

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146552.4 + phase: 0
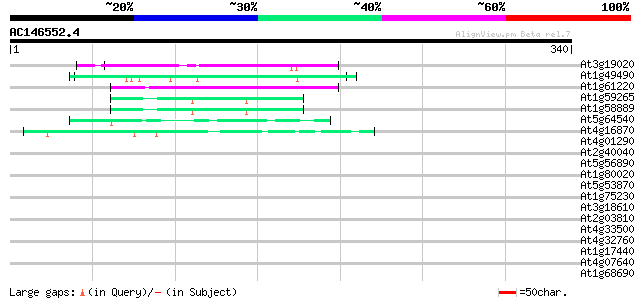
(340 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g19020 hypothetical protein 50 1e-06
At1g49490 hypothetical protein 50 1e-06
At1g61220 unknown protein 44 2e-04
At1g59265 polyprotein, putative 42 4e-04
At1g58889 polyprotein, putative 42 4e-04
At5g64540 unknown protein 42 5e-04
At4g16870 retrotransposon like protein 41 8e-04
At4g01290 hypothetical protein 41 0.001
At2g40040 unknown protein 40 0.002
At5g56890 unknown protein 39 0.005
At1g80020 hypothetical protein 39 0.005
At5g53870 predicted GPI-anchored protein 38 0.007
At1g75230 3-methyladenine DNA glycosylase like protein 38 0.009
At3g18610 unknown protein 37 0.012
At2g03810 unknown protein 37 0.012
At4g33500 unknown protein 37 0.020
At4g32760 unknown protein 37 0.020
At1g17440 unknown protein 37 0.020
At4g07640 putative athila transposon protein 36 0.026
At1g68690 protein kinase, putative 36 0.026
>At3g19020 hypothetical protein
Length = 951
Score = 50.4 bits (119), Expect = 1e-06
Identities = 38/161 (23%), Positives = 66/161 (40%), Gaps = 10/161 (6%)
Query: 41 PTHPLYPDNPEPVSVQQPQPNPEPTTNSPHNSTTQKASEVASDATTSETPQHQEYSTLHN 100
P+ P ++P+P QQP P PE ++ P N K ++ +E P+ + S
Sbjct: 427 PSKPKPEESPKP---QQPSPKPETPSHEPSNPKEPKPESPKQESPKTEQPKPKPESPKQE 483
Query: 101 LEKHLGGEMQPTPTKASKTVPEKTVLETQTETQTIPEQTVQEQTASEQVAPDQTTSDQHI 160
K + P P + K PE E+ + PE++ + + + +P Q
Sbjct: 484 SPK----QEAPKP-EQPKPKPESPKQESSKQEPPKPEESPKPEPPKPEESPKPQPPKQET 538
Query: 161 PSDQTTEQQQQP--DSPTIIDLTSDQPSISNTTQTEPSPIP 199
P + + + Q P ++P + QP T + E SP P
Sbjct: 539 PKPEESPKPQPPKQETPKPEESPKPQPPKQETPKPEESPKP 579
Score = 42.4 bits (98), Expect = 4e-04
Identities = 34/149 (22%), Positives = 52/149 (34%), Gaps = 12/149 (8%)
Query: 58 PQPNPEPTTNSPHNSTTQKASEVASDATTSETPQHQEYSTLHNLEKHLGGEMQPTPTKAS 117
P+P P P P E S E+P+ Q+ S H +P+ K
Sbjct: 404 PKPTPTPKAPEPKKEINPPNLEEPSKPKPEESPKPQQPSPKPETPSH-----EPSNPKEP 458
Query: 118 KTVPEKTVLETQTETQTIPEQTVQEQTASEQVAPDQTTSDQHIPSDQTTEQQ-------Q 170
K K + + PE QE E P+Q P ++++Q+
Sbjct: 459 KPESPKQESPKTEQPKPKPESPKQESPKQEAPKPEQPKPKPESPKQESSKQEPPKPEESP 518
Query: 171 QPDSPTIIDLTSDQPSISNTTQTEPSPIP 199
+P+ P + QP T + E SP P
Sbjct: 519 KPEPPKPEESPKPQPPKQETPKPEESPKP 547
Score = 31.2 bits (69), Expect = 0.85
Identities = 37/173 (21%), Positives = 59/173 (33%), Gaps = 16/173 (9%)
Query: 37 FNLPPTHPLYPDNPEPVSVQQPQPN--PEPTTNSPHNSTTQKASEVASDATTSETPQHQE 94
F+ PP P+Y P PV P + P P +SP V S P H
Sbjct: 735 FSPPPPAPIYSPPPPPVHSPPPPVHSPPPPPVHSPPPPVHSPPPPVHS----PPPPVHSP 790
Query: 95 YSTLHNLEKHLGGEMQPTPTKASKTVPEKTVLETQTETQTIPEQTVQE--QTASEQVAPD 152
+H+ P P + P + + P + E + ++ AP
Sbjct: 791 PPPVHSPPPPSPIYSPPPPVFSPPPKPVTPLPPATSPMANAPTPSSSESGEISTPVQAPT 850
Query: 153 QTTSDQHIPSDQT------TEQQQQPDSPTIID--LTSDQPSISNTTQTEPSP 197
+ D PSD + PDS ++ + S +P + Q+E +P
Sbjct: 851 PDSEDIEAPSDSNHSPVFKSSPAPSPDSEPEVEAPVPSSEPEVEAPKQSEATP 903
>At1g49490 hypothetical protein
Length = 847
Score = 50.4 bits (119), Expect = 1e-06
Identities = 43/181 (23%), Positives = 73/181 (39%), Gaps = 7/181 (3%)
Query: 37 FNLPPTHPLYPDNPEPVSVQQPQPNPEPTTNSP---HNSTTQKA---SEVASDATTSETP 90
F+ PP P+Y P S P +P P T SP HN+ + + +SET
Sbjct: 607 FSPPPPSPVYSPPPPSHSPPPPVYSPPPPTFSPPPTHNTNQPPMGAPTPTQAPTPSSETT 666
Query: 91 QHQEYSTLHNLEKHLGGEMQPTPTKASKTVPEKTVLETQTETQTIPEQTVQEQTASEQV- 149
Q S+ + + L PTP ++S E T + T + +++ + A V
Sbjct: 667 QVPTPSSESDQSQILSPVQAPTPVQSSTPSSEPTQVPTPSSSESYQAPNLSPVQAPTPVQ 726
Query: 150 APDQTTSDQHIPSDQTTEQQQQPDSPTIIDLTSDQPSISNTTQTEPSPIPDHILESEYIE 209
AP ++ +P+ + Q +PT I P+ ++ P+P + + E E
Sbjct: 727 APTTSSETSQVPTPSSESNQSPSQAPTPILEPVHAPTPNSKPVQSPTPSSEPVSSPEQSE 786
Query: 210 E 210
E
Sbjct: 787 E 787
Score = 43.5 bits (101), Expect = 2e-04
Identities = 51/197 (25%), Positives = 74/197 (36%), Gaps = 39/197 (19%)
Query: 40 PPTHPLYPDNPEPVSVQQPQPNPEPTTNSPHNS----------TTQKASEVASDATTSET 89
PP H P P PV+ P P+P P +SP + S V S S +
Sbjct: 568 PPPHVYSP--PPPVA-SPPPPSPPPPVHSPPPPPVFSPPPPVFSPPPPSPVYSPPPPSHS 624
Query: 90 PQHQEYS---------TLHNLEKHLGGEMQPT--PTKASKTVPEKTVLETQTETQTIPEQ 138
P YS HN + G PT PT +S+T T ++Q +
Sbjct: 625 PPPPVYSPPPPTFSPPPTHNTNQPPMGAPTPTQAPTPSSETTQVPTPSSESDQSQIL--S 682
Query: 139 TVQEQTASEQVAPDQTTSDQHIPSDQTTEQQQQPD-----------SPTIIDLTSDQPSI 187
VQ T + P ++ +P+ ++E Q P+ +PT TS P+
Sbjct: 683 PVQAPTPVQSSTP--SSEPTQVPTPSSSESYQAPNLSPVQAPTPVQAPTTSSETSQVPTP 740
Query: 188 SNTTQTEPSPIPDHILE 204
S+ + PS P ILE
Sbjct: 741 SSESNQSPSQAPTPILE 757
Score = 37.4 bits (85), Expect = 0.012
Identities = 41/181 (22%), Positives = 66/181 (35%), Gaps = 21/181 (11%)
Query: 58 PQPNPEPTTNSPHNSTTQKASEVASDATTSETPQHQEYSTLHNLEKHLGGEMQPTPTKAS 117
P PNP P T+ P S + SD++ ETP+ E + +P P K
Sbjct: 394 PSPNP-PRTSEPKPSKPEPVMPKPSDSSKPETPKTPEQPS-----------PKPQPPKHE 441
Query: 118 KTVPE----KTVLETQTETQTIPEQTVQEQTASEQVAPDQTTSDQHIPSDQTTEQQQQPD 173
PE K L Q E+ ++ EQ P+++ + + T+ P+
Sbjct: 442 SPKPEEPENKHELPKQKESPKPQPSKPEDSPKPEQPKPEESPKPEQPQIPEPTKPVSPPN 501
Query: 174 SPTIIDLTSDQPSISNTTQTEPSPIPDHILESEYIEEQLIRLSDEIQALILRRTVPAPPI 233
T D P ++ + SP P + ++ Q S + I P PP+
Sbjct: 502 EAQ--GPTPDDPYDASPVKNRRSPPPPKVEDTRVPPPQPPMPSPSPPSPIYS---PPPPV 556
Query: 234 H 234
H
Sbjct: 557 H 557
Score = 34.7 bits (78), Expect = 0.077
Identities = 39/160 (24%), Positives = 62/160 (38%), Gaps = 34/160 (21%)
Query: 44 PLYPDNPEPVSVQQPQPNPEPTTNS------PHNSTTQKASEVASDATTSETPQHQEYST 97
P+ P S +P PT +S P+ S Q + V + T+SET Q
Sbjct: 683 PVQAPTPVQSSTPSSEPTQVPTPSSSESYQAPNLSPVQAPTPVQAPTTSSETSQ------ 736
Query: 98 LHNLEKHLGGEMQPTPTKASKTVPEKTVLETQTETQTIPEQTVQEQTASEQVAPDQTTSD 157
PTP+ S P +Q T + + V T + + T S
Sbjct: 737 ------------VPTPSSESNQSP------SQAPTPIL--EPVHAPTPNSKPVQSPTPSS 776
Query: 158 QHIPSDQTTEQQQQPDSPTIIDLTSDQPSISNTTQTEPSP 197
+ + S + +E+ + P+ PT ++ S PS S +T T P
Sbjct: 777 EPVSSPEQSEEVEAPE-PTPVN-PSSVPSSSPSTDTSIPP 814
Score = 28.1 bits (61), Expect = 7.2
Identities = 27/139 (19%), Positives = 46/139 (32%), Gaps = 22/139 (15%)
Query: 35 EKFNLPPTHPLYPDNPEPVSVQQPQPNPEPTTNSPHNSTTQKASEVASDATTSETPQHQE 94
E + P P+ P + + PT +S N + +A + + TP +
Sbjct: 709 ESYQAPNLSPVQAPTPVQAPTTSSETSQVPTPSSESNQSPSQAPTPILEPVHAPTPNSKP 768
Query: 95 YSTLHNLEKHLGGEMQPTPTKASKTVPEKTVLETQTETQTIPEQTVQEQTASEQVAPDQT 154
+ PTP+ + PE Q+E PE T ++ +P
Sbjct: 769 VQS-------------PTPSSEPVSSPE------QSEEVEAPEPTPVNPSSVPSSSPSTD 809
Query: 155 TSDQHIPSDQTTEQQQQPD 173
TS IP + + D
Sbjct: 810 TS---IPPPENNDDDDDGD 825
>At1g61220 unknown protein
Length = 873
Score = 43.5 bits (101), Expect = 2e-04
Identities = 32/138 (23%), Positives = 59/138 (42%), Gaps = 2/138 (1%)
Query: 62 PEPTTNSPHNSTTQKASEVASDATTSETPQHQEYSTLHNLEKHLGGEMQPTPTKASKTVP 121
P +T S T S+V+ TS+ + + + + +M+ + T P
Sbjct: 403 PVTSTRPDRTSATNLTSDVSG--VTSKRQTRTSPAPVMPMILNQTTKMKSDEPSITSTWP 460
Query: 122 EKTVLETQTETQTIPEQTVQEQTASEQVAPDQTTSDQHIPSDQTTEQQQQPDSPTIIDLT 181
++T T + + Q +T+ V P + I SD+ +PD P+ +LT
Sbjct: 461 DRTSATDLTSDVSGVISSRQTRTSPAPVMPMKLNQKTKIKSDEPPITSTRPDRPSATNLT 520
Query: 182 SDQPSISNTTQTEPSPIP 199
SD+ +++T Q + SP P
Sbjct: 521 SDESPVTSTRQAKTSPAP 538
Score = 36.6 bits (83), Expect = 0.020
Identities = 29/120 (24%), Positives = 49/120 (40%), Gaps = 4/120 (3%)
Query: 81 ASDATTSETPQHQEYSTLHNLEKHLGGEMQPTPTKASKTVPEKTVLETQTETQTIPE-QT 139
+ D S T H S E+ L +Q ++ +P K T + T +P T
Sbjct: 320 SEDKAASATVVHSSNSVE---EEPLTASVQTVSMMPTQVMPVKLDQATNSTTVDVPVLST 376
Query: 140 VQEQTASEQVAPDQTTSDQHIPSDQTTEQQQQPDSPTIIDLTSDQPSISNTTQTEPSPIP 199
+ ++ +V P D + +D +PD + +LTSD +++ QT SP P
Sbjct: 377 RRTKSTPVRVMPVVLGRDTSMATDTPPVTSTRPDRTSATNLTSDVSGVTSKRQTRTSPAP 436
>At1g59265 polyprotein, putative
Length = 1466
Score = 42.4 bits (98), Expect = 4e-04
Identities = 31/128 (24%), Positives = 48/128 (37%), Gaps = 19/128 (14%)
Query: 62 PEPTTNSPHNSTTQKASEVASDATTSETPQHQEYSTLHNLEKHLGGEM--QPTPTKASKT 119
P P+ + PH++ T +S A P + NL+ P PT +
Sbjct: 786 PAPSCSDPHHAATPPSSPSA--------PFRNSQVSSSNLDSSFSSSFPSSPEPTAPRQN 837
Query: 120 VPEKTVLETQTETQTIPEQTVQE---------QTASEQVAPDQTTSDQHIPSDQTTEQQQ 170
P+ T TQT+TQT Q + Q A P Q++S P+ +
Sbjct: 838 GPQPTTQPTQTQTQTHSSQNTSQNNPTNESPSQLAQSLSTPAQSSSSSPSPTTSASSSST 897
Query: 171 QPDSPTII 178
P P+I+
Sbjct: 898 SPTPPSIL 905
Score = 34.7 bits (78), Expect = 0.077
Identities = 23/93 (24%), Positives = 39/93 (41%), Gaps = 18/93 (19%)
Query: 46 YPDNPEPVSVQQ--PQPNPEPTTNSPHNSTTQKASEVASDATTSETPQHQEYSTLHNLEK 103
+P +PEP + +Q PQP +PT ++Q S+ + T+E+P S
Sbjct: 825 FPSSPEPTAPRQNGPQPTTQPTQTQTQTHSSQNTSQ---NNPTNESPSQLAQSL------ 875
Query: 104 HLGGEMQPTPTKASKTVPEKTVLETQTETQTIP 136
TP ++S + P T + + T P
Sbjct: 876 -------STPAQSSSSSPSPTTSASSSSTSPTP 901
>At1g58889 polyprotein, putative
Length = 1466
Score = 42.4 bits (98), Expect = 4e-04
Identities = 31/128 (24%), Positives = 48/128 (37%), Gaps = 19/128 (14%)
Query: 62 PEPTTNSPHNSTTQKASEVASDATTSETPQHQEYSTLHNLEKHLGGEM--QPTPTKASKT 119
P P+ + PH++ T +S A P + NL+ P PT +
Sbjct: 786 PAPSCSDPHHAATPPSSPSA--------PFRNSQVSSSNLDSSFSSSFPSSPEPTAPRQN 837
Query: 120 VPEKTVLETQTETQTIPEQTVQE---------QTASEQVAPDQTTSDQHIPSDQTTEQQQ 170
P+ T TQT+TQT Q + Q A P Q++S P+ +
Sbjct: 838 GPQPTTQPTQTQTQTHSSQNTSQNNPTNESPSQLAQSLSTPAQSSSSSPSPTTSASSSST 897
Query: 171 QPDSPTII 178
P P+I+
Sbjct: 898 SPTPPSIL 905
Score = 34.7 bits (78), Expect = 0.077
Identities = 23/93 (24%), Positives = 39/93 (41%), Gaps = 18/93 (19%)
Query: 46 YPDNPEPVSVQQ--PQPNPEPTTNSPHNSTTQKASEVASDATTSETPQHQEYSTLHNLEK 103
+P +PEP + +Q PQP +PT ++Q S+ + T+E+P S
Sbjct: 825 FPSSPEPTAPRQNGPQPTTQPTQTQTQTHSSQNTSQ---NNPTNESPSQLAQSL------ 875
Query: 104 HLGGEMQPTPTKASKTVPEKTVLETQTETQTIP 136
TP ++S + P T + + T P
Sbjct: 876 -------STPAQSSSSSPSPTTSASSSSTSPTP 901
>At5g64540 unknown protein
Length = 440
Score = 42.0 bits (97), Expect = 5e-04
Identities = 42/165 (25%), Positives = 64/165 (38%), Gaps = 44/165 (26%)
Query: 37 FNLPPTHPLYPDNPEPVSVQQPQP-------NPEPTTNSPHNSTTQKASEVASDATTSET 89
+ PP PL P P+P ++ + N PTT+S TT +S++ + TTS++
Sbjct: 244 YQRPPLFPLLPHGPDPYTLDSQKDFPPILSGNQTPTTSSHSVPTTPSSSQL--NTTTSDS 301
Query: 90 PQHQEYSTLHNLEKHLGGEMQPTPTKASKTVPEKTVLETQTETQTIPEQTVQEQTASEQV 149
P H TP + TV L T T T T ++ +
Sbjct: 302 PVH-------------------TPISTTLTVD----LNTSTTTSTHSSSPSVDRLDTVAQ 338
Query: 150 APDQTTSDQHIPSDQTTEQQQQPDSPTIIDLTSDQPSISNTTQTE 194
PD ++S PSD Q+PD + PS T+T+
Sbjct: 339 IPDLSSS----PSDGLETMTQKPDLAS--------PSAGRETETQ 371
>At4g16870 retrotransposon like protein
Length = 1474
Score = 41.2 bits (95), Expect = 8e-04
Identities = 57/252 (22%), Positives = 88/252 (34%), Gaps = 58/252 (23%)
Query: 9 TKLTDLGTLDWEQ------TQIEFSKNRIKLCEKFNLPPTHPLYPDNPEPVSVQQPQPNP 62
T L T+ +EQ T I S + + C H P P S QP
Sbjct: 756 TSQNSLPTVTFEQSSSPLVTPILSSSSVLPSCLSSPCTVLHQQQPPVTTPNSPHSSQPTT 815
Query: 63 EPTTNSPHNSTT---------------QKASEVASDATTS-----------------ETP 90
P SPH STT +S + S+ T P
Sbjct: 816 SPAPLSPHRSTTMDFQVPQVRSSSPLLSSSSSLNSEPTAPNENGPEPEAQSPPIGPLSNP 875
Query: 91 QHQEY-STLHNLEKHLGGEMQPTPTKASKTVPEKTVLETQTETQTIPEQTVQEQTASEQV 149
H+ + L N ++ E++PTP K V T T T P +T + +
Sbjct: 876 THEAFIGPLPNPNRNPTNEIEPTPAPHPKPV-------KPTTTTTTPNRTTVSDASHQPT 928
Query: 150 APDQTTSDQHIPSDQTTEQQQQPDSPTIIDLTSDQPSISNTTQTEPSPIPDHILESEYIE 209
AP Q +QH + ++P+ T LT+ P N + +EP+ + + + ++
Sbjct: 929 APQQ---NQHNMKTRAKNNIKKPN--TKFSLTATLP---NRSPSEPTNVTQALKDKKW-- 978
Query: 210 EQLIRLSDEIQA 221
+SDE A
Sbjct: 979 --RFAMSDEFDA 988
Score = 38.9 bits (89), Expect = 0.004
Identities = 29/105 (27%), Positives = 43/105 (40%), Gaps = 10/105 (9%)
Query: 41 PTH-----PLYPDNPEPVSVQQPQPNPEPTTNSPHNSTTQKASEVASDATTSETPQHQEY 95
PTH PL N P + +P P P P P +TT SDA+ T Q
Sbjct: 875 PTHEAFIGPLPNPNRNPTNEIEPTPAPHPKPVKPTTTTTTPNRTTVSDASHQPTAPQQNQ 934
Query: 96 STLHNLEKHLGGEMQPTPTKAS--KTVPEKTVLETQTETQTIPEQ 138
HN++ ++ TK S T+P ++ E TQ + ++
Sbjct: 935 ---HNMKTRAKNNIKKPNTKFSLTATLPNRSPSEPTNVTQALKDK 976
>At4g01290 hypothetical protein
Length = 736
Score = 40.8 bits (94), Expect = 0.001
Identities = 35/150 (23%), Positives = 64/150 (42%), Gaps = 10/150 (6%)
Query: 61 NPEPTTNSPHNS-TTQKASEVASDATTSETPQHQEYSTLHNLEKHL----GGEMQPTPTK 115
NP+ +T+ P T++ +++S +TT T +LE+ + G P P
Sbjct: 204 NPDLSTDFPFQGHATKRTDQLSSTSTTKSVTAVPPVLTCEDLEQSILSEVGDSYHPPPPP 263
Query: 116 ASKTVPEKTVLETQTETQTIPEQTVQEQTASEQVAPDQTTSDQHIPSDQTTEQQQQPDSP 175
+ +V T+ ++ +Q Q + Q + D + D + S TE++ P S
Sbjct: 264 VDQDTSVPSVKMTKQRKTSVDDQASQHLLSLLQRSSDPKSQDTQLLS--ATERRPPPPS- 320
Query: 176 TIIDLTSDQPSISNTTQTEPSPIPDHILES 205
+ T+ PS+ +TT E P LE+
Sbjct: 321 --MKTTTPPPSVKSTTAGEADPGKSLTLEN 348
>At2g40040 unknown protein
Length = 839
Score = 40.0 bits (92), Expect = 0.002
Identities = 35/155 (22%), Positives = 61/155 (38%), Gaps = 20/155 (12%)
Query: 46 YPDNPEPVSVQQPQPNPEPTTNSPHNSTTQKASEVASDATTSETPQHQEYSTLHNLEKHL 105
YPD E + + P P+ N N+ DAT P +E S N
Sbjct: 698 YPDRAEEF-IDKYFTKPRPSGNRDRNN---------QDAT----PPGEEQSQPPNQSIGN 743
Query: 106 GGE------MQPTPTKASKTVPEKTVLETQTETQTIPEQTVQEQTASEQVAPDQTTSDQH 159
GG+ +P++ P + ++ ++TQ+ + Q Q+ S+ + Q+ S
Sbjct: 744 GGDDFQTQTQSQSPSQTRAQSPSQAQAQSPSQTQSQSQSQSQSQSQSQSQSQSQSQSQSQ 803
Query: 160 IPSDQTTEQQQQPDSPTIIDLTSDQPSISNTTQTE 194
S + Q Q SP+ + PS + +QT+
Sbjct: 804 SQSQSQSPSQTQTQSPSQTQAQAQSPSSQSPSQTQ 838
>At5g56890 unknown protein
Length = 1113
Score = 38.5 bits (88), Expect = 0.005
Identities = 41/171 (23%), Positives = 59/171 (33%), Gaps = 17/171 (9%)
Query: 40 PPTHPLYPDNPEPVSVQQPQPNPEPTTNSPHNS--TTQKASEVASDATTSETPQHQEYST 97
PP HP+ P S P P PT SP + TT + S A +P H
Sbjct: 210 PPVHPVIPKLTPSSS---PVPTSTPTKGSPRRNPPTTHPVFPIESPAV---SPDHAANPV 263
Query: 98 LHNLEKHLGGEMQ-PTPTKASKTVPEKTVLETQTETQTIPEQTVQEQTASEQVAPDQTTS 156
H G + + P A++T V + +I + S +P T
Sbjct: 264 KHPPPSDNGDDSKSPGAAPANETAKPLPVFPHKASPPSIAPSAPKFNRHSHHTSPSTTPP 323
Query: 157 DQHIPSDQTTEQQQQPDSPTIIDLTS----DQPSISNTTQTEPSPIPDHIL 203
PS+ P SP+ L+S Q PSP+P H++
Sbjct: 324 PDSTPSN----VHHHPSSPSPPPLSSHHQHHQERKKIADSPAPSPLPPHLI 370
Score = 28.1 bits (61), Expect = 7.2
Identities = 19/74 (25%), Positives = 29/74 (38%), Gaps = 6/74 (8%)
Query: 40 PPTHPLYPDNPEPVSVQQPQPNPEPTTNSPHNSTTQKASEVASDATTSETPQHQEYSTLH 99
P T P P + P +V +P P S H+ Q+ ++A S P H +
Sbjct: 318 PSTTP--PPDSTPSNVHHHPSSPSPPPLSSHHQHHQERKKIADSPAPSPLPPH----LIS 371
Query: 100 NLEKHLGGEMQPTP 113
+ + G M P P
Sbjct: 372 PKKSNRKGSMTPPP 385
>At1g80020 hypothetical protein
Length = 1076
Score = 38.5 bits (88), Expect = 0.005
Identities = 43/165 (26%), Positives = 67/165 (40%), Gaps = 22/165 (13%)
Query: 59 QPNPEPTTNSPHNSTTQKASEVASDATTSETPQHQEYSTLHNLEKHLGGEMQPTPTKASK 118
Q N EPTT T D T ET +Q + + E L E P ++ S+
Sbjct: 653 QSNGEPTTAQESRLT---------DETMEETHANQSAEEMAH-ETQLVEEF-PCESQPSE 701
Query: 119 TVPEKTVLETQTETQTIPEQTVQEQTASEQVAPDQTTSDQHIPSDQTTEQQQQPDSPTII 178
VP ++ +T+P + + E T + + TS P ++ + QQ D
Sbjct: 702 NVPLQSQSGEGNVPETLPMEEIVEDTQPVEEVMQEETSQVSRPVEEIPLENQQVDDK--- 758
Query: 179 DLTSD-QPSISNTTQTEPSPIPDHILESEYIEEQLIRLSDEIQAL 222
D+T D QP T+P DH E Q + + D+IQ++
Sbjct: 759 DITHDVQPVEEMLEDTQPVEGVDH-------EAQELEVHDDIQSV 796
>At5g53870 predicted GPI-anchored protein
Length = 370
Score = 38.1 bits (87), Expect = 0.007
Identities = 37/165 (22%), Positives = 59/165 (35%), Gaps = 21/165 (12%)
Query: 40 PPTHPLYPDNPEPVSVQQPQPNPEPTTNSPHNSTTQKASEVASDATTSETPQHQEYSTLH 99
P P + + P + +P P ++SP +S S + T S +P H +
Sbjct: 192 PALSPSHATSHSPATPSPSPKSPSPVSHSPSHSPAHTPSHSPAH-TPSHSPAHAPSHSPA 250
Query: 100 NLEKHLGGEM-------QPTPTKASKTVPEKTVLETQTETQTIPEQTVQEQTASEQVAPD 152
+ H P+ + A+ P + Q+ P T Q + +PD
Sbjct: 251 HAPSHSPAHAPSHSPAHSPSHSPATPKSPSPSSSPAQSPATPSP-MTPQSPSPVSSPSPD 309
Query: 153 QTTSDQHIPSDQTTEQQQQPDSPTIIDLTSDQPSISNTTQTEPSP 197
Q+ + PSDQ+T P T P+ N T PSP
Sbjct: 310 QSAA----PSDQSTPLAPSPSETT--------PTADNITAPAPSP 342
>At1g75230 3-methyladenine DNA glycosylase like protein
Length = 391
Score = 37.7 bits (86), Expect = 0.009
Identities = 32/158 (20%), Positives = 62/158 (38%), Gaps = 7/158 (4%)
Query: 40 PPTHPLYPDNPEPVSVQQPQPNPEPTTNSPHNSTTQKASEVASDATTSETPQHQEYSTLH 99
P +H L P+ PE + + P P P P TN ++++ S +TT E PQ E +
Sbjct: 9 PSSHTLPPNQPESPNHETPNPIP-PETNDDDSASSAGVSGSIVSSTTIEAPQVTELGNVS 67
Query: 100 NLEKHLGGEMQPTPTKASKTVPEKTVLETQTETQTIPEQTVQEQTASEQVAPDQTTSDQH 159
+ ++ P K K P+ + + + T + +++ +T +
Sbjct: 68 SPPT----KIPLRPRKIRKLSPDDDASDGFNPEHNLSQMTTTKPATKSKLSQSRTVTVPR 123
Query: 160 IPSDQTTEQQQQPDSPTIIDLTSDQPSISNTTQTEPSP 197
I + T + + + L S P +++ P P
Sbjct: 124 IQARSLTCEGEL--EAALHHLRSVDPLLASLIDIHPPP 159
>At3g18610 unknown protein
Length = 636
Score = 37.4 bits (85), Expect = 0.012
Identities = 28/126 (22%), Positives = 56/126 (44%), Gaps = 20/126 (15%)
Query: 51 EPVSVQQPQPNPEPTTNSPHNSTTQKASEVASDATTSETPQHQEYSTLHNLEKHLGGEMQ 110
E S +P P +PT + A A D+++SE +E S + +
Sbjct: 271 ESSSDDEPTPAKKPTV-------VKNAKPAAKDSSSSEEDSDEEES-----------DDE 312
Query: 111 PTPTKASKTVPEKTVLETQTETQTIPEQTVQEQTASEQVAPDQTTSDQHIPSDQTTEQQQ 170
PTK +K + + E+ ++ + +++ +E++ E+V P + SD + + +
Sbjct: 313 KPPTKKAKVSSKTSKQESSSDESS--DESDKEESKDEKVTPKKKDSDVEMVDAEQKSNAK 370
Query: 171 QPDSPT 176
QP +PT
Sbjct: 371 QPKTPT 376
Score = 28.5 bits (62), Expect = 5.5
Identities = 22/103 (21%), Positives = 47/103 (45%), Gaps = 14/103 (13%)
Query: 115 KASKTVPEK--------TVLETQTETQTIPEQTVQEQTASEQVAPDQTTSDQHIPSDQTT 166
KA KTVP+K + + + +T+ P + E ++ E+ D ++SD+ I
Sbjct: 53 KAEKTVPKKVESSSSDASDSDEEEKTKETPSKLKDESSSEEE---DDSSSDEEI---APA 106
Query: 167 EQQQQPDSPTIIDLTSDQPSISNTTQTEPSPIPDHILESEYIE 209
+++ +P ++ +S ++ +T P +LE +E
Sbjct: 107 KKRPEPIKKAKVESSSSDDDSTSDEETAPVKKQPAVLEKAKVE 149
>At2g03810 unknown protein
Length = 439
Score = 37.4 bits (85), Expect = 0.012
Identities = 35/162 (21%), Positives = 62/162 (37%), Gaps = 25/162 (15%)
Query: 18 DWEQTQIEFSKNRIKLCEKFNLPPTHPLYPDNPEP-------VSVQQPQPNPEPTTNSPH 70
D E T +++ + + E+ PTH L P EP V++ Q + E T
Sbjct: 194 DAEGTSSNYNQEHLIVTEEVKASPTHGLSPSEIEPDENSKDEVAISQDNDSKECLTLGDI 253
Query: 71 NSTTQKASEVASDATTSETPQHQEYSTLHNLEKHLGGEMQPTPTKASKTVPEKTVLETQT 130
S + + D +S++ + Q S L + EK E T +ET+
Sbjct: 254 LSREDEQKSLNQDNISSDSHEEQSPSQLQDKEKRS---------------LETTAIETEL 298
Query: 131 ETQTIP---EQTVQEQTASEQVAPDQTTSDQHIPSDQTTEQQ 169
E P E+ + + + P++T ++ P + QQ
Sbjct: 299 EKTEEPKQGEEKLSSVSTTTSQEPNKTCNEPEKPETENHHQQ 340
>At4g33500 unknown protein
Length = 724
Score = 36.6 bits (83), Expect = 0.020
Identities = 39/173 (22%), Positives = 69/173 (39%), Gaps = 16/173 (9%)
Query: 88 ETPQHQEYSTLHNLEKHLGGEMQPTPTKASKTVPEKTVLETQTETQTIPEQTVQEQTASE 147
ET +QE L + + TP + V E E + E V E TA++
Sbjct: 244 ETNGYQENRMEVQARPSLSTQQEITPVSTIEIDDNLDVTEKPIEAE---ENLVAEPTATD 300
Query: 148 QVAPDQTTSDQHIPSDQTTEQQQQPDSPTIIDLTSDQPSISNTTQTEPSPIPDHILESEY 207
++PD+ S E Q+P +ID + + ++ NT + E +P+ + +
Sbjct: 301 DLSPDELLSTSEATHRSVDEIAQKP----VID--TSEENLLNTFEAEENPVVEPTATAAV 354
Query: 208 IEEQLIRLS-------DEIQALILRRTVPAPPIHYYDQWMDLQKSFDELLDQL 253
++LI S DEI + T P+ + + + S DE ++L
Sbjct: 355 SSDELISTSEATRHSVDEIAQKPIIDTSEKNPMETFVEPEAVHSSVDESTEKL 407
>At4g32760 unknown protein
Length = 675
Score = 36.6 bits (83), Expect = 0.020
Identities = 41/164 (25%), Positives = 63/164 (38%), Gaps = 28/164 (17%)
Query: 44 PLYPDNPEPVSVQQPQPNPEPTTNSPHNSTTQKASEVASDATTSETPQHQEYSTLHNLEK 103
P+ +NP P Q PQ P NSP+ Q VA+ + + PQ+ Y + +
Sbjct: 498 PVNNNNPYP---QIPQTGPPVNNNSPYAQMPQTGQAVANISPYPQIPQNGVYMP-NQPNQ 553
Query: 104 HLGGEMQPTPTKASKTVPEKTVLETQTETQTIPEQTVQEQTASEQVAPDQTTSDQHIPSD 163
LG QP + Q + Q + Q +Q +Q Q +Q
Sbjct: 554 ALGSGYQP---------------QQQQQQQMMMAQYYAQQQQLQQQQQQQAYGNQMGGYG 598
Query: 164 QTTEQQQQPDSP---------TIIDLTSDQPSISNTTQTEPSPI 198
QQQQ SP ++ D TS Q + S++T + P+
Sbjct: 599 YGYNQQQQGSSPYLDQQMYGLSMRDQTSHQVASSSSTTSYLPPM 642
>At1g17440 unknown protein
Length = 683
Score = 36.6 bits (83), Expect = 0.020
Identities = 45/187 (24%), Positives = 76/187 (40%), Gaps = 30/187 (16%)
Query: 44 PLYPDNPEPVSVQQP---QPNPEPTTNSPHNSTTQK-------ASEVASDATTSETPQHQ 93
P+ + P S+Q P +P+P +T P +S+ Q+ ++ V S A+ + T
Sbjct: 4 PIPSSSLSPKSLQSPNPMEPSPASSTPLPSSSSQQQQLMTAPISNSVNSAASPAMTVTTT 63
Query: 94 EYSTLHNLEKHLGGEMQPT----------PTKASKTVPEKTVLETQTETQTIPEQT---- 139
E + N + PT P+ + + P + L+ QT+TQ + +QT
Sbjct: 64 EGIVIQNNSQPNISSPNPTSSNPPIGAQIPSPSPLSHPSSS-LDQQTQTQQLVQQTQQLP 122
Query: 140 VQEQTASEQVA----PDQTTSDQHIPSDQTTEQQQQP-DSPTIIDLTSDQPSISNTTQTE 194
Q+Q +Q++ P + Q I Q QQ P S I PS+S +Q +
Sbjct: 123 QQQQQIMQQISSSPIPQLSPQQQQILQQQHMTSQQIPMSSYQIAQSLQRSPSLSRLSQIQ 182
Query: 195 PSPIPDH 201
H
Sbjct: 183 QQQQQQH 189
>At4g07640 putative athila transposon protein
Length = 866
Score = 36.2 bits (82), Expect = 0.026
Identities = 34/144 (23%), Positives = 58/144 (39%), Gaps = 27/144 (18%)
Query: 66 TNSPHNSTTQKASEVASDATTSETPQHQE--------YSTLHNLEKHLGGEMQPTPTKAS 117
TN+P+ S +++ +A+ PQ Q+ Y+ ++ G QP P
Sbjct: 299 TNNPNLS--YRSTNIANPQDQVYPPQQQQVQNKPFVPYNQGFVPKQQFQGNYQPPPPPGG 356
Query: 118 KTVPEKTVLETQTETQTIPEQTVQEQTASEQVAPDQTTSDQHIPSDQTTEQQQQPDSPTI 177
K +P T+ E +T+ E + + + DQ P DQ+ EQ
Sbjct: 357 KALP------TREEPKTVTEDSEDQDGEDLSLEKDQADKPHEQPLDQSLEQP-------- 402
Query: 178 IDLTSDQP---SISNTTQTEPSPI 198
+DL+ +QP + N T+ PI
Sbjct: 403 LDLSLEQPLDLPLDNVTRPTTRPI 426
>At1g68690 protein kinase, putative
Length = 708
Score = 36.2 bits (82), Expect = 0.026
Identities = 37/164 (22%), Positives = 54/164 (32%), Gaps = 8/164 (4%)
Query: 40 PPTHPLYPDNPEPVSVQQPQPNPEPTTNSPHNSTTQKASEVASDATTSETPQHQEYSTLH 99
PP P P P + P P+P SP ST+ V S +P L
Sbjct: 61 PPVANGAPPPPLPKPPESSSPPPQPVIPSPPPSTSPPPQPVIPSPPPSASPPPALVPPLP 120
Query: 100 NLEKHLGGEMQPTPTKASKTV---PEKTVLETQTETQTIPEQTVQEQTASEQVAPDQTTS 156
+ P P+ + + P +V Q+ ++ Q +P
Sbjct: 121 SSPPPPASVPPPRPSPSPPILVRSPPPSVRPIQSPPPPPSDRPTQSPPPPSPPSPPSERP 180
Query: 157 DQHIPSDQTTEQQQQPDSPTIIDLTSDQPSISNTTQTEPSPIPD 200
Q PS + Q P P+ SD+PS Q+ P P D
Sbjct: 181 TQSPPSPPSERPTQSPPPPSPPSPPSDRPS-----QSPPPPPED 219
Score = 28.9 bits (63), Expect = 4.2
Identities = 39/168 (23%), Positives = 55/168 (32%), Gaps = 19/168 (11%)
Query: 39 LPPTHPLYPDNPEPVSVQQPQPNPEPTTNSPHNST--TQKASEVASDATTSETPQHQEYS 96
L P P P P V +P P+P SP S Q SD T P S
Sbjct: 115 LVPPLPSSPPPPASVPPPRPSPSPPILVRSPPPSVRPIQSPPPPPSDRPTQSPPPPSPPS 174
Query: 97 TLHNLEKHLGGEMQPTPTKASKTVPEKTVLETQTETQTIPEQTVQEQTASEQVAPDQTTS 156
PT++ + P + TQ+ P ++ + P + T
Sbjct: 175 -----------PPSERPTQSPPSPPSER--PTQSPPPPSPPSPPSDRPSQSPPPPPEDTK 221
Query: 157 DQ---HIPSDQTTEQQQQPDS-PTIIDLTSDQPSISNTTQTEPSPIPD 200
Q P+ P S P I+ S+ PS +N T P P+
Sbjct: 222 PQPPRRSPNSPPPTFSSPPRSPPEILVPGSNNPSQNNPTLRPPLDAPN 269
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.308 0.126 0.352
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,006,866
Number of Sequences: 26719
Number of extensions: 370789
Number of successful extensions: 2609
Number of sequences better than 10.0: 232
Number of HSP's better than 10.0 without gapping: 40
Number of HSP's successfully gapped in prelim test: 196
Number of HSP's that attempted gapping in prelim test: 2128
Number of HSP's gapped (non-prelim): 518
length of query: 340
length of database: 11,318,596
effective HSP length: 100
effective length of query: 240
effective length of database: 8,646,696
effective search space: 2075207040
effective search space used: 2075207040
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC146552.4


