLAKENQTSFEEYLSNTPNADPGIDLTVTVLTTGFWPSYKSFDLNLPAEMVKCVEVFK 540
D+ K ++ + + + P G L V VLTTG WP+ + NLPAE+ E F+
Sbjct: 467 DM---KTSEDTMRGFYGSHPELSEGPTLIVQVLTTGSWPTQPAVPCNLPAEVSVLCEKFR 523
Query: 541 EFYSTKTKHRKLTWIYSLGTCNISGKFDP-KTVELVVTTYQASALLLFNSSDRLSYSEIM 599
+Y R+L+W ++GT +I F + EL V+T+Q L+LFN+SDRLSY EI
Sbjct: 524 SYYLGTHTGRRLSWQTNMGTADIKAIFGKGQKHELNVSTFQMCVLMLFNNSDRLSYKEIE 583
Query: 600 TQLNLLDEDVIRLLHSLSCAKYK-ILIKEPNTKTILPTDYFEFNAKFTDKMRRIKIPLPP 658
+ D+ R L SL+C K K ++ KEP +K I D F N KFT K ++KI
Sbjct: 584 QATEIPAADLKRCLQSLACVKGKNVIKKEPMSKDIGEEDLFVVNDKFTSKFYKVKIGTVV 643
Query: 659 VD-----EKKKVIEDVDKDRRYAIDASIVRIMKSRKVLGYQQLVMECVEQLGRMFKPDVK 713
EK++ + V++DR+ I+A+IVRIMKSRK+L + ++ E +QL F +
Sbjct: 644 AQKETEPEKQETRQRVEEDRKPQIEAAIVRIMKSRKILDHNNIIAEVTKQLQPRFLANPT 703
Query: 714 AI*KRIE 720
I KRIE
Sbjct: 704 EIKKRIE 710
>At1g69670 putative cullin
Length = 732
Score = 374 bits (961), Expect = e-104
Identities = 231/687 (33%), Positives = 378/687 (54%), Gaps = 37/687 (5%)
Query: 43 LYTTIYNMCTQKPPNDYSQPLYDKYKEAFEEYIVSTVLPSLREKHDEFMLRELVKRWANH 102
LY YNM K Y LY ++ + S+ E L L ++W +H
Sbjct: 52 LYRNAYNMVLHK----YGDKLYTGLVTTMTFHL-KEICKSIEEAQGGAFLELLNRKWNDH 106
Query: 103 KIMVRWLSRFFHYLDRYFIARRSLPPLNEVGLACFRD-LVYK-ELHGKMRDAIISLIDQE 160
++ + Y+DR +++ ++E+GL +RD +VY ++ ++ + ++ L+ +E
Sbjct: 107 NKALQMIRDILMYMDRTYVSTTKKTHVHELGLHLWRDNVVYSSKIQTRLLNTLLDLVHKE 166
Query: 161 REGEQIDRALLKNVLDIFVEIGMGKMDHYENDFEADMLKDTSAYYSRKASNWILEDSCPD 220
R GE IDR L++NV+ +F+++G Y++DFE L+ ++ +Y ++ +I C +
Sbjct: 167 RTGEVIDRVLMRNVIKMFMDLGESV---YQDDFEKPFLEASAEFYKVESMEFIESCDCGE 223
Query: 221 YMLKAEECLRREKDRVAHYLHSSSEPKLLEKVQNELLSVYASQLLEKEHSGCHALLRDDK 280
Y+ KAE+ L E +RV +YL + SE K+ V+ E+++ + +L+ E+SG +L +DK
Sbjct: 224 YLKKAEKPLVEEVERVVNYLDAKSEAKITSVVEREMIANHVQRLVHMENSGLVNMLLNDK 283
Query: 281 CEDLSRMFRLFSKIPRGLDPVSSIFKQHVTTEGMALVKHAEDAASNKKAEKRDIVGTQEQ 340
ED+ RM+ LF ++ GL V + H+ G LV E + +D V
Sbjct: 284 YEDMGRMYSLFRRVANGLVTVRDVMTLHLREMGKQLVTDPE--------KSKDPV----- 330
Query: 341 VFVRKVIELHDKYLAYVNSCFQNHTLFHKALKEAFEVFCNKGVAGNSSAELLATFCDNIL 400
FV+++++ DKY +N F N F AL +FE F N S E ++ F D+ L
Sbjct: 331 EFVQRLLDERDKYDRIINMAFNNDKTFQNALNSSFEYFVN---LNTRSPEFISLFVDDKL 387
Query: 401 KKGGSEKLSDEAIEETLEKVVKLLAYISDKDLFAEFYRKKLARRLLFDKSANDDHERSIL 460
+K G + + +E ++ L+KV+ L Y+ +KD+F ++Y++ LA+RLL K+ +DD ER+++
Sbjct: 388 RK-GLKGVGEEDVDLILDKVMMLFRYLQEKDVFEKYYKQHLAKRLLSGKTVSDDAERNLI 446
Query: 461 TKLKQQCGGQFTSKMEGMVTDLTLAKENQTSFEEYLSNTPNADPGIDLTVTVLTTGFWPS 520
KLK +CG QFTSK+EGM TD+ K + + + ++ P G L V VLTTG WP+
Sbjct: 447 VKLKTECGYQFTSKLEGMFTDM---KTSHDTLLGFYNSHPELSEGPTLVVQVLTTGSWPT 503
Query: 521 YKSFDLNLPAEMVKCVEVFKEFYSTKTKHRKLTWIYSLGTCNISGKFDP-KTVELVVTTY 579
+ NLPAE+ E F+ +Y R+L+W ++GT +I F + EL V+T+
Sbjct: 504 QPTIQCNLPAEVSVLCEKFRSYYLGTHTGRRLSWQTNMGTADIKAVFGKGQKHELNVSTF 563
Query: 580 QASALLLFNSSDRLSYSEIMTQLNLLDEDVIRLLHSLSCAKYK-ILIKEPNTKTILPTDY 638
Q L+LFN+SDRLSY EI + D+ R L S++C K K +L KEP +K I D+
Sbjct: 564 QMCVLMLFNNSDRLSYKEIEQATEIPTPDLKRCLQSMACVKGKNVLRKEPMSKEIAEEDW 623
Query: 639 FEFNAKFTDKMRRIKIPLPPVD-----EKKKVIEDVDKDRRYAIDASIVRIMKSRKVLGY 693
F N +F K ++KI EK++ + V++DR+ I+A+IVRIMKSR+VL +
Sbjct: 624 FVVNDRFASKFYKVKIGTVVAQKETEPEKQETRQRVEEDRKPQIEAAIVRIMKSRRVLDH 683
Query: 694 QQLVMECVEQLGRMFKPDVKAI*KRIE 720
++ E +QL F + I KRIE
Sbjct: 684 NNIIAEVTKQLQTRFLANPTEIKKRIE 710
>At5g46210 cullin
Length = 792
Score = 342 bits (877), Expect = 4e-94
Identities = 230/692 (33%), Positives = 363/692 (52%), Gaps = 46/692 (6%)
Query: 39 DYMMLYTTIYNMCTQKPPNDYSQPLYDKYKEAFEEYIVSTVLPSLREKHDEF--MLRELV 96
D LY + N+C K LYD+ ++ EE+I S L SL ++ + L +
Sbjct: 115 DLESLYQAVDNLCLHK----LDGKLYDQIEKECEEHI-SAALQSLVGQNTDLTVFLSRVE 169
Query: 97 KRWANHKIMVRWLSRFFHYLDR-YFIARRSLPPLNEVGLACFRD--LVYKELHGKMRDAI 153
K W + + + LDR Y I ++ L E+GL FR + E+ + +
Sbjct: 170 KCWQDFCDQMLMIRSIALTLDRKYVIQNPNVRSLWEMGLQLFRKHLSLAPEVEQRTVKGL 229
Query: 154 ISLIDQEREGEQIDRALLKNVLDIFVEIGMGKMDHYENDFEADMLKDTSAYYSRKASNWI 213
+S+I++ER E ++R LL ++L +F +G+ Y FE L+ TS +Y+ + ++
Sbjct: 230 LSMIEKERLAEAVNRTLLSHLLKMFTALGI-----YMESFEKPFLEGTSEFYAAEGMKYM 284
Query: 214 LEDSCPDYMLKAEECLRREKDRVAHYLHSSSEPKLLEKVQNELLSVYASQLLEKEHSGCH 273
+ P+Y+ E L E +R Y+ + + L+ V+ +LL + +LEK G
Sbjct: 285 QQSDVPEYLKHVEGRLHEENERCILYIDAVTRKPLITTVERQLLERHILVVLEK---GFT 341
Query: 274 ALLRDDKCEDLSRMFRLFSKIPRGLDPVSSIFKQHVTTEGMALVKHAEDAASNKKAEKRD 333
L+ + EDL RM LFS++ L+ + +V G +V E
Sbjct: 342 TLMDGRRTEDLQRMQTLFSRV-NALESLRQALSSYVRKTGQKIVMDEE------------ 388
Query: 334 IVGTQEQVFVRKVIELHDKYLAYVNSCFQNHTLFHKALKEAFEVFCNKGVAGNSSAELLA 393
+++ V+ +++ F + F +K++FE N + N AEL+A
Sbjct: 389 ----KDKDMVQSLLDFKASLDIIWEESFYKNESFGNTIKDSFEHLIN--LRQNRPAELIA 442
Query: 394 TFCDNILKKGGSEKLSDEAIEETLEKVVKLLAYISDKDLFAEFYRKKLARRLLFDKSAND 453
F D L+ G++ S+E +E LEKV+ L +I KD+F FY+K LA+RLL KSA+
Sbjct: 443 KFLDEKLR-AGNKGTSEEELESVLEKVLVLFRFIQGKDVFEAFYKKDLAKRLLLGKSASI 501
Query: 454 DHERSILTKLKQQCGGQFTSKMEGMVTDLTLAKENQTSFEEYLSNTPNADPGIDLTVTVL 513
D E+S+++KLK +CG QFT+K+EGM D+ L+KE SF++ GI+++V VL
Sbjct: 502 DAEKSMISKLKTECGSQFTNKLEGMFKDIELSKEINESFKQSSQARTKLPSGIEMSVHVL 561
Query: 514 TTGFWPSYKSFDLNLPAEMVKCVEVFKEFYSTKTKHRKLTWIYSLGTCNISGKFDPKTVE 573
TTG+WP+Y D+ LP E+ ++FKEFY +K R+L W SLG C + F E
Sbjct: 562 TTGYWPTYPPMDVKLPHELNVYQDIFKEFYLSKYSGRRLMWQNSLGHCVLKADFSKGKKE 621
Query: 574 LVVTTYQASALLLFNSSDRLSYSEIMTQLNLLDEDVIRLLHSLSCAKYKILIKEPNTKTI 633
L V+ +QA L+LFN + +LS+ +I ++ D+++ R L SL+C K ++L K P + +
Sbjct: 622 LAVSLFQAVVLMLFNDAMKLSFEDIKDSTSIEDKELRRTLQSLACGKVRVLQKNPKGRDV 681
Query: 634 LPTDYFEFNAKFTDKMRRIKIPL----PPVDEKKKVIEDVDKDRRYAIDASIVRIMKSRK 689
D FEFN +F + RIK+ V+E E V +DR+Y IDA+IVRIMK+RK
Sbjct: 682 EDGDEFEFNDEFAAPLYRIKVNAIQMKETVEENTSTTERVFQDRQYQIDAAIVRIMKTRK 741
Query: 690 VLGYQQLVMECVEQLGRMFKP-DVKAI*KRIE 720
VL + L+ E +QL KP D+K KRIE
Sbjct: 742 VLSHTLLITELFQQLKFPIKPADLK---KRIE 770
>At1g59800 hypothetical protein
Length = 255
Score = 275 bits (704), Expect = 5e-74
Identities = 134/244 (54%), Positives = 172/244 (69%), Gaps = 4/244 (1%)
Query: 7 IDLEQGWDFMHRGIMKLKNILEGLPEPQFSPEDYMMLYTTIYNMCTQKPPNDYSQPLYDK 66
I E W + +G KL ++EG EP F+ E MM++T Y +C K P Q LYDK
Sbjct: 7 IKFEVEWSNIQQGFTKLIRMIEGESEPAFNQEIMMMMHTATYRICAYKNP----QQLYDK 62
Query: 67 YKEAFEEYIVSTVLPSLREKHDEFMLRELVKRWANHKIMVRWLSRFFHYLDRYFIARRSL 126
Y+E E Y + TVLPSLREKHDE MLREL KRW HK++VR SR YLD F++++ L
Sbjct: 63 YRELIENYAIQTVLPSLREKHDECMLRELAKRWNAHKLLVRLFSRRLVYLDDSFLSKKGL 122
Query: 127 PPLNEVGLACFRDLVYKELHGKMRDAIISLIDQEREGEQIDRALLKNVLDIFVEIGMGKM 186
P L EVGL CFRD VY+E+ +AI++LI +EREGEQIDR L++NV+D+FVE GMG +
Sbjct: 123 PSLREVGLNCFRDQVYREMQSMAAEAILALIHKEREGEQIDRELVRNVIDVFVENGMGTL 182
Query: 187 DHYENDFEADMLKDTSAYYSRKASNWILEDSCPDYMLKAEECLRREKDRVAHYLHSSSEP 246
YE DFE ML+DT++YYS KAS WI E+SC DY LK ++CL+RE++RV HYLH ++EP
Sbjct: 183 KKYEEDFERLMLQDTASYYSSKASRWIQEESCLDYTLKPQQCLQRERERVTHYLHPTTEP 242
Query: 247 KLLE 250
KL E
Sbjct: 243 KLFE 246
>At2g04660 unknown protein
Length = 865
Score = 64.3 bits (155), Expect = 2e-10
Identities = 48/183 (26%), Positives = 79/183 (42%), Gaps = 10/183 (5%)
Query: 423 LLAYISDKDLFAEFYRKKLARRLLFDKSANDDHERSILTKLKQQCGGQFTSKMEGMVTDL 482
L+ I K+ YR LA +LL + D E + LK G + E M+ DL
Sbjct: 543 LVDIIGSKEQLVNEYRVMLAEKLLNKTDYDIDTEIRTVELLKIHFGEASMQRCEIMLNDL 602
Query: 483 TLAKENQTSFE-------EYLSNTPNADPGIDLTVTVLTTGFWPSYKSFDLNLPAEMVKC 535
+K T+ + E N + D LT T+L+T FWP + L LP + K
Sbjct: 603 IDSKRVNTNIKKASQTGAELRENELSVDT---LTSTILSTNFWPPIQDEPLELPGPVDKL 659
Query: 536 VEVFKEFYSTKTKHRKLTWIYSLGTCNISGKFDPKTVELVVTTYQASALLLFNSSDRLSY 595
+ + Y RKL W +LGT + +F+ + ++ V+ A+ ++ F +Y
Sbjct: 660 LSDYANRYHEIKTPRKLLWKKNLGTVKLELQFEDRAMQFTVSPTHAAIIMQFQEKKSWTY 719
Query: 596 SEI 598
++
Sbjct: 720 KDL 722
>At4g12100 unknown protein (At4g12100)
Length = 434
Score = 58.9 bits (141), Expect = 1e-08
Identities = 83/389 (21%), Positives = 157/389 (40%), Gaps = 60/389 (15%)
Query: 3 ERKTIDLEQGWDFMHRGIMKLKNILEGLPEPQFSPEDYMMLYTTIYNMCTQKPPND--YS 60
+R + L + WD + I + L + ++ ++ +TTI+ + D S
Sbjct: 71 DRNKLYLAKAWDLLKPAIKII------LDDDEYKKPGDVLCFTTIFRAVKRACLGDPRQS 124
Query: 61 QPLYDKYKEAFEEYIVSTVLPSLREK-----HDEFMLRELVKRWANHKIMVRWLSRFFHY 115
+ +++ K E +I ++ SL + L + RW + K + +S Y
Sbjct: 125 ELVFNLVKHECEPHIAE-LIQSLEKNCSGSDDPSVFLPHVYNRWLDFKRKMSLVSDVAMY 183
Query: 116 LDRYFIARRSLPPLNEVGLACFRDLVYK--ELHGKMRDAIISLIDQEREGEQIDRA--LL 171
+ L +VG F + +L ++ I+ LI ER G+ + LL
Sbjct: 184 QTLNGLT------LWDVGQKLFHKQLSMAPQLQDQVITGILRLITDERLGKAANNTSDLL 237
Query: 172 KNVLDIFVEIGMGKMDHYENDFEADMLKDTSAYYSRKASNWILEDSCPDYMLKAEECLRR 231
KN++D+F + Y++ F L TS +Y+ +A + Y+ E
Sbjct: 238 KNLMDMF-RMQWQCTYVYKDPF----LDSTSKFYAEEAEQVLQRSDISHYLKYVERTFLA 292
Query: 232 EKDRVA--HYLHSSSEPKLLEKVQNELLSVYASQLLEKEHSGCHALLRDDKCEDLSRMFR 289
E+++ ++ SSS +L++ ++++LL ++S L E G L+ + +DL RM+R
Sbjct: 293 EEEKCDKHYFFFSSSRSRLMKVLKSQLLEAHSSFLEE----GFMLLMDESLIDDLRRMYR 348
Query: 290 LFSKIPRGLDPVSSIFKQHVTTEGMALVKHAEDAASNKKAEKRDIVGTQEQVFVRKVIEL 349
LFS + D + I + ++ +G E A + EL
Sbjct: 349 LFSMVD-SEDYIDRILRAYILAKG-------EGARQEGSLQ-----------------EL 383
Query: 350 HDKYLAYVNSCFQNHTLFHKALKEAFEVF 378
H + CF L K +++ FE F
Sbjct: 384 HTSIDKIWHQCFGQDDLLDKTIRDCFEGF 412
>At2g02090 putative helicase
Length = 763
Score = 33.9 bits (76), Expect = 0.33
Identities = 31/96 (32%), Positives = 43/96 (44%), Gaps = 17/96 (17%)
Query: 65 DKYKEAFEEYIVSTV--LPSLREKHDEFMLRELVKRW-----------ANHKIMVRWLSR 111
D YKEA EEY ++ L L K + + L KR ANH +++R R
Sbjct: 466 DAYKEAIEEYRAASQARLVKLSSKSLNSLAKALPKRQISNYFTQFRKIANHPLLIR---R 522
Query: 112 FFHYLDRYFIARRSLPPLNEVGLACFRDLVYKELHG 147
+ D IAR+ L P+ G C D V +E+ G
Sbjct: 523 IYSDEDVIRIARK-LHPIGAFGFECSLDRVIEEVKG 557
>At1g36390 putative heat shock protein
Length = 279
Score = 33.1 bits (74), Expect = 0.56
Identities = 45/167 (26%), Positives = 72/167 (42%), Gaps = 14/167 (8%)
Query: 555 IYSLGTCNISGKFDPKTVELVVTTYQASALLLFNSSDRLSYSEIMTQL-------NLLDE 607
I+S N S + + K V T ++ L N D S +EI T N +D+
Sbjct: 55 IFSAHQTNNSEEANSKQQADVKTLIRSYKQALLNG-DETSVTEIETMFCKIEKEKNKMDQ 113
Query: 608 DVIRLLHSLSCAK-YKILIKEPNTKTILPTDYFEFNAKFTDKMRRIKIPLPPVD--EKKK 664
V+ L ++ K KI ++ T D + + K++ +K LP +D EK K
Sbjct: 114 KVLSLSMKIASEKEMKIRLQADFDNTRKKLDKDRLSTESNAKVQILKSLLPIIDSFEKAK 173
Query: 665 VIEDVDKDRRYAIDASIVRIMKS-RKVLGYQQLVMECVEQLGRMFKP 710
+ VD D+ ID S I + +VL Y L + + +G+ F P
Sbjct: 174 LQVRVDTDKEKKIDTSYQGIYRQFVEVLRY--LRVSVIATVGKPFDP 218
>At1g78340 GST7 like protein
Length = 218
Score = 32.7 bits (73), Expect = 0.73
Identities = 16/40 (40%), Positives = 22/40 (55%), Gaps = 2/40 (5%)
Query: 206 SRKASNWILEDSCPDYMLKAEECLRREKDRVAHYLHSSSE 245
S K +N+ +E CP M A+ CL+RE V LH S +
Sbjct: 167 SEKLANFSIEPECPTLMASAKRCLQRES--VVQSLHDSEK 204
>At5g47360 putative protein
Length = 477
Score = 31.6 bits (70), Expect = 1.6
Identities = 25/98 (25%), Positives = 48/98 (48%), Gaps = 3/98 (3%)
Query: 240 LHSSSEPKLLEKVQNELLSVYASQLLEKEHSGCHALLRDDKCEDLSRMFRLFSKIPRGLD 299
L + + K L K+ ++L+ + L E S +L+R + E+ ++FRL + RG+
Sbjct: 320 LENDEDVKALSKLIDKLVKLGGVSLSECFSSATVSLIRMKRWEEAEKIFRLM--LVRGVR 377
Query: 300 PVSSIFKQHVTTEGMALVKHAEDAASNKKAEKRDIVGT 337
P + HV E L ++ + ++ EK+D+ T
Sbjct: 378 P-DGLACSHVFRELCLLERYLDCFLLYQEIEKKDVKST 414
>At1g60340
Length = 371
Score = 31.6 bits (70), Expect = 1.6
Identities = 24/76 (31%), Positives = 33/76 (42%), Gaps = 10/76 (13%)
Query: 495 YLSNTPNADPGIDLTVTVLTTGFWPSYKSFDLNL--PAEMVKCVEVFKEFYSTKTKHRKL 552
YLSNT D T+ WPSY + ++ P E+V +V + Y
Sbjct: 180 YLSNTQEEDEFYLKTIMTSEGNDWPSYVTNNVYCLHPLELVDLQDVMFQSYG-------- 231
Query: 553 TWIYSLGTCNISGKFD 568
T I++ GTC S K D
Sbjct: 232 TCIFANGTCGESDKCD 247
>At1g48090 unknown protein
Length = 4099
Score = 31.2 bits (69), Expect = 2.1
Identities = 21/80 (26%), Positives = 43/80 (53%), Gaps = 3/80 (3%)
Query: 191 NDFEADMLKDTSAYYSRKASNWILEDSCPDYMLKAEECLRREKDRVAHYLHSSSEPKLLE 250
N +EA + + +S+ S+ S W +++ ++ +++ L+R R+A + S +PK E
Sbjct: 2384 NIYEASVTRSSSSKSSQLWSIWKVDNQACTFLARSD--LKRPPSRMAFAVGESVKPKTQE 2441
Query: 251 KVQNEL-LSVYASQLLEKEH 269
V E+ L ++ LL+ H
Sbjct: 2442 NVNAEIKLRCFSLTLLDGLH 2461
>At1g03830 unknown protein
Length = 991
Score = 31.2 bits (69), Expect = 2.1
Identities = 39/179 (21%), Positives = 74/179 (40%), Gaps = 17/179 (9%)
Query: 302 SSIFKQHVTTEGMALV----KHAEDAASNKKAEKRDIVGTQEQVFVRKVIELH--DKYLA 355
+++ +Q TE +A + K A +AE ++ +E++ K +L ++ LA
Sbjct: 528 NTVSEQKAVTEKIAAMEEKLKQASTTEDGLRAEFSRVLDEKEKIITEKAAKLATLEQQLA 587
Query: 356 YVNSCFQNHTLF-------HKALKEAFEVFCNKGVAGNSSAELLATFCDNILKKGGSEKL 408
+ + L K ++ + K + S++ELL T + + ++
Sbjct: 588 STRAELKKSALKVDECSSEAKDVRLQMSLLNEKYESVKSASELLETETETLKREKDELDK 647
Query: 409 SDEAIEETLEKVVKLLAYISDKDLFAEFYRKKLARRLLFDKSANDDHERSILTKLKQQC 467
E LEK+V L + + L A KKL L + A D+E + T L ++C
Sbjct: 648 KCHIHLEELEKLVLRLTNVESEALEA----KKLVDSLKLEAEAARDNENKLQTSLVERC 702
>At5g18475 putative protein
Length = 537
Score = 30.8 bits (68), Expect = 2.8
Identities = 45/173 (26%), Positives = 73/173 (42%), Gaps = 28/173 (16%)
Query: 151 DAIISLIDQEREGEQIDRALLKNVLDIFVEIGMGKMDHYENDFEADMLKDTSAYYSRKAS 210
++ +SL+ +ER+ + VLDIF + K ++ N + +L + + A
Sbjct: 88 ESAVSLMKRERDPQ--------GVLDIFNKASQQKGFNHNNATYSVLLDNLVRHKKFLAV 139
Query: 211 NWILEDSCPDYMLKAEECLRREK---DRVAHYLHSSSEPKLLEKVQNELLSVYASQLLEK 267
+ IL + +K E C +E + + H+ S K++E L+ V A
Sbjct: 140 DAIL------HQMKYETCRFQESLFLNLMRHFSRSDLHDKVMEMFN--LIQVIARVKPSL 191
Query: 268 EH-SGCHALLRDDKCEDLSRMFRLFSKIPRGLDPVSSIFKQHVTTEGMALVKH 319
S C LL D +LSR L++K GL P + IF LVKH
Sbjct: 192 NAISTCLNLLIDSGEVNLSRKLLLYAKHNLGLQPNTCIFN--------ILVKH 236
>At4g03390 SRF3 (LRR receptor-like protein kinase like protein)
Length = 776
Score = 30.4 bits (67), Expect = 3.6
Identities = 21/84 (25%), Positives = 37/84 (44%), Gaps = 3/84 (3%)
Query: 64 YDKYKEAFEEYIVSTVLPSLREKHDEFMLRELVKRWANHKIMVRWLSRFFHYLDRYFIAR 123
YD+ + E+++V +P L HD L ++V N + + LS F + R +
Sbjct: 689 YDRDRSRGEQFLVRWAIPQL---HDIDALGKMVDPSLNGQYPAKSLSHFADIISRCVQSE 745
Query: 124 RSLPPLNEVGLACFRDLVYKELHG 147
PL + D++ +E HG
Sbjct: 746 PEFRPLMSEVVQDLLDMIRRERHG 769
>At1g03080 unknown protein
Length = 1744
Score = 30.4 bits (67), Expect = 3.6
Identities = 45/215 (20%), Positives = 89/215 (40%), Gaps = 13/215 (6%)
Query: 229 LRREKDRVAHYLHSSSE--PKLLEK---VQNEL------LSVYASQLLEKEHSGCHALLR 277
L EKD + L S++E KL E+ ++N L L S+L E S CH LL
Sbjct: 711 LHSEKDMLISRLQSATENSKKLSEENMVLENSLFNANVELEELKSKLKSLEES-CH-LLN 768
Query: 278 DDKCEDLSRMFRLFSKIPRGLDPVSSIFKQHVTTEGMALVKHAEDAASNKKAEKRDIVGT 337
DDK S L S I + + K+H + L E +S +K E+ +
Sbjct: 769 DDKTTLTSERESLLSHIDTMRKRIEDLEKEHAELKVKVLELATERESSLQKIEELGVSLN 828
Query: 338 QEQVFVRKVIELHDKYLAYVNSCFQNHTLFHKALKEAFEVFCNKGVAGNSSAELLATFCD 397
+ ++ + + + S + ++ ++V ++ + +L
Sbjct: 829 AKDCEYASFVQFSESRMNGMESTIHHLQDENQCRVREYQVELDRAHDAHIEIIVLQKCLQ 888
Query: 398 NILKKGGSEKLSDEAIEETLEKVVKLLAYISDKDL 432
+ L+K S ++ I+E + + KL++ + ++++
Sbjct: 889 DWLEKSSSLIAENQDIKEASKLLEKLVSELEEENI 923
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.321 0.137 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,043,445
Number of Sequences: 26719
Number of extensions: 708150
Number of successful extensions: 2055
Number of sequences better than 10.0: 28
Number of HSP's better than 10.0 without gapping: 11
Number of HSP's successfully gapped in prelim test: 17
Number of HSP's that attempted gapping in prelim test: 2006
Number of HSP's gapped (non-prelim): 35
length of query: 720
length of database: 11,318,596
effective HSP length: 106
effective length of query: 614
effective length of database: 8,486,382
effective search space: 5210638548
effective search space used: 5210638548
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 64 (29.3 bits)
Medicago: description of AC146552.7


