

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146552.2 - phase: 0 /pseudo
(392 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
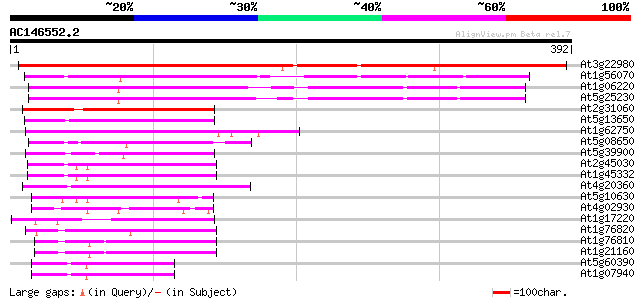
Score E
Sequences producing significant alignments: (bits) Value
At3g22980 eukaryotic translation elongation factor 2, putative 543 e-155
At1g56070 elongation factor like protein 220 9e-58
At1g06220 elongation factor like protein 165 3e-41
At5g25230 translation Elongation Factor 2 - like protein 158 4e-39
At2g31060 putative GTP-binding protein 106 3e-23
At5g13650 GTP-binding protein typA (tyrosine phosphorylated prot... 103 2e-22
At1g62750 elongation factor G, putative 101 6e-22
At5g08650 GTP-binding protein LepA homolog 97 2e-20
At5g39900 GTP-binding protein-like 91 1e-18
At2g45030 putative mitochondrial translation elongation factor G 86 3e-17
At1g45332 mitochondrial elongation factor, putative 86 3e-17
At4g20360 translation elongation factor EF-Tu precursor, chlorop... 69 4e-12
At5g10630 putative protein 60 2e-09
At4g02930 mitochondrial elongation factor Tu 57 2e-08
At1g17220 putative translation initiation factor IF2 53 3e-07
At1g76820 putative translation initiation factor IF-2 49 6e-06
At1g76810 translation initiation factor IF-2 like protein 49 6e-06
At1g21160 transcription factor, putative 49 6e-06
At5g60390 translation elongation factor eEF-1 alpha chain (gene A4) 47 2e-05
At1g07940 elongation factor 1-alpha 47 2e-05
>At3g22980 eukaryotic translation elongation factor 2, putative
Length = 1015
Score = 543 bits (1399), Expect = e-155
Identities = 282/390 (72%), Positives = 324/390 (82%), Gaps = 11/390 (2%)
Query: 7 DNNDRKKIRNICILAHVDHGKTTLADQLIAAASGGMVHPKVAGKVRFMDYLDEEQRRAIT 66
D ++ +K+RNICILAHVDHGKTTLAD LIA++ GG++HP++AGK+RFMDYLDEEQRRAIT
Sbjct: 2 DESEGRKVRNICILAHVDHGKTTLADHLIASSGGGVLHPRLAGKLRFMDYLDEEQRRAIT 61
Query: 67 MKSSSISLHYNHHTVNLIDSPGHIDFCGEVSTAARLSDGALLLVDAVEGVHIQTHAVLRQ 126
MKSSSISL Y +++NLIDSPGH+DFC EVSTAARLSDGAL+LVDAVEGVHIQTHAVLRQ
Sbjct: 62 MKSSSISLKYKDYSLNLIDSPGHMDFCSEVSTAARLSDGALVLVDAVEGVHIQTHAVLRQ 121
Query: 127 CWTEMLEPCLVLNKMDRLITELNLTPLEAYTRLLRIVHEVNGIWSAYNSEKYLSDVDALL 186
W E L PCLVLNK+DRLI EL L+P+EAYTRL+RIVHEVNGI SAY SEKYLSDVD++L
Sbjct: 122 AWIEKLTPCLVLNKIDRLIFELRLSPMEAYTRLIRIVHEVNGIVSAYKSEKYLSDVDSIL 181
Query: 187 AGG----TAAGGEVMEDYDDVEDKFQPQKGNVVFACALDGWGFGIHEFAEIYASKLGGSA 242
A +A E++E DD E FQPQKGNVVF CALDGWGFGI EFA YASKLG SA
Sbjct: 182 ASPSGELSAESLELLE--DDEEVTFQPQKGNVVFVCALDGWGFGIAEFANFYASKLGASA 239
Query: 243 SVGALLRALWGPWYYNPKTKMIVGKKGIS-GSKARPMFVQFVLEPLWQVYQGAL--GGGK 299
+ AL ++LWGP YY PKTKMIVGKK +S GSKA+PMFVQFVLEPLWQVY+ AL GG K
Sbjct: 240 T--ALQKSLWGPRYYIPKTKMIVGKKNLSAGSKAKPMFVQFVLEPLWQVYEAALDPGGDK 297
Query: 300 GMVEKVIKSFNLQIQARELQNKDSKVVLQAVMSRWLPLSDAILSMVLKCLPDPVEGQKSR 359
++EKVIKSFNL I RELQNKD K VLQ+VMSRWLPLSDA+LSM +K LPDP+ Q R
Sbjct: 298 AVLEKVIKSFNLSIPPRELQNKDPKNVLQSVMSRWLPLSDAVLSMAVKHLPDPIAAQAYR 357
Query: 360 ISRLIPERKVGSENGVDRRVVEESELVRKS 389
I RL+PERK+ + VD V+ E+ELVRKS
Sbjct: 358 IPRLVPERKIIGGDDVDSSVLAEAELVRKS 387
>At1g56070 elongation factor like protein
Length = 843
Score = 220 bits (561), Expect = 9e-58
Identities = 135/369 (36%), Positives = 196/369 (52%), Gaps = 46/369 (12%)
Query: 11 RKKIRNICILAHVDHGKTTLADQLIAAASGGMVHPKVAGKVRFMDYLDEEQRRAITMKSS 70
+ IRN+ ++AHVDHGK+TL D L+AAA G++ +VAG VR D +E R IT+KS+
Sbjct: 16 KHNIRNMSVIAHVDHGKSTLTDSLVAAA--GIIAQEVAGDVRMTDTRADEAERGITIKST 73
Query: 71 SISLHY----------------NHHTVNLIDSPGHIDFCGEVSTAARLSDGALLLVDAVE 114
ISL+Y N + +NLIDSPGH+DF EV+ A R++DGAL++VD +E
Sbjct: 74 GISLYYEMTDESLKSFTGARDGNEYLINLIDSPGHVDFSSEVTAALRITDGALVVVDCIE 133
Query: 115 GVHIQTHAVLRQCWTEMLEPCLVLNKMDRLITELNLTPLEAYTRLLRIVHEVNGIWSAYN 174
GV +QT VLRQ E + P L +NKMDR EL + EAY R++ N I + Y
Sbjct: 134 GVCVQTETVLRQALGERIRPVLTVNKMDRCFLELQVDGEEAYQTFSRVIENANVIMATY- 192
Query: 175 SEKYLSDVDALLAGGTAAGGEVMEDYDDVEDKFQPQKGNVVFACALDGWGFGIHEFAEIY 234
+ L DV + P+KG V F+ L GW F + FA++Y
Sbjct: 193 EDPLLGDV-----------------------QVYPEKGTVAFSAGLHGWAFTLTNFAKMY 229
Query: 235 ASKLGGSASVGALLRALWGPWYYNPKTKMIVGKKGISGSKARPMFVQFVLEPLWQVYQGA 294
ASK G S ++ LWG +++P T+ GK S + R FVQF EP+ Q+
Sbjct: 230 ASKFGVVES--KMMERLWGENFFDPATRKWSGKNTGSPTCKRG-FVQFCYEPIKQIIATC 286
Query: 295 LGGGKGMVEKVIKSFNLQIQARELQNKDSKVVLQAVMSRWLPLSDAILSMVLKCLPDPVE 354
+ K + ++ + ++ E + K +++ VM WLP S A+L M++ LP P
Sbjct: 287 MNDQKDKLWPMLAKLGVSMKNDE-KELMGKPLMKRVMQTWLPASTALLEMMIFHLPSPHT 345
Query: 355 GQKSRISRL 363
Q+ R+ L
Sbjct: 346 AQRYRVENL 354
>At1g06220 elongation factor like protein
Length = 987
Score = 165 bits (418), Expect = 3e-41
Identities = 103/353 (29%), Positives = 171/353 (48%), Gaps = 33/353 (9%)
Query: 14 IRNICILAHVDHGKTTLADQLIAAASG-GMVHPKVAGKVRFMDYLDEEQRRAITMKSSSI 72
+RN+ ++ H+ HGKT D L+ + K +++ D +EQ R I++K+ +
Sbjct: 138 VRNVALVGHLQHGKTVFMDMLVEQTHHMSTFNAKNEKHMKYTDTRVDEQERNISIKAVPM 197
Query: 73 SL-----HYNHHTVNLIDSPGHIDFCGEVSTAARLSDGALLLVDAVEGVHIQTHAVLRQC 127
SL + N++D+PGH++F E++ + RL+DGA+L+VDA EGV + T +R
Sbjct: 198 SLVLEDSRSKSYLCNIMDTPGHVNFSDEMTASLRLADGAVLIVDAAEGVMVNTERAIRHA 257
Query: 128 WTEMLEPCLVLNKMDRLITELNLTPLEAYTRLLRIVHEVNGIWSAYNSEKYLSDVDALLA 187
+ L +V+NK+DRLITEL L P +AY +L + +N + + A
Sbjct: 258 IQDHLPIVVVINKVDRLITELKLPPRDAYYKLRHTIEVIN---------------NHISA 302
Query: 188 GGTAAGGEVMEDYDDVEDKFQPQKGNVVFACALDGWGFGIHEFAEIYASKLGGSASVGAL 247
T AG + D P GNV FA GW F + FA++YA G + V
Sbjct: 303 ASTTAGDLPLID---------PAAGNVCFASGTAGWSFTLQSFAKMYAKLHGVAMDVDKF 353
Query: 248 LRALWGPWYYNPKTKMIVGKKGISGSKARPMFVQFVLEPLWQVYQGALGGGKGMVEKVIK 307
LWG YY+ T++ + G + FVQF+LEPL+++Y +G K VE +
Sbjct: 354 ASRLWGDVYYHSDTRVFKRSPPVGGGER--AFVQFILEPLYKIYSQVIGEHKKSVETTLA 411
Query: 308 SFNLQIQARELQNKDSKVVLQAVMSRWLPLSDAILSMVLKCLPDPVEGQKSRI 360
+ + + + + +L+ S + M++K +P P E ++
Sbjct: 412 ELGVTL-SNSAYKLNVRPLLRLACSSVFGSASGFTDMLVKHIPSPREAAARKV 463
>At5g25230 translation Elongation Factor 2 - like protein
Length = 973
Score = 158 bits (400), Expect = 4e-39
Identities = 102/353 (28%), Positives = 169/353 (46%), Gaps = 33/353 (9%)
Query: 14 IRNICILAHVDHGKTTLADQLIAAASGGMVHPKVAGK-VRFMDYLDEEQRRAITMKSSSI 72
+RN+ ++ H+ HGKT D L+ K +R+ D +EQ R I++K+ +
Sbjct: 124 VRNVALVGHLQHGKTVFMDMLVEQTHRMSTFNAENDKHMRYTDTRVDEQERNISIKAVPM 183
Query: 73 SL-----HYNHHTVNLIDSPGHIDFCGEVSTAARLSDGALLLVDAVEGVHIQTHAVLRQC 127
SL + N++D+PG+++F E++ + RL+DGA+ +VDA +GV + T +R
Sbjct: 184 SLVLEDSRSKSYLCNIMDTPGNVNFSDEMTASLRLADGAVFIVDAAQGVMVNTERAIRHA 243
Query: 128 WTEMLEPCLVLNKMDRLITELNLTPLEAYTRLLRIVHEVNGIWSAYNSEKYLSDVDALLA 187
+ L +V+NK+DRLITEL L P +AY +L + +N SA A
Sbjct: 244 IQDHLPIVVVINKVDRLITELKLPPRDAYYKLRYTIEVINNHISA--------------A 289
Query: 188 GGTAAGGEVMEDYDDVEDKFQPQKGNVVFACALDGWGFGIHEFAEIYASKLGGSASVGAL 247
AA +++ P GNV FA GW F + FA +YA G + V
Sbjct: 290 STNAADLPLID----------PAAGNVCFASGTAGWSFTLQSFARMYAKLHGVAMDVDKF 339
Query: 248 LRALWGPWYYNPKTKMIVGKKGISGSKARPMFVQFVLEPLWQVYQGALGGGKGMVEKVIK 307
LWG YY+P T++ + G + FVQF+LEPL+++Y +G K VE +
Sbjct: 340 ASRLWGDVYYHPDTRVFNTSPPVGGGER--AFVQFILEPLYKIYSQVIGEHKKSVETTLA 397
Query: 308 SFNLQIQARELQNKDSKVVLQAVMSRWLPLSDAILSMVLKCLPDPVEGQKSRI 360
+ + + + + +L+ S + M++K +P P E ++
Sbjct: 398 ELGVTL-SNSAYKLNVRPLLRLACSSVFGSASGFTDMLVKHIPSPREAAARKV 449
>At2g31060 putative GTP-binding protein
Length = 664
Score = 106 bits (264), Expect = 3e-23
Identities = 55/134 (41%), Positives = 81/134 (60%), Gaps = 6/134 (4%)
Query: 10 DRKKIRNICILAHVDHGKTTLADQLIAAASGGMVHPKVAGKVRFMDYLDEEQRRAITMKS 69
D ++RN+ ++AHVDHGKTTL D+L+ + H R MD ++ E+ R IT+ S
Sbjct: 55 DPNRLRNVAVIAHVDHGKTTLMDRLLRQCGADIPHE------RAMDSINLERERGITISS 108
Query: 70 SSISLHYNHHTVNLIDSPGHIDFCGEVSTAARLSDGALLLVDAVEGVHIQTHAVLRQCWT 129
S+ + + +N++D+PGH DF GEV + +GA+L+VDA EG QT VL +
Sbjct: 109 KVTSIFWKDNELNMVDTPGHADFGGEVERVVGMVEGAILVVDAGEGPLAQTKFVLAKALK 168
Query: 130 EMLEPCLVLNKMDR 143
L P L+LNK+DR
Sbjct: 169 YGLRPILLLNKVDR 182
>At5g13650 GTP-binding protein typA (tyrosine phosphorylated protein
A)
Length = 609
Score = 103 bits (256), Expect = 2e-22
Identities = 58/133 (43%), Positives = 79/133 (58%), Gaps = 2/133 (1%)
Query: 11 RKKIRNICILAHVDHGKTTLADQLIAAASGGMVHPKVAGKVRFMDYLDEEQRRAITMKSS 70
R +RNI I+AHVDHGKTTL D ++ A + + R MD D E+ R IT+ S
Sbjct: 13 RDNVRNIAIVAHVDHGKTTLVDSMLRQAK--VFRDNQVMQERIMDSNDLERERGITILSK 70
Query: 71 SISLHYNHHTVNLIDSPGHIDFCGEVSTAARLSDGALLLVDAVEGVHIQTHAVLRQCWTE 130
+ S+ Y + VN+ID+PGH DF GEV + DG LL+VD+VEG QT VL++
Sbjct: 71 NTSITYKNTKVNIIDTPGHSDFGGEVERVLNMVDGVLLVVDSVEGPMPQTRFVLKKALEF 130
Query: 131 MLEPCLVLNKMDR 143
+V+NK+DR
Sbjct: 131 GHAVVVVVNKIDR 143
>At1g62750 elongation factor G, putative
Length = 783
Score = 101 bits (252), Expect = 6e-22
Identities = 68/217 (31%), Positives = 108/217 (49%), Gaps = 26/217 (11%)
Query: 12 KKIRNICILAHVDHGKTTLADQLIAAASGGMVHPKVAGKVRFMDYLDEEQRRAITMKSSS 71
K RNI I+AH+D GKTT ++++ +V MD++++EQ R IT+ S++
Sbjct: 94 KDYRNIGIMAHIDAGKTTTTERILYYTGRNYKIGEVHEGTATMDWMEQEQERGITITSAA 153
Query: 72 ISLHYNHHTVNLIDSPGHIDFCGEVSTAARLSDGALLLVDAVEGVHIQTHAVLRQCWTEM 131
+ ++ H +N+ID+PGH+DF EV A R+ DGA+ L D+V GV Q+ V RQ
Sbjct: 154 TTTFWDKHRINIIDTPGHVDFTLEVERALRVLDGAICLFDSVAGVEPQSETVWRQADKYG 213
Query: 132 LEPCLVLNKMDRL-----------ITELNLTPL---------EAYTRLLRIVHEVNGIWS 171
+ +NKMDRL +T L PL + + ++ +V +WS
Sbjct: 214 VPRICFVNKMDRLGANFFRTRDMIVTNLGAKPLVLQIPIGAEDVFKGVVDLVRMKAIVWS 273
Query: 172 A------YNSEKYLSDVDALLAGGTAAGGEVMEDYDD 202
++ E D++ L AA E++ D DD
Sbjct: 274 GEELGAKFSYEDIPEDLEDLAQEYRAAMMELIVDLDD 310
>At5g08650 GTP-binding protein LepA homolog
Length = 675
Score = 97.1 bits (240), Expect = 2e-20
Identities = 64/160 (40%), Positives = 91/160 (56%), Gaps = 15/160 (9%)
Query: 14 IRNICILAHVDHGKTTLADQLIAAASGGMVHPKVAGKVRFMDYLDEEQRRAITMKSSSIS 73
IRN I+AH+DHGK+TLAD+L+ G V + K +F+D +D E+ R IT+K +
Sbjct: 80 IRNFSIIAHIDHGKSTLADKLLQVT--GTVQNRDM-KEQFLDNMDLERERGITIKLQAAR 136
Query: 74 LHYNHHT----VNLIDSPGHIDFCGEVSTAARLSDGALLLVDAVEGVHIQTHAVLRQCWT 129
+ Y + +NLID+PGH+DF EVS + +GALL+VDA +GV QT A +
Sbjct: 137 MRYVYEDTPFCLNLIDTPGHVDFSYEVSRSLAACEGALLVVDASQGVEAQTLANVYLALE 196
Query: 130 EMLEPCLVLNKMDRLITELNLTPLEAYTRLLRIVHEVNGI 169
LE VLNK+D P ++LR + EV G+
Sbjct: 197 NNLEIIPVLNKID--------LPGAEPEKVLREIEEVIGL 228
>At5g39900 GTP-binding protein-like
Length = 661
Score = 90.9 bits (224), Expect = 1e-18
Identities = 55/141 (39%), Positives = 82/141 (58%), Gaps = 14/141 (9%)
Query: 12 KKIRNICILAHVDHGKTTLADQLIAAASGGMVHPKVAGKVRFMDYLDEEQRRAITMKSSS 71
+KIRN I+AH+DHGK+TLAD+L+ K G+ +++D L E R IT+K+ +
Sbjct: 64 EKIRNFSIIAHIDHGKSTLADRLMELTG---TIKKGHGQPQYLDKLQRE--RGITVKAQT 118
Query: 72 ISLHYNH---------HTVNLIDSPGHIDFCGEVSTAARLSDGALLLVDAVEGVHIQTHA 122
++ Y + + +NLID+PGH+DF EVS + GALL+VDA +GV QT A
Sbjct: 119 ATMFYENKVEDQEASGYLLNLIDTPGHVDFSYEVSRSLSACQGALLVVDAAQGVQAQTVA 178
Query: 123 VLRQCWTEMLEPCLVLNKMDR 143
+ L V+NK+D+
Sbjct: 179 NFYLAFEANLTIVPVINKIDQ 199
>At2g45030 putative mitochondrial translation elongation factor G
Length = 754
Score = 86.3 bits (212), Expect = 3e-17
Identities = 52/137 (37%), Positives = 83/137 (59%), Gaps = 7/137 (5%)
Query: 13 KIRNICILAHVDHGKTTLADQLIAAASGGMVHP--KVAGKVRF---MDYLDEEQRRAITM 67
K+RNI I AH+D GKTTL ++++ G +H +V G+ MD +D E+ + IT+
Sbjct: 64 KLRNIGISAHIDSGKTTLTERVLFYT--GRIHEIHEVRGRDGVGAKMDSMDLEREKGITI 121
Query: 68 KSSSISLHYNHHTVNLIDSPGHIDFCGEVSTAARLSDGALLLVDAVEGVHIQTHAVLRQC 127
+S++ + + VN+ID+PGH+DF EV A R+ DGA+L++ +V GV Q+ V RQ
Sbjct: 122 QSAATYCTWKDYKVNIIDTPGHVDFTIEVERALRVLDGAILVLCSVGGVQSQSITVDRQM 181
Query: 128 WTEMLEPCLVLNKMDRL 144
+ +NK+DR+
Sbjct: 182 RRYEVPRVAFINKLDRM 198
>At1g45332 mitochondrial elongation factor, putative
Length = 754
Score = 86.3 bits (212), Expect = 3e-17
Identities = 52/137 (37%), Positives = 83/137 (59%), Gaps = 7/137 (5%)
Query: 13 KIRNICILAHVDHGKTTLADQLIAAASGGMVHP--KVAGKVRF---MDYLDEEQRRAITM 67
K+RNI I AH+D GKTTL ++++ G +H +V G+ MD +D E+ + IT+
Sbjct: 64 KLRNIGISAHIDSGKTTLTERVLFYT--GRIHEIHEVRGRDGVGAKMDSMDLEREKGITI 121
Query: 68 KSSSISLHYNHHTVNLIDSPGHIDFCGEVSTAARLSDGALLLVDAVEGVHIQTHAVLRQC 127
+S++ + + VN+ID+PGH+DF EV A R+ DGA+L++ +V GV Q+ V RQ
Sbjct: 122 QSAATYCTWKDYKVNIIDTPGHVDFTIEVERALRVLDGAILVLCSVGGVQSQSITVDRQM 181
Query: 128 WTEMLEPCLVLNKMDRL 144
+ +NK+DR+
Sbjct: 182 RRYEVPRVAFINKLDRM 198
>At4g20360 translation elongation factor EF-Tu precursor,
chloroplast
Length = 476
Score = 68.9 bits (167), Expect = 4e-12
Identities = 52/161 (32%), Positives = 78/161 (48%), Gaps = 4/161 (2%)
Query: 10 DRKKIR-NICILAHVDHGKTTLADQLIAAASGGMVHPKVAGKVRFMDYLDEEQRRAITMK 68
+RKK NI + HVDHGKTTL L A + + VA K +D EE+ R IT+
Sbjct: 74 ERKKPHVNIGTIGHVDHGKTTLTAALTMALAS--IGSSVAKKYDEIDAAPEERARGITIN 131
Query: 69 SSSISLHYNHHTVNLIDSPGHIDFCGEVSTAARLSDGALLLVDAVEGVHIQT-HAVLRQC 127
++++ + +D PGH D+ + T A DGA+L+V +G QT +L
Sbjct: 132 TATVEYETENRHYAHVDCPGHADYVKNMITGAAQMDGAILVVSGADGPMPQTKEHILLAK 191
Query: 128 WTEMLEPCLVLNKMDRLITELNLTPLEAYTRLLRIVHEVNG 168
+ + + LNK D++ L +E R L +E NG
Sbjct: 192 QVGVPDMVVFLNKEDQVDDAELLELVELEVRELLSSYEFNG 232
>At5g10630 putative protein
Length = 804
Score = 60.5 bits (145), Expect = 2e-09
Identities = 50/151 (33%), Positives = 72/151 (47%), Gaps = 26/151 (17%)
Query: 16 NICILAHVDHGKTTLADQLI---AAASGGMVHP-----KVAGKVRF-----MDYLDEEQR 62
N+ I+ HVD GK+TL+ +L+ S +H K+ GK F +D EE+
Sbjct: 378 NLAIVGHVDSGKSTLSGRLLHLLGRISQKQMHKYEKEAKLQGKGSFAYAWALDESAEERE 437
Query: 63 RAITMKSSSISLHYNHHTVNLIDSPGHIDFCGEVSTAARLSDGALLLVDAVEGV------ 116
R ITM + + H V L+DSPGH DF + A +D A+L++DA G
Sbjct: 438 RGITMTVAVAYFNSKRHHVVLLDSPGHKDFVPNMIAGATQADAAILVIDASVGAFEAGFD 497
Query: 117 ----HIQTHA-VLRQCWTEMLEPCLVLNKMD 142
+ HA VLR E + + +NKMD
Sbjct: 498 NLKGQTREHARVLRGFGVEQV--IVAINKMD 526
>At4g02930 mitochondrial elongation factor Tu
Length = 454
Score = 57.0 bits (136), Expect = 2e-08
Identities = 46/138 (33%), Positives = 69/138 (49%), Gaps = 23/138 (16%)
Query: 16 NICILAHVDHGKTTLADQLIAAASGGMVHPKVAGKVRF--MDYLDEEQRRAITMKSSSIS 73
N+ + HVDHGKTTL AA + + A + F +D EE++R IT+ ++ +
Sbjct: 69 NVGTIGHVDHGKTTLT----AAITKVLAEEGKAKAIAFDEIDKAPEEKKRGITIATAHVE 124
Query: 74 L-----HYNHHTVNLIDSPGHIDFCGEVSTAARLSDGALLLVDAVEGVHIQT--HAVLRQ 126
HY H +D PGH D+ + T A DG +L+V +G QT H +L +
Sbjct: 125 YETAKRHYAH-----VDCPGHADYVKNMITGAAQMDGGILVVSGPDGPMPQTKEHILLAR 179
Query: 127 CWTEMLEPCLV--LNKMD 142
++ P LV LNK+D
Sbjct: 180 ---QVGVPSLVCFLNKVD 194
>At1g17220 putative translation initiation factor IF2
Length = 1026
Score = 52.8 bits (125), Expect = 3e-07
Identities = 40/151 (26%), Positives = 69/151 (45%), Gaps = 28/151 (18%)
Query: 2 EESTSDNNDRKKIRN----ICILAHVDHGKTTLAD-----QLIAAASGGMVHPKVAGKVR 52
+ T D D K+ + I I+ HVDHGKTTL D ++ A+ +GG+ A KV
Sbjct: 485 KRQTFDEEDLDKLEDRPPVITIMGHVDHGKTTLLDYIRKSKVAASEAGGITQGIGAYKV- 543
Query: 53 FMDYLDEEQRRAITMKSSSISLHYNHHTVNLIDSPGHIDFCGEVSTAARLSDGALLLVDA 112
S+ + + +D+PGH F + AR++D A+++V A
Sbjct: 544 ------------------SVPVDGKLQSCVFLDTPGHEAFGAMRARGARVTDIAIIVVAA 585
Query: 113 VEGVHIQTHAVLRQCWTEMLEPCLVLNKMDR 143
+G+ QT+ + + + +NK+D+
Sbjct: 586 DDGIRPQTNEAIAHAKAAAVPIVIAINKIDK 616
>At1g76820 putative translation initiation factor IF-2
Length = 1146
Score = 48.5 bits (114), Expect = 6e-06
Identities = 43/137 (31%), Positives = 60/137 (43%), Gaps = 8/137 (5%)
Query: 12 KKIRNI--CILAHVDHGKTTLADQLIAAASGGMVHPKVAGKVRFMDYLDEEQRRAITMKS 69
+K+R+I CI+ HVD GKT L D + G V AG + + I ++
Sbjct: 570 EKLRSIICCIMGHVDSGKTKLLDCI----RGTNVQEGEAGGITQQIGATYFPAKNIRERT 625
Query: 70 SSISLHYNHHTVNL--IDSPGHIDFCGEVSTAARLSDGALLLVDAVEGVHIQTHAVLRQC 127
+ L ID+PGH F S + L D A+L+VD G+ QT L
Sbjct: 626 RELKADAKLKVPGLLVIDTPGHESFTNLRSRGSSLCDLAILVVDITHGLQPQTIESLNLL 685
Query: 128 WTEMLEPCLVLNKMDRL 144
E + LNK+DRL
Sbjct: 686 RMRNTEFIIALNKVDRL 702
>At1g76810 translation initiation factor IF-2 like protein
Length = 1280
Score = 48.5 bits (114), Expect = 6e-06
Identities = 43/130 (33%), Positives = 56/130 (43%), Gaps = 8/130 (6%)
Query: 18 CILAHVDHGKTTLADQLIAAASGGMVHPKVAGKVRFM---DYLDEEQRRAITMKSSSISL 74
CI+ HVD GKT L D + G V AG + Y E R T K
Sbjct: 695 CIMGHVDTGKTKLLDCI----RGTNVQEGEAGGITQQIGATYFPAENIRERT-KELKADA 749
Query: 75 HYNHHTVNLIDSPGHIDFCGEVSTAARLSDGALLLVDAVEGVHIQTHAVLRQCWTEMLEP 134
+ +ID+PGH F S + L D A+L+VD + G+ QT L E
Sbjct: 750 KLKVPGLLVIDTPGHESFTNLRSRGSSLCDLAILVVDIMHGLEPQTIESLNLLRMRNTEF 809
Query: 135 CLVLNKMDRL 144
+ LNK+DRL
Sbjct: 810 IVALNKVDRL 819
>At1g21160 transcription factor, putative
Length = 1088
Score = 48.5 bits (114), Expect = 6e-06
Identities = 41/130 (31%), Positives = 58/130 (44%), Gaps = 8/130 (6%)
Query: 18 CILAHVDHGKTTLADQLIAAASGGMVHPKVAGKVRFM---DYLDEEQRRAITMKSSSISL 74
CI+ HVD GKT L D + G V AG + + E R T K +
Sbjct: 497 CIMGHVDSGKTKLLDCI----RGTNVQEGEAGGITQQIGATFFPAENIRERT-KELQANA 551
Query: 75 HYNHHTVNLIDSPGHIDFCGEVSTAARLSDGALLLVDAVEGVHIQTHAVLRQCWTEMLEP 134
+ +ID+PGH F S + L D A+L+VD + G+ QT L ++
Sbjct: 552 KLKVPGILVIDTPGHESFTNLRSRGSNLCDLAILVVDIMRGLEPQTIESLNLLRRRNVKF 611
Query: 135 CLVLNKMDRL 144
+ LNK+DRL
Sbjct: 612 IIALNKVDRL 621
>At5g60390 translation elongation factor eEF-1 alpha chain (gene A4)
Length = 449
Score = 46.6 bits (109), Expect = 2e-05
Identities = 31/115 (26%), Positives = 50/115 (42%), Gaps = 17/115 (14%)
Query: 16 NICILAHVDHGKTTLADQLIAAASGGMVHPKVAGKVR---------------FMDYLDEE 60
NI ++ HVD GK+T LI G + +V + +D L E
Sbjct: 9 NIVVIGHVDSGKSTTTGHLIYKLGG--IDKRVIERFEKEAAEMNKRSFKYAWVLDKLKAE 66
Query: 61 QRRAITMKSSSISLHYNHHTVNLIDSPGHIDFCGEVSTAARLSDGALLLVDAVEG 115
+ R IT+ + + +ID+PGH DF + T +D A+L++D+ G
Sbjct: 67 RERGITIDIALWKFETTKYYCTVIDAPGHRDFIKNMITGTSQADCAVLIIDSTTG 121
>At1g07940 elongation factor 1-alpha
Length = 449
Score = 46.6 bits (109), Expect = 2e-05
Identities = 31/115 (26%), Positives = 50/115 (42%), Gaps = 17/115 (14%)
Query: 16 NICILAHVDHGKTTLADQLIAAASGGMVHPKVAGKVR---------------FMDYLDEE 60
NI ++ HVD GK+T LI G + +V + +D L E
Sbjct: 9 NIVVIGHVDSGKSTTTGHLIYKLGG--IDKRVIERFEKEAAEMNKRSFKYAWVLDKLKAE 66
Query: 61 QRRAITMKSSSISLHYNHHTVNLIDSPGHIDFCGEVSTAARLSDGALLLVDAVEG 115
+ R IT+ + + +ID+PGH DF + T +D A+L++D+ G
Sbjct: 67 RERGITIDIALWKFETTKYYCTVIDAPGHRDFIKNMITGTSQADCAVLIIDSTTG 121
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.136 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,788,878
Number of Sequences: 26719
Number of extensions: 367132
Number of successful extensions: 998
Number of sequences better than 10.0: 39
Number of HSP's better than 10.0 without gapping: 33
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 923
Number of HSP's gapped (non-prelim): 44
length of query: 392
length of database: 11,318,596
effective HSP length: 101
effective length of query: 291
effective length of database: 8,619,977
effective search space: 2508413307
effective search space used: 2508413307
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC146552.2


