

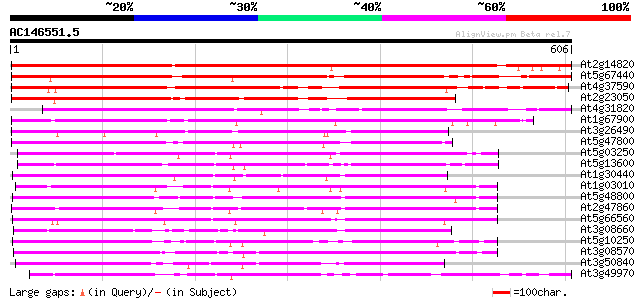
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146551.5 + phase: 0
(606 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g14820 hypothetical protein 691 0.0
At5g67440 photoreceptor-interacting protein-like 524 e-149
At4g37590 unknown protein 521 e-148
At2g23050 unknown protein 489 e-138
At4g31820 unknown protein 402 e-112
At1g67900 unknown protein 395 e-110
At3g26490 non-phototropic hypocotyl, putative 338 5e-93
At5g47800 photoreceptor-interacting protein-like 337 1e-92
At5g03250 photoreceptor-interacting protein - like 317 9e-87
At5g13600 photoreceptor-interacting protein-like; non-phototropi... 306 2e-83
At1g30440 non-phototropic hypocotyl, putative 302 3e-82
At1g03010 hypothetical protein 301 5e-82
At5g48800 non-phototropic hypocotyl-like protein 293 2e-79
At2g47860 unknown protein 287 1e-77
At5g66560 unknown protein 280 2e-75
At3g08660 putative non-phototropic hypocotyl 273 2e-73
At5g10250 non-phototropic hypocotyl 3-like protein 267 1e-71
At3g08570 hypothetical protein 265 7e-71
At3g50840 putative protein 257 1e-68
At3g49970 putative protein 250 1e-66
>At2g14820 hypothetical protein
Length = 634
Score = 691 bits (1782), Expect = 0.0
Identities = 370/625 (59%), Positives = 475/625 (75%), Gaps = 32/625 (5%)
Query: 3 RYVASELASDIIVSIGDTKFYLHKFPLISKSSHLQKLISLNNEENINEIQISDIPGGASA 62
RYV +ELASDI V + ++F LHKFPL+SK + LQKL+S ++ NI++I IS IPGG +A
Sbjct: 21 RYVENELASDISVDVEGSRFCLHKFPLLSKCACLQKLLSSTDKNNIDDIDISGIPGGPTA 80
Query: 63 FEICAKFCYGMTVTLNAYNVIATRCAAEYLGMHENIEKGNLLFKIDVFLSSSIFRSWKDS 122
FE CAKFCYGMTVTL+AYNV+ATRCAAEYLGMHE +EKGNL++KIDVFLSSS+FRSWKDS
Sbjct: 81 FETCAKFCYGMTVTLSAYNVVATRCAAEYLGMHETVEKGNLIYKIDVFLSSSLFRSWKDS 140
Query: 123 IILLQTTKSMSPLDEEQKVVNRCIESIANKACVDVSKVDWSYTYNRKKLPEENGFESNQN 182
II+LQTTK PL E+ K+V+ CI++IA KACVDVS V+WSYTYN+KKL EEN + +
Sbjct: 141 IIVLQTTKPFLPLSEDLKLVSLCIDAIATKACVDVSHVEWSYTYNKKKLAEEN---NGAD 197
Query: 183 GVRTRNVPKDWWVEDLCELEVDMYKSVITNIKTKEIQSNDVIGEALKAYAYRKLPNFSKG 242
++ R+VP DWWVEDLCELE+D YK VI NIKTK I +VIGEALKAY YR+L F+KG
Sbjct: 198 SIKARDVPHDWWVEDLCELEIDYYKRVIMNIKTKCILGGEVIGEALKAYGYRRLSGFNKG 257
Query: 243 MIPCEDVSKHRLIVETIVKLLPSDKGSVSCRFLVKLLKAVIFVESEDRIRDVLVKRIGQQ 302
++ D+ KH+ I+ET+V LLP++K SVSC FL+KLLKAV V S + +++ LV+RIGQQ
Sbjct: 258 VMEQGDLVKHKTIIETLVWLLPAEKNSVSCGFLLKLLKAVTMVNSGEVVKEQLVRRIGQQ 317
Query: 303 LEEASVNDILIKAPDGEITMYDVGIVQKIVREFLMKDHNSEIEL---VGGGELEGIRK-P 358
LEEAS+ ++LIK+ G T+YDV +VQKIV EF+ +D NSEIE+ G E++ +RK P
Sbjct: 318 LEEASMAELLIKSHQGSETLYDVDLVQKIVMEFMRRDKNSEIEVQDDEDGFEVQEVRKLP 377
Query: 359 GILSDASKLMVAKLIDGYLAEIAKDPNLPLLNFVNLAELVSRISRPSHDGLYRAIDTYLK 418
GILS+ASKLMVAK+ID YL EIAKDPNLP F+++AE V+ I RP+HD LYRAID +LK
Sbjct: 378 GILSEASKLMVAKVIDSYLTEIAKDPNLPASKFIDVAESVTSIPRPAHDALYRAIDMFLK 437
Query: 419 EHPGISKGEKKSICQLMDCRKLSVDASLHAVQNERLPTRVIVQVLHFEQVRTAASSGTST 478
EHPGI+KGEKK +C+LMDCRKLSV+A +HAVQN+RLP RV+VQVL FEQVR AASSG+ST
Sbjct: 438 EHPGITKGEKKRMCKLMDCRKLSVEACMHAVQNDRLPLRVVVQVLFFEQVRAAASSGSST 497
Query: 479 PDIAKGI-KDL-SIGSNGSSRSGTTNPEYDTDGAGTAEELKALRKELASLRQSNGVGNSF 536
PD+ +G+ ++L S G+ GSSRS T E + + T EE++AL+ E+A+L+
Sbjct: 498 PDLPRGMGRELRSCGTYGSSRSVPTVMEDEWEAVATEEEMRALKSEIAALK--------L 549
Query: 537 KDGDAKPNMDKA---VIGKMKGLLKSKKSF---IKLWASKGGH----GENSGSDSSESIG 586
++ + +MD+A I K++ L+ SKK F ++L + GG G GSDSSES+G
Sbjct: 550 QEESGRKSMDRAGVTAISKIRSLIMSKKIFGKKVQLQSKGGGEKNNGGGGGGSDSSESLG 609
Query: 587 SVNPEE---VKSTPSRN--RRNSVS 606
S+N E +TPSRN RR SVS
Sbjct: 610 SMNAAEETAKTATPSRNLTRRVSVS 634
>At5g67440 photoreceptor-interacting protein-like
Length = 579
Score = 524 bits (1349), Expect = e-149
Identities = 303/615 (49%), Positives = 403/615 (65%), Gaps = 67/615 (10%)
Query: 3 RYVASELASDIIVSIGDTKFYLHKFPLISKSSHLQKLISL---NNEENINEIQISDIPGG 59
RYVA+ELA+D++V +GD KF+LHKFPL+SKS+ LQKLI+ + + + +EI+I DIPGG
Sbjct: 21 RYVATELATDVVVIVGDVKFHLHKFPLLSKSARLQKLIATTTTDEQSDDDEIRIPDIPGG 80
Query: 60 ASAFEICAKFCYGMTVTLNAYNVIATRCAAEYLGMHENIEKGNLLFKIDVFLSSSIFRSW 119
AFEICAKFCYGM VTLNAYNV+A RCAAEYL M+E+IE GNL++K++VFL+SS+ RSW
Sbjct: 81 PPAFEICAKFCYGMAVTLNAYNVVAVRCAAEYLEMYESIENGNLVYKMEVFLNSSVLRSW 140
Query: 120 KDSIILLQTTKSMSPLDEEQKVVNRCIESIANKACVDVSKVDWSYTYNRKKL--PEENGF 177
KDSII+LQTT+S P E+ K+ RC+ESIA KA +D ++VDWSYTYNR+KL PE N
Sbjct: 141 KDSIIVLQTTRSFYPWSEDVKLDVRCLESIALKAAMDPARVDWSYTYNRRKLLPPEMN-- 198
Query: 178 ESNQNGVRTRNVPKDWWVEDLCELEVDMYKSVITNIKTKEIQSNDVIGEALKAYAYRKLP 237
+VP+DWWVEDL EL +D++K V++ I+ K +VIGEAL+ YA +++P
Sbjct: 199 --------NNSVPRDWWVEDLAELSIDLFKRVVSTIRRKGGVLPEVIGEALEVYAAKRIP 250
Query: 238 NF---SKGMIPCEDVSKHRLIVETIVKLLPSDKGSVSCRFLVKLLKAVIFVESEDRIRDV 294
F + EDV + R ++ET+V +LPS+K SVSC FL+KLLK+ + E + R
Sbjct: 251 GFMIQNDDNDDEEDVMEQRSLLETLVSMLPSEKQSVSCGFLIKLLKSSVSFECGEEERKE 310
Query: 295 LVKRIGQQLEEASVNDILIKAPDGEITMYDVGIVQKIVREFLMKDHNSEIELVGGGELEG 354
L +RIG++LEEA+V D+LI+AP+G T+YD+ IV+ ++ EF+ + + EL +
Sbjct: 311 LSRRIGEKLEEANVGDLLIRAPEGGETVYDIDIVETLIDEFVTQTEKRD-ELDCSDD--- 366
Query: 355 IRKPGILSDASKLMVAKLIDGYLAEIAK-DPNLPLLNFVNLAELVSRISRPSHDGLYRAI 413
++D+SK VAKLIDGYLAEI++ + NL F+ +AE VS R SHDG+YRAI
Sbjct: 367 ------INDSSKANVAKLIDGYLAEISRIETNLSTTKFITIAEKVSTFPRQSHDGVYRAI 420
Query: 414 DTYLKEHPGISKGEKKSICQLMDCRKLSVDASLHAVQNERLPTRVIVQVLHFEQVRTAAS 473
D +LK+HPGI+K EKKS +LMDCRKLS +A HAVQNERLP RV+VQ+L FEQVR
Sbjct: 421 DMFLKQHPGITKSEKKSSSKLMDCRKLSPEACAHAVQNERLPLRVVVQILFFEQVR---- 476
Query: 474 SGTSTPDIAKGIKDLSIGSNGSSRSGTTNPEYDTDGAGTAEELKALRKELASLRQSNGVG 533
T+ P + GS+GSSR TT E T E R + +S ++
Sbjct: 477 -ATTKPSLPPS------GSHGSSR--TTTEEECESVTATEETTTTTRDKTSSSEKT---- 523
Query: 534 NSFKDGDAKPNMDKAVIGKMKGLLKSKKSFIKLWA--SKGGHGENSGSDSSESIGSVNPE 591
K KG++ S + F KLW K G G+ S SD+SES GSV
Sbjct: 524 ------------------KAKGVVMS-RIFSKLWTGKDKDGVGDVSSSDTSESPGSVTTV 564
Query: 592 EVKSTPSRNRRNSVS 606
KSTPS RR S S
Sbjct: 565 GDKSTPSTRRRRSSS 579
>At4g37590 unknown protein
Length = 580
Score = 521 bits (1343), Expect = e-148
Identities = 300/619 (48%), Positives = 411/619 (65%), Gaps = 77/619 (12%)
Query: 3 RYVASELASDIIVSIGDTKFYLHKFPLISKSSHLQKLI----SLNNEENI-------NEI 51
RYV+SELA+D+IV IGD KFYLHKFPL+SKS+ LQKLI S +NEEN +EI
Sbjct: 21 RYVSSELATDVIVIIGDVKFYLHKFPLLSKSARLQKLITTSTSSSNEENQIHHHHHEDEI 80
Query: 52 QISDIPGGASAFEICAKFCYGMTVTLNAYNVIATRCAAEYLGMHENIEKGNLLFKIDVFL 111
+I++IPGG ++FEICAKFCYGMTVTLNAYNV+A RCAAE+L M+E +EKGNL++KI+VFL
Sbjct: 81 EIAEIPGGPASFEICAKFCYGMTVTLNAYNVVAARCAAEFLEMYETVEKGNLVYKIEVFL 140
Query: 112 SSSIFRSWKDSIILLQTTKSMSPLDEEQKVVNRCIESIANKACVDVSKVDWSYTYNRKKL 171
+SSI +SWKDSII+LQTT+++SP EE K+ RC++SIA++A +D SKV+WSYTY++KK
Sbjct: 141 NSSILQSWKDSIIVLQTTRALSPYSEELKLTGRCLDSIASRASIDTSKVEWSYTYSKKK- 199
Query: 172 PEENGFESNQNGVR-TRNVPKDWWVEDLCELEVDMYKSVITNIKTKEIQSNDVIGEALKA 230
+ NG+R + VP+DWWVEDLC+L +D+YK + I+ + S DVIGEAL A
Sbjct: 200 -------NLDNGLRKPQAVPRDWWVEDLCDLHIDLYKRALATIEARGNVSADVIGEALHA 252
Query: 231 YAYRKLPNFSK-GMIPCEDVSKHRLIVETIVKLLPSDKGSVSCRFLVKLLKAVIFVESED 289
YA +++P FSK + D +K+R + ++I++L+P +K SVS FL KLL+A IF+ ++
Sbjct: 253 YAIKRIPGFSKSSSVQVTDFAKYRALADSIIELIPDEKRSVSSSFLTKLLRASIFLGCDE 312
Query: 290 RIRDVLVKRIGQQLEEASVNDILIKAPDGEITMYDVGIVQKIVREFLMKDHNSEIELVGG 349
L R+G++L+EA++ D+L +YDV ++Q +V FL
Sbjct: 313 VAG--LKNRVGERLDEANLGDVL---------LYDVELMQSLVEVFLKS----------- 350
Query: 350 GELEGIRKPGILSDASKLMVAKLIDGYLAEIAKD-PNLPLLNFVNLAELVSRISRPSHDG 408
R P +K VAKL+DGYLAE ++D NLPL F++LAE+VS R SHDG
Sbjct: 351 ------RDPREDDVTAKASVAKLVDGYLAEKSRDSDNLPLQKFLSLAEMVSSFPRQSHDG 404
Query: 409 LYRAIDTYLKEHPGISKGEKKSICQLMDCRKLSVDASLHAVQNERLPTRVIVQVLHFEQV 468
+YRAID +LKEHP ++K EKK IC+LMDCRKLS +A HAVQNERLP RV+VQVL FEQV
Sbjct: 405 VYRAIDMFLKEHPEMNKSEKKRICRLMDCRKLSAEACAHAVQNERLPMRVVVQVLFFEQV 464
Query: 469 RT----AASSGTSTPDIAKGIKDLSIGSNGSSRSGTTNPEYDTDGAGTAEELKALRKELA 524
R ++S+G STP++ +SRS T + DT+ T E++KALR ELA
Sbjct: 465 RANNNGSSSTGNSTPEVIP-----------ASRSTNTTDQEDTECWDT-EDIKALRGELA 512
Query: 525 SLRQSNGVGNSFKDGDAKPNMDKAVIGKMKGLLKSKKSFIKLWASKGGHGENSGSDSSES 584
+LR + K+ N K + G G L + F KLW+ K G+ S + S
Sbjct: 513 NLRLA-------KNQQESHNQGKLMKG---GGLGVSRVFSKLWSGKERSGDMISSSGTSS 562
Query: 585 IGSVNPEEVKSTPSRNRRN 603
GSVN ++ KS+ S ++++
Sbjct: 563 PGSVN-DDSKSSSSTSKKH 580
>At2g23050 unknown protein
Length = 481
Score = 489 bits (1259), Expect = e-138
Identities = 267/485 (55%), Positives = 341/485 (70%), Gaps = 48/485 (9%)
Query: 3 RYVASELASDIIVSIGDTKFYLHKFPLISKSSHLQKLI--SLNNEEN---INEIQISDIP 57
RYV +EL ++II+ IG+ KFYLHKFPL+SKS LQK I S N EE I+EI IS+IP
Sbjct: 21 RYVTNELETEIIIIIGNVKFYLHKFPLLSKSGFLQKHIATSKNEEEKKNQIDEIDISEIP 80
Query: 58 GGASAFEICAKFCYGMTVTLNAYNVIATRCAAEYLGMHENIEKGNLLFKIDVFLSSSIFR 117
GG+ AFEIC KFCYG+TVTLNAYNV+A RCAAE+L M+E EK NL++KIDVFL+S+IFR
Sbjct: 81 GGSVAFEICVKFCYGITVTLNAYNVVAVRCAAEFLEMNETFEKSNLVYKIDVFLNSTIFR 140
Query: 118 SWKDSIILLQTTKSMSPLDEEQKVVNRCIESIANKACVDVSKVDWSYTYNRKKLPEENGF 177
SWKDSII+LQTTK + D+ +++V RC+ SIA+ A +D SKV WSYTYNRKK E+
Sbjct: 141 SWKDSIIVLQTTKDLLS-DDSEELVKRCLGSIASTASIDTSKVKWSYTYNRKKKLEK--V 197
Query: 178 ESNQNGVRTRNVPKDWWVEDLCELEVDMYKSVITNIKTKEIQSNDVIGEALKAYAYRKLP 237
++GV PKDWWVEDLCEL +D+YK I IK + ++VIGEAL AYA R++
Sbjct: 198 RKPEDGV-----PKDWWVEDLCELHIDLYKQAIKAIKNRGKVPSNVIGEALHAYAIRRIA 252
Query: 238 NFSKGMIPCEDVSKHRLIVETIVKLLPSDKGSVSCRFLVKLLKAVIFVESEDRIRDVLVK 297
FSK + D R ++ TI++LLP +KG++S FL KL +A IF+ E+ +++ L K
Sbjct: 253 GFSKESMQLID----RSLINTIIELLPDEKGNISSSFLTKLHRASIFLGCEETVKEKLKK 308
Query: 298 RIGQQLEEASVNDILIKAPDGEITMYDVGIVQKIVREFLMKDHNSEIELVGGGELEGIRK 357
R+ +QLEE +VNDIL MYD+ +VQ +V+EF+ +D +
Sbjct: 309 RVSEQLEETTVNDIL---------MYDLDMVQSLVKEFMNRDPKTH-------------- 345
Query: 358 PGILSDASKLMVAKLIDGYLAEIAKDPNLPLLNFVNLAELVSRISRPSHDGLYRAIDTYL 417
SK+ VAKLIDGYLAE ++DPNLPL NF++LAE +S R SHD LYRAID +L
Sbjct: 346 -------SKVSVAKLIDGYLAEKSRDPNLPLQNFLSLAETLSSFPRHSHDVLYRAIDMFL 398
Query: 418 KEHPGISKGEKKSICQLMDCRKLSVDASLHAVQNERLPTRVIVQVLHFEQVR-TAASSGT 476
KEH GISK EKK +C LMDCRKLS +A HAVQNERLP RVIVQVL FEQ+R +S+G
Sbjct: 399 KEHSGISKSEKKRVCGLMDCRKLSAEACEHAVQNERLPMRVIVQVLFFEQIRANGSSTGY 458
Query: 477 STPDI 481
STP++
Sbjct: 459 STPEL 463
>At4g31820 unknown protein
Length = 518
Score = 402 bits (1032), Expect = e-112
Identities = 237/574 (41%), Positives = 335/574 (58%), Gaps = 59/574 (10%)
Query: 36 LQKLISLNNEENINEIQISDIPGGASAFEICAKFCYGMTVTLNAYNVIATRCAAEYLGMH 95
+Q+L+ +EE +EI I D+PGG AFEICAKFCYGMTVTLNAYN+ A RCAAEYL M
Sbjct: 1 MQRLVFEASEEKTDEITILDMPGGYKAFEICAKFCYGMTVTLNAYNITAVRCAAEYLEMT 60
Query: 96 ENIEKGNLLFKIDVFLSSSIFRSWKDSIILLQTTKSMSPLDEEQKVVNRCIESIANKACV 155
E+ ++GNL++KI+VFL+S IFRSWKDSII+LQTT+S+ P E+ K+V RCI+S++ K V
Sbjct: 61 EDADRGNLIYKIEVFLNSGIFRSWKDSIIVLQTTRSLLPWSEDLKLVGRCIDSVSAKILV 120
Query: 156 DVSKVDWSYTYNRKKLPEENGFESNQNGVRTRNVPKDWWVEDLCELEVDMYKSVITNIKT 215
+ + WSYT+NRK + E ++ +PKDWWVED+CELE+DM+K VI+ +K+
Sbjct: 121 NPETITWSYTFNRKLSGPDKIVEYHREKREENVIPKDWWVEDVCELEIDMFKRVISVVKS 180
Query: 216 KEIQSNDVIGEALKAYAYRKLPNFSKGMIPCEDVSKHRLIVETIVKLLPSDKGSV---SC 272
+N VI EAL+ Y R LP + + + S ++ +VET+V LLP ++ SC
Sbjct: 181 SGRMNNGVIAEALRYYVARWLPESMESL--TSEASSNKDLVETVVFLLPKVNRAMSYSSC 238
Query: 273 RFLVKLLKAVIFVESEDRIRDVLVKRIGQQLEEASVNDILIKAPDGEITMYDVGIVQKIV 332
FL+KLLK I V +++ +R+ LV+ + +L EASV D+LI ++V +V +IV
Sbjct: 239 SFLLKLLKVSILVGADETVREDLVENVSLKLHEASVKDLLI---------HEVELVHRIV 289
Query: 333 REFLMKDHNSEIELVGGGELEGIRKPGILSDASKLMVAKLIDGYLAEIAKDPNLPLLNFV 392
+F M D E + K +L + L V +LID YL A + L L +FV
Sbjct: 290 DQF-MADEKRVSE-------DDRYKEFVLGNGILLSVGRLIDAYL---ALNSELTLSSFV 338
Query: 393 NLAELVSRISRPSHDGLYRAIDTYLKEHPGISKGEKKSICQLMDCRKLSVDASLHAVQNE 452
L+ELV +RP HDGLY+AIDT++KEHP ++K EKK +C LMD RKL+ +AS HA QNE
Sbjct: 339 ELSELVPESARPIHDGLYKAIDTFMKEHPELTKSEKKRLCGLMDVRKLTNEASTHAAQNE 398
Query: 453 RLPTRVIVQVLHFEQVRTAASSGTSTPDIAKGIKDLSIGSNGSSRSGTTNPEYDTDGAGT 512
RLP RV+VQVL+FEQ+R S + S +S + G
Sbjct: 399 RLPLRVVVQVLYFEQLRANHS---------------PVASVAASSHSPVEKTEENKGEEA 443
Query: 513 AEELKALRKELASLRQSNGVGNSFKDGDAKPNMDKAVIGKMKGLLKSKKSFIKLWASKGG 572
++++ +K S +G G +S++ F K+W K G
Sbjct: 444 TKKVELSKKSRGSKSTRSGGGAQLMPS------------------RSRRIFEKIWPGK-G 484
Query: 573 HGENSGSDSSESIGSVNPEEVKSTPSRNRRNSVS 606
N S+ S P + S+ SR RR+S+S
Sbjct: 485 EISNKSSEVSSGSSQSPPAKSSSSSSRRRRHSIS 518
>At1g67900 unknown protein
Length = 631
Score = 395 bits (1016), Expect = e-110
Identities = 236/612 (38%), Positives = 352/612 (56%), Gaps = 60/612 (9%)
Query: 3 RYVASELASDIIVSIGDTKFYLHKFPLISKSSHLQKLISLNNEENINEIQISDIPGGASA 62
R V+SE++SD + + +++ LHKFPL+SK LQ++ S + E + IQ+ + PGG A
Sbjct: 20 RCVSSEVSSDFTIEVSGSRYLLHKFPLLSKCLRLQRMCSESPE---SIIQLPEFPGGVEA 76
Query: 63 FEICAKFCYGMTVTLNAYNVIATRCAAEYLGMHENIEKGNLLFKIDVFLSSSIFRSWKDS 122
FE+CAKFCYG+T+T++AYN++A RCAAEYL M E +EKGNL++K++VF +S I W+DS
Sbjct: 77 FELCAKFCYGITITISAYNIVAARCAAEYLQMSEEVEKGNLVYKLEVFFNSCILNGWRDS 136
Query: 123 IILLQTTKSMSPLDEEQKVVNRCIESIANKACVDVSKVDWSYTYNRKKLPEENGFESNQN 182
I+ LQTTK+ E+ + +RCIE+IA+K SKV S++++R+ ++ SN+
Sbjct: 137 IVTLQTTKAFPLWSEDLAITSRCIEAIASKVLSHPSKVSLSHSHSRRV--RDDDMSSNRA 194
Query: 183 GVRTRNVPKDWWVEDLCELEVDMYKSVITNIKTKEIQSNDVIGEALKAYAYRKLPNFSKG 242
+R WW ED+ EL +D+Y + IK+ +IG+AL+ YA + LP +
Sbjct: 195 AASSRG----WWAEDIAELGIDLYWRTMIAIKSGGKVPASLIGDALRVYASKWLPTLQRN 250
Query: 243 M------------IPCEDVSKHRLIVETIVKLLPSDKGSVSCRFLVKLLKAVIFVESEDR 290
+ SKHRL++E+I+ LLP++KG+VSC FL+KLLKA + +
Sbjct: 251 RKVVKKKEDSDSDSDTDTSSKHRLLLESIISLLPAEKGAVSCSFLLKLLKAANILNASTS 310
Query: 291 IRDVLVKRIGQQLEEASVNDILIKAPDGEITM-YDVGIVQKIVREFLMKDHNSEIELVGG 349
+ L +R+ QLEEA+V+D+LI + + YDV IV I+ +F+++ S
Sbjct: 311 SKMELARRVALQLEEATVSDLLIPPMSYKSELLYDVDIVATILEQFMVQGQTSPPTSPLR 370
Query: 350 G-------------------ELEGIRKPGILSDASKLMVAKLIDGYLAEIAKDPNLPLLN 390
G E + R+ S +SKL VAKL+DGYL +IA+D NLPL
Sbjct: 371 GKKGMMDRRRRSRSAENIDLEFQESRRSSSASHSSKLKVAKLVDGYLQQIARDVNLPLSK 430
Query: 391 FVNLAELVSRISRPSHDGLYRAIDTYLKEHPGISKGEKKSICQLMDCRKLSVDASLHAVQ 450
FV LAE V SR HD LYRAID YLK H ++K E+K +C+++DC+KLS++A +HA Q
Sbjct: 431 FVTLAESVPEFSRLDHDDLYRAIDIYLKAHKNLNKSERKRVCRVLDCKKLSMEACMHAAQ 490
Query: 451 NERLPTRVIVQVLHFEQVRTAASSGT---STPDIAKGIKDLSIGSN------GSSRSGTT 501
NE LP RV+VQVL +EQ R AA++ +T ++ IK L N ++ TT
Sbjct: 491 NEMLPLRVVVQVLFYEQARAAAATNNGEKNTTELPSNIKALLAAHNIDPSNPNAAAFSTT 550
Query: 502 NPEYDTDGAGTAEELKALRKEL-----ASLRQSNGVGNSFKDGDA--KPNMDKAVIGKMK 554
+ + LK+ + +L + L + V F +GD P+ KA +
Sbjct: 551 TSIAAPEDRWSVSGLKSPKSKLSGTLRSKLAEDEEVDERFMNGDGGRTPSRFKAFCA-IP 609
Query: 555 GLLKSKKSFIKL 566
G + KK F KL
Sbjct: 610 G--RPKKMFSKL 619
>At3g26490 non-phototropic hypocotyl, putative
Length = 588
Score = 338 bits (867), Expect = 5e-93
Identities = 197/506 (38%), Positives = 294/506 (57%), Gaps = 47/506 (9%)
Query: 3 RYVASELASDIIVSIGDTKFYLHKFPLISKSSHLQKLISLNNEENINE---IQISDIPGG 59
R V+S+L +D+++ + TK+ LHKFP++SK L+ L+S E E IQ+ D PG
Sbjct: 20 RSVSSDLLNDLVIQVKSTKYLLHKFPMLSKCLRLKNLVSSQETETSQEQQVIQLVDFPGE 79
Query: 60 ASAFEICAKFCYGMTVTLNAYNVIATRCAAEYLGMHENIEKG---NLLFKIDVFLSSSIF 116
AFE+CAKFCYG+T+TL A+NV+A RCAAEYLGM E +E G NL+ ++++FL++ +F
Sbjct: 80 TEAFELCAKFCYGITITLCAHNVVAVRCAAEYLGMTEEVELGETENLVQRLELFLTTCVF 139
Query: 117 RSWKDSIILLQTTKSMSPLDEEQKVVNRCIESIANKACVDV-----SKVDWSYTYNRKKL 171
+SW+DS + LQTTK + E+ + NRCIE+IAN V ++++ NR ++
Sbjct: 140 KSWRDSYVTLQTTKVLPLWSEDLGITNRCIEAIANGVTVSPGEDFSTQLETGLLRNRSRI 199
Query: 172 PEENGFESNQNGVRTRNVPKDWWVEDLCELEVDMYKSVITNIKTKEIQ-SNDVIGEALKA 230
+ + G + ++ WW EDL EL +D+Y+ + IK+ + S +IG AL+
Sbjct: 200 RRDEILCNGGGGSKAESLR--WWGEDLAELGLDLYRRTMVAIKSSHRKISPRLIGNALRI 257
Query: 231 YAYRKLPNFSKGMIPCEDVSKHRLIVETIVKLLPSDKGSVSCRFLVKLLKAVIFVESEDR 290
YA + LP+ E + L++E+++ LLP +K SV C FL++LLK +
Sbjct: 258 YASKWLPSIQ------ESSADSNLVLESVISLLPEEKSSVPCSFLLQLLKMANVMNVSHS 311
Query: 291 IRDVLVKRIGQQLEEASVNDILIKAPDGEITMYDVGIVQKIVREFLMK------------ 338
+ L + G QL++A+V+++LI D +YDV +V+ +V++FL
Sbjct: 312 SKMELAIKAGNQLDKATVSELLIPLSDKSGMLYDVDVVKMMVKQFLSHISPEIRPTRTRT 371
Query: 339 DHN----------SEIELVGGGELEGIRKPGILSDASKLMVAKLIDGYLAEIAKDPNLPL 388
+H EI+ V G P +LS VAKL+D YL EIA+D NL +
Sbjct: 372 EHRRSRSEENINLEEIQEVRGSLSTSSSPPPLLSK-----VAKLVDSYLQEIARDVNLTV 426
Query: 389 LNFVNLAELVSRISRPSHDGLYRAIDTYLKEHPGISKGEKKSICQLMDCRKLSVDASLHA 448
FV LAE + SR HD LY AID YL+ H I K E+K +C+++DC+KLSV+AS A
Sbjct: 427 SKFVELAETIPDTSRICHDDLYNAIDVYLQVHKKIEKCERKRLCRILDCKKLSVEASKKA 486
Query: 449 VQNERLPTRVIVQVLHFEQVRTAASS 474
QNE LP RVIVQ+L EQ R ++
Sbjct: 487 AQNELLPLRVIVQILFVEQARATLAT 512
>At5g47800 photoreceptor-interacting protein-like
Length = 559
Score = 337 bits (864), Expect = 1e-92
Identities = 183/487 (37%), Positives = 292/487 (59%), Gaps = 34/487 (6%)
Query: 3 RYVASELASDIIVSIGDTKFYLHKFPLISKSSHLQKLISLNNEENINEIQISDIPGGASA 62
R + ++ +D+++ I +T ++LH+ L+ K L++L + E + I+++DIPGGA A
Sbjct: 20 RILITDTPNDLVIRINNTTYHLHRSCLVPKCGLLRRLCTDLEESDTVTIELNDIPGGADA 79
Query: 63 FEICAKFCYGMTVTLNAYNVIATRCAAEYLGMHENIEKGNLLFKIDVFLSSSIFRSWKDS 122
FE+CAKFCY +T+ L+A+N++ CA+++L M ++++KGNLL K++ F S I + WKDS
Sbjct: 80 FELCAKFCYDITINLSAHNLVNALCASKFLRMSDSVDKGNLLPKLEAFFHSCILQGWKDS 139
Query: 123 IILLQTTKSMSPLDEEQKVVNRCIESIANKACVDVSKVDWSYTYNRKKLPEENGFESNQN 182
I+ LQ+T + E +V +CI+SI K S+V WS+TY R G+ Q+
Sbjct: 140 IVTLQSTTKLPEWCENLGIVRKCIDSIVEKILTPTSEVSWSHTYTRP------GYAKRQH 193
Query: 183 GVRTRNVPKDWWVEDLCELEVDMYKSVITNIKTKEIQSNDVIGEALKAYAYRKLPNFS-- 240
+VP+DWW ED+ +L++D+++ VIT ++ +IGEAL Y R LP F
Sbjct: 194 ----HSVPRDWWTEDISDLDLDLFRCVITAARSTFTLPPQLIGEALHVYTCRWLPYFKSN 249
Query: 241 --KGMIPCED---VSKHRLIVETIVKLLPSDKGSVSCRFLVKLLKAVIFVESEDRIRDVL 295
G E+ + +HR +V T+V ++P+DKGSVS FL++L+ +V + + L
Sbjct: 250 SHSGFSVKENEAALERHRRLVNTVVNMIPADKGSVSEGFLLRLVSIASYVRASLTTKTEL 309
Query: 296 VKRIGQQLEEASVNDILIKA-PDGEITMYDVGIVQKIVREFLM---KDHNSEIELVGGGE 351
+++ QLEEA++ D+L+ + + YD +V ++ FLM + ++ +
Sbjct: 310 IRKSSLQLEEATLEDLLLPSHSSSHLHRYDTDLVATVLESFLMLWRRQSSAHLSSNNTQL 369
Query: 352 LEGIRKPGILSDASKLMVAKLIDGYLAEIAKDPNLPLLNFVNLAELVSRISRPSHDGLYR 411
L IRK VAKLID YL +A+D ++P+ FV+L+E V I+R SHD LY+
Sbjct: 370 LHSIRK-----------VAKLIDSYLQAVAQDVHMPVSKFVSLSEAVPDIARQSHDRLYK 418
Query: 412 AIDTYLKEHPGISKGEKKSICQLMDCRKLSVDASLHAVQNERLPTRVIVQVLHFEQVRTA 471
AI+ +LK HP ISK EKK +C+ +DC+KLS HAV+NER+P R +VQ L F+Q +
Sbjct: 419 AINIFLKVHPEISKEEKKRLCRSLDCQKLSAQVRAHAVKNERMPLRTVVQALFFDQ--ES 476
Query: 472 ASSGTST 478
S G S+
Sbjct: 477 GSKGASS 483
>At5g03250 photoreceptor-interacting protein - like
Length = 592
Score = 317 bits (813), Expect = 9e-87
Identities = 194/542 (35%), Positives = 304/542 (55%), Gaps = 35/542 (6%)
Query: 9 LASDIIVSIGDTKFYLHKFPLISKSSHLQKLISLNNEENINE--IQISDIPGGASAFEIC 66
L SD+ + +GD KF+LHKFPL+S+S L++LI ++ ++ + + + +IPGG FE+
Sbjct: 26 LVSDVTIEVGDMKFHLHKFPLLSRSGLLERLIEESSTDDGSGCVLSLDEIPGGGKTFELV 85
Query: 67 AKFCYGMTVTLNAYNVIATRCAAEYLGMHENIEKGNLLFKIDVFLSSSIFRSWKDSIILL 126
KFCYG+ + L A+NV++ RCAAEYL M +N +GNL+ + FL+ +F +W DSI L
Sbjct: 86 TKFCYGVKIELTAFNVVSLRCAAEYLEMTDNYGEGNLVGMTETFLNE-VFGNWTDSIKAL 144
Query: 127 QTTKSMSPLDEEQKVVNRCIESIANKACVDVSKVDWSYTYNRKKLPEENGFESNQ----- 181
QT + + E+ +++RC++S+A KAC D S +W + +N + +
Sbjct: 145 QTCEEVIDYAEDLHIISRCVDSLAVKACADPSLFNWPVGGGKNATSGQNTEDESHLWNGI 204
Query: 182 --NGVRTRNVPKDWWVEDLCELEVDMYKSVITNIKTKEIQSNDVIGEALKAYAYRKLP-- 237
+G ++ +DWW +D L + ++K +IT I+ + ++ + I A+ Y + +P
Sbjct: 205 SASGKMLQHTGEDWWFDDASFLSLPLFKRLITAIEARGMKLEN-IAMAVMYYTRKHVPLM 263
Query: 238 ----NFSKGMIPCEDVSKH--RLIVETIVKLLPSDKGSVSCRFLVKLLKAVIFVESEDRI 291
N + +I + S+ + +E IV LLPS KG +FL++LL+ + + +
Sbjct: 264 NRQVNMDEQVIETPNPSEEDQKTCLEEIVGLLPSKKGVNPTKFLLRLLQTAMVLHASQSS 323
Query: 292 RDVLVKRIGQQLEEASVNDILIKAPDGEITMYDVGIVQKIVREFLMKDHNSEIE----LV 347
R+ L +RIG QL++A++ D+LI T+YDV V +++ +F+ + I ++
Sbjct: 324 RENLERRIGNQLDQAALVDLLIPNMGYSETLYDVECVLRMIEQFVSSTEQAGIVPSPCII 383
Query: 348 GGGELEGIRKPGILSDASKLMVAKLIDGYLAEIAKDPNLPLLNFVNLAELVSRISRPSHD 407
G L K G +VA L+DGYLAE+A D NL L F +A + +RP D
Sbjct: 384 EEGHLV---KDGADLLTPTTLVATLVDGYLAEVAPDVNLKLAKFEAIAAAIPDYARPLDD 440
Query: 408 GLYRAIDTYLKEHPGISKGEKKSICQLMDCRKLSVDASLHAVQNERLPTRVIVQVLHFEQ 467
G+Y AID YLK HP I+ E++ IC+LM+C+KLS++AS HA QNERLP RVIVQVL FEQ
Sbjct: 441 GVYHAIDVYLKAHPWITDSEREHICRLMNCQKLSLEASTHAAQNERLPLRVIVQVLFFEQ 500
Query: 468 VRTAAS-SGTSTPDIAKGIKDLSIGSNGSSRSGTTNPEYDTDGAGTAEELKALRKELASL 526
+R S SG D G+NG G P G E + L KE ++
Sbjct: 501 LRLRTSVSGWFFVSENLDNPDNQHGANG----GLLKPR----GENVRERVSELEKECMNM 552
Query: 527 RQ 528
+Q
Sbjct: 553 KQ 554
>At5g13600 photoreceptor-interacting protein-like; non-phototropic
hypocotyl-like protein
Length = 591
Score = 306 bits (784), Expect = 2e-83
Identities = 199/542 (36%), Positives = 302/542 (55%), Gaps = 43/542 (7%)
Query: 9 LASDIIVSIGDTKFYLHKFPLISKSSHLQKLISLNNEENINEIQISDIPGGASAFEICAK 68
L D+++ + D F+LHKFPL+S+S +L+ L S +E Q+ DIPGG F + AK
Sbjct: 26 LKPDVMIQVVDESFHLHKFPLLSRSGYLETLFSKASETTC-VAQLHDIPGGPETFLLVAK 84
Query: 69 FCYGMTVTLNAYNVIATRCAAEYLGMHENIEKGNLLFKIDVFLSSSIFRSWKDSIILLQT 128
FCYG+ + + N ++ RCAAEYL M EN NL++ + FL+ +F +W+DSI L+
Sbjct: 85 FCYGVRIEVTPENAVSLRCAAEYLQMSENYGDANLIYLTESFLNDHVFVNWEDSIKALEK 144
Query: 129 T--KSMSPLDEEQKVVNRCIESIANKACVD--VSKVDWSYTYNRKKLPEENGFESNQ-NG 183
+ + PL EE +V+RCI S+A KAC + S +W + LPE + NG
Sbjct: 145 SCEPKVLPLAEELHIVSRCIGSLAMKACAEDNTSFFNWPIS-----LPEGTTTTTIYWNG 199
Query: 184 VRTRNVPKDWWVEDLCE-LEVDMYKSVITNIKTKEIQSNDVIGEALKAYAYRKLPNFS-- 240
++T+ ++WW D+ L++ MYK I ++++ + + +I ++ YA R LP
Sbjct: 200 IQTKATSENWWFNDVSSFLDLPMYKRFIKTVESRGVNAG-IIAASVTHYAKRNLPLLGCS 258
Query: 241 -KGMIPCEDVSKH-----------RLIVETIVKLLPSDKGSVSCRFLVKLLKAVIFVESE 288
K P E+ + + R ++E IV+LLP K S +FL++LL+ + + +
Sbjct: 259 RKSGSPSEEGTNYGDDMYYSHEEQRSLLEEIVELLPGKKCVTSTKFLLRLLRTSMVLHAS 318
Query: 289 DRIRDVLVKRIGQQLEEASVNDILIKAP--DGEITMYDVGIVQKIVREFLMKDHNSEIEL 346
++ L KRIG QL+EA++ D+LI GE T+YD VQ+I+ F++ +S +E
Sbjct: 319 QVTQETLEKRIGMQLDEAALEDLLIPNMKYSGE-TLYDTDSVQRILDHFMLTFDSSIVE- 376
Query: 347 VGGGELEGIRKPGILSDASKLMVAKLIDGYLAEIAKDPNLPLLNFVNLAELVSRISRPSH 406
++ G P L +K VA LIDGYLAE+A D NL L F L L+ RP
Sbjct: 377 --EKQMMGDSHP--LKSITK--VASLIDGYLAEVASDENLKLSKFQALGALIPEDVRPMD 430
Query: 407 DGLYRAIDTYLKEHPGISKGEKKSICQLMDCRKLSVDASLHAVQNERLPTRVIVQVLHFE 466
DG+YRAID Y+K HP +++ E++ +C LM+C+KLS++A HA QNERLP RVIVQVL FE
Sbjct: 431 DGIYRAIDIYIKAHPWLTESEREQLCLLMNCQKLSLEACTHAAQNERLPLRVIVQVLFFE 490
Query: 467 QVRTAASSGTSTPDIAKGIKDLSIGSNGSSRSGTTNPEYDTDGAGTAEELKALRKELASL 526
Q+R TS G ++ + + G N + G E + L KE S+
Sbjct: 491 QMRLR----TSIAGWLFGSEENN--DTSGALEGNKNTNANMVMHGMRERVFELEKECMSM 544
Query: 527 RQ 528
+Q
Sbjct: 545 KQ 546
>At1g30440 non-phototropic hypocotyl, putative
Length = 665
Score = 302 bits (774), Expect = 3e-82
Identities = 186/500 (37%), Positives = 282/500 (56%), Gaps = 38/500 (7%)
Query: 4 YVASELASDIIVSIGDTKFYLHKFPLISKSSHLQKLISLNNEENINE--IQISDIPGGAS 61
+ + L SDI+V +G+ F+LHKFPL+S+S +++ I+ ++E ++ I+ISD+PGG
Sbjct: 21 FCTTGLPSDIVVEVGEMSFHLHKFPLLSRSGVMERRIAEASKEGDDKCLIEISDLPGGDK 80
Query: 62 AFEICAKFCYGMTVTLNAYNVIATRCAAEYLGMHENIEKGNLLFKIDVFLSSSIFRSWKD 121
FE+ AKFCYG+ + L A NV+ RCAAE+L M E +GNL+ + + F + + +SWKD
Sbjct: 81 TFELVAKFCYGVKLELTASNVVYLRCAAEHLEMTEEHGEGNLISQTETFFNQVVLKSWKD 140
Query: 122 SIILLQTTKSMSPLDEEQKVVNRCIESIANKACVDVSKVDWSYTYNRKKLPEENG---FE 178
SI L + + +E + +CIES+A +A D + W + + G +
Sbjct: 141 SIKALHSCDEVLEYADELNITKKCIESLAMRASTDPNLFGWPVVEHGGPMQSPGGSVLWN 200
Query: 179 SNQNGVRTRNVPKDWWVEDLCELEVDMYKSVITNIKTKEIQSNDVIGEALKAYAYRKLPN 238
G R ++ DWW ED L ++K +IT ++++ I+ D+I +L Y + LP
Sbjct: 201 GISTGARPKHTSSDWWYEDASMLSFPLFKRLITVMESRGIRE-DIIAGSLTYYTRKHLPG 259
Query: 239 FSK-----------------GMIPCEDVSKHRLIVETIVKLLPSDKGSVSCRFLVKLLK- 280
+ G + E+ K+ ++E I +LL KG V +F V +L+
Sbjct: 260 LKRRRGGPESSGRFSTPLGSGNVLSEEEQKN--LLEEIQELLRMQKGLVPTKFFVDMLRI 317
Query: 281 AVIFVESEDRIRDVLVKRIGQQLEEASVNDILIKAPDGEI-TMYDVGIVQKIVREFLMKD 339
A I S D I + L KRIG QL++A++ D+++ + + T+YDV VQ+I+ FL D
Sbjct: 318 AKILKASPDCIAN-LEKRIGMQLDQAALEDLVMPSFSHTMETLYDVDSVQRILDHFLGTD 376
Query: 340 H------NSEIELVGGGELEGIRKPGILSDASKLMVAKLIDGYLAEIAKDPNLPLLNFVN 393
S V G L G + S VAKLIDGYLAE+A D NL L F
Sbjct: 377 QIMPGGVGSPCSSVDDGNLIGSPQ----SITPMTAVAKLIDGYLAEVAPDVNLKLPKFQA 432
Query: 394 LAELVSRISRPSHDGLYRAIDTYLKEHPGISKGEKKSICQLMDCRKLSVDASLHAVQNER 453
LA + +R DGLYRAID YLK HP +++ E++++C+L+DC+KLS++A HA QNER
Sbjct: 433 LAASIPEYARLLDDGLYRAIDIYLKHHPWLAETERENLCRLLDCQKLSLEACTHAAQNER 492
Query: 454 LPTRVIVQVLHFEQVRTAAS 473
LP R+IVQVL FEQ++ S
Sbjct: 493 LPLRIIVQVLFFEQLQLRTS 512
>At1g03010 hypothetical protein
Length = 634
Score = 301 bits (772), Expect = 5e-82
Identities = 203/552 (36%), Positives = 300/552 (53%), Gaps = 53/552 (9%)
Query: 7 SELASDIIVSIGDTKFYLHKFPLISKSSHLQKLISLNNEENINEIQISDIPGGASAFEIC 66
S+++SD+ V +G + F LHKFPL+S+S ++KL++ + I+ + +S+ PGG+ AFE+
Sbjct: 34 SDVSSDLTVQVGSSSFCLHKFPLVSRSGKIRKLLA---DPKISNVCLSNAPGGSEAFELA 90
Query: 67 AKFCYGMTVTLNAYNVIATRCAAEYLGMHENIEKGNLLFKIDVFLSSSIFRSWKDSIILL 126
AKFCYG+ + +N N+ RCA+ YL M E+ + NL K + FL +IF S +SII+L
Sbjct: 91 AKFCYGINIEINLLNIAKLRCASHYLEMTEDFSEENLASKTEHFLKETIFPSILNSIIVL 150
Query: 127 QTTKSMSPLDEEQKVVNRCIESIANKACVD-----VSKVDWSYTYNRKKLPEENGFESNQ 181
+++ P+ E+ +VNR I ++AN AC + + K+D+S++ +
Sbjct: 151 HHCETLIPVSEDLNLVNRLIIAVANNACKEQLTSGLLKLDYSFSGTNIE----------- 199
Query: 182 NGVRTRNVPKDWWVEDLCELEVDMYKSVITNIKTKEIQSNDVIGEALKAYAYRKL----- 236
P DWW + L L +D ++ VI+ +K+K + DVI + L +Y + L
Sbjct: 200 -----PQTPLDWWGKSLAVLNLDFFQRVISAVKSKGL-IQDVISKILISYTNKSLQGLIV 253
Query: 237 --PNFSKG-MIPCEDVSKHRLIVETIVKLLPSD--KGSVSCRFLVKLLKAVIFVESE--- 288
P K ++ E K RLIVETIV+LLP+ + SV FL LLK VI S
Sbjct: 254 RDPKLEKERVLDSEGKKKQRLIVETIVRLLPTQGRRSSVPMAFLSSLLKMVIATSSSAST 313
Query: 289 DRIRDVLVKRIGQQLEEASVNDILI--KAPDGEITMYDVGIVQKIVREFLMKDHNSEIE- 345
R L +RIG QL++A + D+LI TMYD+ + +I FL D + E E
Sbjct: 314 GSCRSDLERRIGLQLDQAILEDVLIPINLNGTNNTMYDIDSILRIFSIFLNLDEDDEEEE 373
Query: 346 ---LVGGGELEGIR---KPGILSDASKLMVAKLIDGYLAEIAKDPNLPLLNFVNLAELVS 399
L E E I PG +S L V+KL+D YLAEIA DPNL F+ LAEL+
Sbjct: 374 HHHLQFRDETEMIYDFDSPGSPKQSSILKVSKLMDNYLAEIAMDPNLTTSKFIALAELLP 433
Query: 400 RISRPSHDGLYRAIDTYLKEHPGISKGEKKSICQLMDCRKLSVDASLHAVQNERLPTRVI 459
+R DGLYRA+D YLK HP I E+ +C+ +D +KLS +A HA QNERLP ++
Sbjct: 434 DHARIISDGLYRAVDIYLKVHPNIKDSERYRLCKTIDSQKLSQEACSHAAQNERLPVQMA 493
Query: 460 VQVLHFEQVR----TAASSGTSTPDIAKGIKDLSIGSNGSSRSGTTNPEYDTDGAGTAEE 515
VQVL+FEQ+R ++S G + S + SG +P + A E
Sbjct: 494 VQVLYFEQIRLRNAMSSSIGPTQFLFNSNCHQFPQRSGSGAGSGAISPR--DNYASVRRE 551
Query: 516 LKALRKELASLR 527
+ L+ E+A +R
Sbjct: 552 NRELKLEVARMR 563
>At5g48800 non-phototropic hypocotyl-like protein
Length = 614
Score = 293 bits (749), Expect = 2e-79
Identities = 177/528 (33%), Positives = 300/528 (56%), Gaps = 24/528 (4%)
Query: 4 YVASELASDIIVSIGDTKFYLHKFPLISKSSHLQKLISLNNEENINEIQISDIPGGASAF 63
++ ++ SDI + + F LHKFPL+S+S ++++++ + + +I+++++ ++PGGA F
Sbjct: 36 WIFRDVPSDITIEVNGGNFALHKFPLVSRSGRIRRIVAEHRDSDISKVELLNLPGGAETF 95
Query: 64 EICAKFCYGMTVTLNAYNVIATRCAAEYLGMHENIEKGNLLFKIDVFLSSSIFRSWKDSI 123
E+ AKFCYG+ + + NV C ++YL M E K NL + + +L S + ++ + +
Sbjct: 96 ELAAKFCYGINFEITSSNVAQLFCVSDYLEMTEEYSKDNLASRTEEYLESIVCKNLEMCV 155
Query: 124 ILLQTTKSMSPLDEEQKVVNRCIESIANKACVDVSKVDWSYTYNRKKLPEENGFESNQNG 183
+L+ ++ + PL +E ++ RCI++IA+KAC + + +++R + ++
Sbjct: 156 QVLKQSEILLPLADELNIIGRCIDAIASKACAE----QIASSFSRLEYSSSGRLHMSRQ- 210
Query: 184 VRTRNVPKDWWVEDLCELEVDMYKSVITNIKTKEIQSNDVIGEALKAYAYRKLPNFSKGM 243
V++ DWW+EDL L +D+Y+ V+ +K + ++ + IG +L +YA R+L S+
Sbjct: 211 VKSSGDGGDWWIEDLSVLRIDLYQRVMNAMKCRGVRP-ESIGASLVSYAERELTKRSE-- 267
Query: 244 IPCEDVSKHRLIVETIVKLLPSDKGSVSCRFLVKLLKAVIFVESEDRIRDVLVKRIGQQL 303
+ IVETIV LLP + V FL LL+ + +++ R L +R+G QL
Sbjct: 268 -------HEQTIVETIVTLLPVENLVVPISFLFGLLRRAVILDTSVSCRLDLERRLGSQL 320
Query: 304 EEASVNDILIKA-PDGEITMYDVGIVQKIVREFLMKDHNSEIELVGGGELEGIRKPGILS 362
+ A+++D+LI + T++D+ V +I+ F + + + E + P S
Sbjct: 321 DMATLDDLLIPSFRHAGDTLFDIDTVHRILVNFSQQGGDDSEDEESVFECDSPHSP---S 377
Query: 363 DASKLMVAKLIDGYLAEIAKDPNLPLLNFVNLAELVSRISRPSHDGLYRAIDTYLKEHPG 422
+ VAKL+D YLAEIA D NL L F+ +AE + +R HDGLYRAID YLK H G
Sbjct: 378 QTAMFKVAKLVDSYLAEIAPDANLDLSKFLLIAEALPPHARTLHDGLYRAIDLYLKAHQG 437
Query: 423 ISKGEKKSICQLMDCRKLSVDASLHAVQNERLPTRVIVQVLHFEQVRTAASSGTSTPD-- 480
+S +KK + +L+D +KLS +A HA QNERLP + IVQVL+FEQ++ +S +S D
Sbjct: 438 LSDSDKKKLSKLIDFQKLSQEAGAHAAQNERLPLQSIVQVLYFEQLKLRSSLCSSYSDEE 497
Query: 481 -IAKGIKDLSIGSNGSSRSGTTNPEYDTDGAGTAEELKALRKELASLR 527
K + S N + S T +P+ + A E + L+ ELA LR
Sbjct: 498 PKPKQQQQQSWRINSGALSATMSPK--DNYASLRRENRELKLELARLR 543
>At2g47860 unknown protein
Length = 635
Score = 287 bits (734), Expect = 1e-77
Identities = 194/552 (35%), Positives = 297/552 (53%), Gaps = 56/552 (10%)
Query: 3 RYVASELASDIIVSIGDTKFYLHKFPLISKSSHLQKLISLNNEENINEIQISDIPGGASA 62
++ S++ SD+ + +G F LHKFPL+S+S ++KL+ + + N+N ++ +PGG+ +
Sbjct: 31 QWPVSDVTSDLTIEVGSATFSLHKFPLVSRSGRIRKLVLESKDTNLN---LAAVPGGSES 87
Query: 63 FEICAKFCYGMTVTLNAYNVIATRCAAEYLGMHENIEKGNLLFKIDVFLSSSIFRSWKDS 122
FE+ AKFCYG+ V N+ N+ A RC A YL M E++ + NL + + +L SIF +S
Sbjct: 88 FELAAKFCYGVGVQYNSSNIAALRCVAHYLEMTEDLSEKNLEARTEAYLKDSIFNDISNS 147
Query: 123 IILLQTTKSMSPLDEEQKVVNRCIESIANKACVD-----VSKVDWSYTYNRKKLPEENGF 177
I +L + + + P+ EE +V R + +IA AC + + K+D S++
Sbjct: 148 ITVLHSCERLLPVAEEINLVGRLVNAIAVNACKEQLASGLLKLDQSFSC----------- 196
Query: 178 ESNQNGVRTRNVPKDWWVEDLCELEVDMYKSVITNIKTKEIQSNDVIGEALKAYAYRKL- 236
GV P DWW L L++D ++ V++ +K+K + ++D+I + L +YA + L
Sbjct: 197 -----GVPETAKPCDWWGRSLPILKLDFFQRVLSAMKSKGL-NHDIISDILMSYARKSLQ 250
Query: 237 ----PNFSKGMIPCEDVSKHRLIVETIVKLLPS--DKGSVSCRFLVKLLKAVIFVESEDR 290
PN K + K R+++E +V LLP+ +K S+ FL LLK I +
Sbjct: 251 IIREPNLVKSDSDLQ--RKQRIVLEAVVGLLPTQANKSSIPISFLSSLLKTAIGSGTSVS 308
Query: 291 IRDVLVKRIGQQLEEASVNDILIKAPDGEITMYDVGIVQKIVREFL-------MKDHNSE 343
R L +RI L++A + DILI A G MYD VQ+I FL D + E
Sbjct: 309 CRSDLERRISHLLDQAILEDILIPANIG--AMYDTDSVQRIFSMFLNLDECEYRDDDDDE 366
Query: 344 IELVGGGEL-----EGIRKPGILSDASKLMVAKLIDGYLAEIAKDPNLPLLNFVNLAELV 398
+ V E+ EG P +S V+KL+D YLAE+A D +LP F+ LAEL+
Sbjct: 367 EDAVDESEMAMYDFEGAESP---KQSSIFKVSKLMDSYLAEVALDSSLPPSKFIALAELL 423
Query: 399 SRISRPSHDGLYRAIDTYLKEHPGISKGEKKSICQLMDCRKLSVDASLHAVQNERLPTRV 458
+R DGLYRA+D +LK HP + E+ +C+ + C+KLS DAS HA QNERLP ++
Sbjct: 424 PDHARVVCDGLYRAVDIFLKVHPHMKDSERYRLCKTVSCKKLSQDASSHAAQNERLPVQI 483
Query: 459 IVQVLHFEQVR--TAASSGTSTPDIAKGIKDLSIGSNGSS-RSGTTNPEYDTDGAGTAEE 515
VQVL +EQ R A +SG T + L +GS SG +P + A E
Sbjct: 484 AVQVLFYEQTRLKNAMTSGGGTGGSNQSQFFLFPNRSGSGMASGAISPR--DNYASVRRE 541
Query: 516 LKALRKELASLR 527
+ LR E+A +R
Sbjct: 542 NRELRLEVARMR 553
>At5g66560 unknown protein
Length = 668
Score = 280 bits (715), Expect = 2e-75
Identities = 192/599 (32%), Positives = 306/599 (51%), Gaps = 86/599 (14%)
Query: 4 YVASELASDIIVSIGDTKFYLHKFPLISKSSHLQKLISLNN------------------- 44
+ + L SDI + + D F+LHKFPL+SKS L +LI+
Sbjct: 14 FCTTGLPSDIEIEVDDMTFHLHKFPLMSKSRKLHRLITEQETRSSSSMALITVIDPKVEE 73
Query: 45 ---------------EENINE----------IQISDIPGGASAFEICAKFCYGMTVTLNA 79
EE + E I++ D PG + +FE+ AKFCYG+ + L+A
Sbjct: 74 TDKKGKGHEIEDDKEEEEVEEQEIEENGYPHIKLEDFPGSSESFEMVAKFCYGVKMDLSA 133
Query: 80 YNVIATRCAAEYLGMHENIEKGNLLFKIDVFLSSSIFRSWKDSIILLQTTKSMSPLDEEQ 139
V+ RCAAE+L M E NL+ K + FLS S+++S ++SI L+ +S+SPL
Sbjct: 134 STVVPLRCAAEHLEMTEEYSPDNLISKTERFLSHSVYKSLRESIKALKACESVSPLAGSL 193
Query: 140 KVVNRCIESIANKAC-VDVSKVDWSYT-------YNRKKLPEENGFESNQNGVRTRNVPK 191
+ +CI+SI ++A D S W + L G + +R+
Sbjct: 194 GITEQCIDSIVSRASSADPSLFGWPVNDGGGRGNISATDLQLIPGGAAKSRKKPSRDSNM 253
Query: 192 DWWVEDLCELEVDMYKSVITNIKTKEIQSNDVIGEALKAYAYRKLPNFSKG-------MI 244
+ W EDL +L + ++K+VI ++++ ++ S+D+I L YA + +P +
Sbjct: 254 ELWFEDLTQLSLPIFKTVILSMRSGDL-SSDIIESCLICYAKKHIPGILRSNRKPPSSSS 312
Query: 245 PCEDVSKHRLIVETIVKLLPSDKGSVSC--RFLVKLLKAVIFVESEDRIRDVLVKRIGQQ 302
++ R ++ETI LP DK S+S RFL LL+ I + + + RD+L ++IG Q
Sbjct: 313 TAVSENEQRELLETITSNLPLDKSSISSTTRFLFGLLRTAIILNAAEICRDLLERKIGSQ 372
Query: 303 LEEASVNDILIKAPDG-EITMYDVGIVQKIVREFLMKDHNSEIELVGGGELEGIRKPGIL 361
LE A+++D+L+ + T+YDV +V++I+ FL S +V E++G
Sbjct: 373 LERATLDDLLVPSYSYLNETLYDVDLVERILGHFLDTLEQSNTAIV---EVDG------- 422
Query: 362 SDASKLMVAKLIDGYLAEIAKDPNLPLLNFVNLAELVSRISRPSHDGLYRAIDTYLKEHP 421
S ++V KLIDG+LAEIA D NL F NLA + +R DGLYRA+D YLK HP
Sbjct: 423 KSPSLMLVGKLIDGFLAEIASDANLKSDKFYNLAISLPDQARLYDDGLYRAVDVYLKAHP 482
Query: 422 GISKGEKKSICQLMDCRKLSVDASLHAVQNERLPTRVIVQVLHFEQV------------- 468
+S+ E++ IC +MDC+KL+++A HA QNERLP R +VQVL FEQ+
Sbjct: 483 WVSEAEREKICGVMDCQKLTLEACTHAAQNERLPLRAVVQVLFFEQLQLRHAIAGTLLAA 542
Query: 469 RTAASSGTSTPDIAKGIKDLSIGSNGSSRSGTTNPEYDTDGAGTAEELKALRKELASLR 527
++ ++S ++ P + I++L+I + T E + LR ++ ++R
Sbjct: 543 QSPSTSQSTEPRPSAAIRNLTITEEDGDEAEGERQVDAGKWKKTVRENQVLRLDMDTMR 601
>At3g08660 putative non-phototropic hypocotyl
Length = 582
Score = 273 bits (699), Expect = 2e-73
Identities = 173/480 (36%), Positives = 273/480 (56%), Gaps = 41/480 (8%)
Query: 5 VASELASDIIVSIGDTKFYLHKFPLISKSSHLQKLISLNNEENINEIQISDIPGGASAFE 64
+ S++A DIIV + F LHKFPL+++S ++K++ + +++ + I++ D PGG S FE
Sbjct: 30 IFSDVAGDIIVVVDGESFLLHKFPLVARSGKMRKMVR-DLKDSSSMIELRDFPGGPSTFE 88
Query: 65 ICAKFCYGMTVTLNAYNVIATRCAAEYLGMHENIEKGNLLFKIDVFLSSSIFRSWKDSII 124
+ KFCYG+ + A+NV++ RCAA YL M E+ ++ NL+F+ + +L +FRS+ +S++
Sbjct: 89 LTMKFCYGINFDITAFNVVSLRCAAGYLEMTEDYKEQNLIFRAENYLDQIVFRSFHESVL 148
Query: 125 LLQTTKSMSPLDEEQKVVNRCIESIANKACVDVSKVDWSYTYNRKKLPEENGFESNQNGV 184
+L + ++ + E ++ +RC+E+IA AC RK+L +G G
Sbjct: 149 VLCSCETQE-IAETYEIPDRCVEAIAMNAC-------------RKQLV--SGLSEELKGR 192
Query: 185 RTRNVPKDWWVEDLCELEVDMYKSVITNIKTKEIQSNDVIGEALKAYAYRKLPNFSKGMI 244
+ W E+L L +D Y V++ + ++S ++ +L YA L KG+I
Sbjct: 193 DCLEM----WTEELSALGIDYYVQVVSAMARLSVRSESIVA-SLVHYAKTSL----KGII 243
Query: 245 PCEDVSKHRLIVETIVKLLPSD-KGSVSCR-----FLVKLLKAVIFVESEDRIRDVLVKR 298
+ + R IVE +V LLP+D KGS S FL +LK ++ E R L +R
Sbjct: 244 D-RNCQEQRKIVEAMVNLLPNDEKGSYSLSIIPLGFLFGMLKVGTIIDIEISCRLELERR 302
Query: 299 IGQQLEEASVNDILIKAPDGEITMYDVGIVQKIVREFLMKDHNSEIELVGGGELEGIRKP 358
IG QLE AS++D+LI + E +MYDV V +I+ FL + + E + G
Sbjct: 303 IGHQLETASLDDLLIPSVQNEDSMYDVDTVHRILTFFLERIEEEDDECGYDSDSTG---- 358
Query: 359 GILSDASKLMVAKLIDGYLAEIAKDPNLPLLNFVNLAELVSRISRPSHDGLYRAIDTYLK 418
+S L V +++D YL EIA DP L L F + E + SR DG+YRAID YLK
Sbjct: 359 ---QHSSLLKVGRIMDAYLVEIAPDPYLSLHKFTAIIETLPEHSRIVDDGIYRAIDMYLK 415
Query: 419 EHPGISKGEKKSICQLMDCRKLSVDASLHAVQNERLPTRVIVQVLHFEQVR-TAASSGTS 477
HP +++ E+K +C +DC+KLS +AS H QN+RLP +++V+VL+ EQ+R A SG S
Sbjct: 416 AHPLLTEEERKKLCNFIDCKKLSQEASNHVAQNDRLPVQMVVRVLYTEQLRLKKALSGDS 475
>At5g10250 non-phototropic hypocotyl 3-like protein
Length = 607
Score = 267 bits (683), Expect = 1e-71
Identities = 179/553 (32%), Positives = 277/553 (49%), Gaps = 85/553 (15%)
Query: 4 YVASELASDIIVSIGDTKFYLHKFPLISKSSHLQKL-ISLNNEENINEIQISDIPGGASA 62
YV S++ +D+ + + D F HKFPLISK ++ + + + EN +++ + PGGA
Sbjct: 45 YVKSQIPTDLSIQVNDITFKAHKFPLISKCGYISSIELKPSTSENGYHLKLENFPGGADT 104
Query: 63 FEICAKFCYGMTVTLNAYNVIATRCAAEYLGMHENIEKGNLLFKIDVFLSSSIFRSWKDS 122
FE KFCY + + LN NV RCA+EYL M E E GNL+ K + F++ + SW+D+
Sbjct: 105 FETILKFCYNLPLDLNPLNVAPLRCASEYLYMTEEFEAGNLISKTEAFITFVVLASWRDT 164
Query: 123 IILLQTTKSMSPLDEEQKVVNRCIESIANKACVDVSKVDWSYTYNRKKLPEENGFESNQN 182
+ +L++ ++SP E ++V RC + +A KAC N +PE+
Sbjct: 165 LTVLRSCTNLSPWAENLQIVRRCCDLLAWKAC------------NDNNIPED-------- 204
Query: 183 GVRTRNVPKDWWVEDLCELEVDMYKSVITNIKTKEIQSNDVIGEALKAYAYRKLP----- 237
V RN + D+ L++D + VIT +K + + + G+ + YA LP
Sbjct: 205 -VVDRN--ERCLYNDIATLDIDHFMRVITTMKARRAKPQ-ITGKIIMKYADNFLPVINDD 260
Query: 238 --------------NFSKGMIPCEDVS-----KHRLIVETIVKLLPSDKGSVSCRFLVKL 278
FS E+ + +H+ +E++V +LP G+VSC FL+++
Sbjct: 261 LEGIKGYGLGKNELQFSVNRGRMEESNSLGCQEHKETIESLVSVLPPQSGAVSCHFLLRM 320
Query: 279 LKAVIFVESEDRIRDVLVKRIGQQLEEASVNDILIKAPDGEITMYDVGIVQKIVREFLMK 338
LK I + + L KR+G LE+A+V D+LI E V I EF +
Sbjct: 321 LKTSIVYSASPALISDLEKRVGMALEDANVCDLLIPNFKNEEQQERVRIF-----EFFLM 375
Query: 339 DHNSEIELVGGGELEGIRKPGILSDASKLMVAKLIDGYLAEIAKDPNLPLLNFVNLAELV 398
++ + KP I +KL+D YLAEIAKDP LP+ F LAE++
Sbjct: 376 HEQQQV----------LGKPSI---------SKLLDNYLAEIAKDPYLPITKFQVLAEML 416
Query: 399 SRISRPSHDGLYRAIDTYLKEHPGISKGEKKSICQLMDCRKLSVDASLHAVQNERLPTRV 458
+ HDGLYRAID +LK HP +S +++ +C+ M+C KLS+DA LHA QN+RLP R
Sbjct: 417 PENAWKCHDGLYRAIDMFLKTHPSLSDHDRRRLCKTMNCEKLSLDACLHAAQNDRLPLRT 476
Query: 459 IV----QVLHFEQVRTAASSGTSTPDIAKGIKDLSIGSNGSSRSGTTNPEYDTDGAGTAE 514
IV QVL EQV+ P+ ++ + G R N T E
Sbjct: 477 IVQINTQVLFSEQVKMRMMMQDKLPE----KEEENSGGREDKRMSRDNEIIKT----LKE 528
Query: 515 ELKALRKELASLR 527
EL+ ++K+++ L+
Sbjct: 529 ELENVKKKMSELQ 541
>At3g08570 hypothetical protein
Length = 612
Score = 265 bits (676), Expect = 7e-71
Identities = 178/538 (33%), Positives = 286/538 (53%), Gaps = 39/538 (7%)
Query: 5 VASELASDIIVSIGDTKFYLHKFPLISKSSHLQKLISLNNEE--NINEIQISDIPGGASA 62
+ S++A DI + + F LHKFPL+++ ++K+++ E N++ ++ D PGG+
Sbjct: 25 IFSDVAGDITIVVDGESFLLHKFPLVARCGKIRKMVAEMKESSSNLSHTELRDFPGGSKT 84
Query: 63 FEICAKFCYGMTVTLNAYNVIATRCAAEYLGMHENIEKGNLLFKIDVFLSSSIFRSWKDS 122
FE+ KFCYG+ + NV+A RCAA YL M E+ ++ NL+ + + +L FRS + S
Sbjct: 85 FELAMKFCYGINFEITISNVVAIRCAAGYLEMTEDFKEENLIARTETYLEQVAFRSLEKS 144
Query: 123 IILLQTTKSMSPLD--EEQKVVNRCIESIANKACVDVSKVDWSYTYNRKKLPEENGFESN 180
+ +L + +++ P D E + +RC+E+IA AC + + S NR
Sbjct: 145 VEVLCSCETLYPQDIAETAHIPDRCVEAIAVNACREQLVLGLS-RLNR----------GT 193
Query: 181 QNGVRTRNVPKDWWVEDLCELEVDMYKSVITNIKTKEIQSNDVIGEALKAYAYRKLPNFS 240
++G R +WW+EDL L +D Y V++ + ++S +I +L YA L
Sbjct: 194 ESGELKRGDSPEWWIEDLSALRIDYYARVVSAMARTGLRSESII-TSLMHYAQESL---- 248
Query: 241 KGMIPCEDVSK---------HRLIVETIVKLLPSDKGSVSCRFLVKLLKAVIFVESEDRI 291
KG+ C++ +K R ++E IV L P+D +V FL +L+ I +
Sbjct: 249 KGIRNCKERTKLDSGTFENEQRNVLEAIVSLFPND--NVPLSFLFGMLRVGITINVAISC 306
Query: 292 RDVLVKRIGQQLEEASVNDILIKAPDGEITMYDVGIVQKIVREFLMK-DHNSEIELVGGG 350
R L +RI QQLE S++D+LI +MYDV V +I+ FL K + E +
Sbjct: 307 RLELERRIAQQLETVSLDDLLIPVVRDGDSMYDVDTVHRILVCFLKKIEEEEEYDEDCCY 366
Query: 351 ELEGIRKPGILSDASKLMVAKLIDGYLAEIAKDPNLPLLNFVNLAELVSRISRPSHDGLY 410
E E G + +S L V +++D YLAEIA DP L L F+ L E++ +R DGLY
Sbjct: 367 ENETENLTGSMCHSSLLKVGRIMDAYLAEIAPDPCLSLHKFMALIEILPDYARVMDDGLY 426
Query: 411 RAIDTYLKEHPGISKGEKKSICQLMDCRKLSVDASLHAVQNERLPTRVIVQVLHFEQVRT 470
RAID +LK HP +++ E KS+C+ +D +KLS +A H QN+RLP +++V+VL+ EQ+R
Sbjct: 427 RAIDMFLKGHPSLNEQECKSLCKFIDTQKLSQEACNHVAQNDRLPMQMVVRVLYSEQLRM 486
Query: 471 A-ASSGTSTPDIAKGIKDLSIGSNGSSRSGTTNPEYDTDGAGTAEELKALRKELASLR 527
SG S + + S SR+ + Y A E + L+ E++ +R
Sbjct: 487 KNVMSGESGEGLL--LSSQKHSSENPSRAVSPRDTY----ASLRRENRELKLEISRVR 538
>At3g50840 putative protein
Length = 567
Score = 257 bits (657), Expect = 1e-68
Identities = 169/482 (35%), Positives = 257/482 (53%), Gaps = 55/482 (11%)
Query: 7 SELASDIIVSIGDTKFYLHKFPLISKSSHLQKLISLNNEENI-NEIQISDIPGGASAFEI 65
+ L SDI + + D F+LHKFPL+SKS L +LI+ + + + I++ + PGG+ FE+
Sbjct: 12 NSLPSDIEIEVDDITFHLHKFPLMSKSKKLHQLITEQEQSKVYSHIKLENFPGGSEIFEM 71
Query: 66 CAKFCYGMTVTLNAYNVIATRCAAEYLGMHENIEKGNLLFKIDVFLSSSIFRSWKDSIIL 125
K YG V ++ + RCAAEYL M E NL+ K + FLS +F + ++SI
Sbjct: 72 VIKISYGFKVDISVSTAVPLRCAAEYLEMTEEYSPENLISKTEKFLSEFVFTNVQESIKA 131
Query: 126 LQTTKSMSPLDEEQKVVNRCIESIANKACVDVSKVDWSYTYNRKKLPEENGFESNQNGVR 185
L+ +S+S L E + +CI+SI +A S D S Y G+ N G+
Sbjct: 132 LKACESVSSLAESLCITEQCIDSIVFQA----SSTDPSSFY---------GWPINNGGIF 178
Query: 186 TRNVPK-------DWWVEDLCELEVDMYKSVITNIKTKEIQSNDVIGEALKAYAYRKLPN 238
T + K + W EDL EL +++ VI ++K+ + S +++ +L YA + +P
Sbjct: 179 TVDRKKQSKDSKTELWFEDLTELSFPIFRRVILSMKSS-VLSPEIVERSLLTYAKKHIPG 237
Query: 239 FSKGMIPCEDVS----------KHRLIVETIVKLLPSDKGSVSCRFLVKLLKAVIFVESE 288
S+ S + R ++ETI LP + + R L LL+A I + +
Sbjct: 238 ISRSSSASSSSSSSSTTIASENQQRELLETITSDLPLT--ATTTRSLFGLLRAAIILNAS 295
Query: 289 DRIRDVLVKRIGQQLEEASVNDILIKAPDG-EITMYDVGIVQKIVREFLMKDHNSEIELV 347
+ R L K+IG LE+A+++D+LI + T+YD+ +V++++R FL
Sbjct: 296 ENCRKFLEKKIGSNLEKATLDDLLIPSYSYLNETLYDIDLVERLLRRFL----------- 344
Query: 348 GGGELEGIRKPGILSDASKLMVAKLIDGYLAEIAKDPNLPLLNFVNLAELVSRISRPSHD 407
E + +S +S +V +LIDG L EIA D NL F NLA L+ +R D
Sbjct: 345 -----ENVA----VSSSSLTVVGRLIDGVLGEIASDANLKPEQFYNLAVLLPVQARVYDD 395
Query: 408 GLYRAIDTYLKEHPGISKGEKKSICQLMDCRKLSVDASLHAVQNERLPTRVIVQVLHFEQ 467
GLYRA+D Y K H I + EK+ IC +MDCRKL+V+ HA QNERLP R +VQVL EQ
Sbjct: 396 GLYRAVDIYFKTHSWILEEEKEKICSVMDCRKLTVEGCTHAAQNERLPLRAVVQVLFLEQ 455
Query: 468 VR 469
++
Sbjct: 456 LQ 457
>At3g49970 putative protein
Length = 526
Score = 250 bits (639), Expect = 1e-66
Identities = 194/603 (32%), Positives = 292/603 (48%), Gaps = 101/603 (16%)
Query: 22 FYLHKFPLISKSSHLQKLISLNNEENINEIQISDIPGGASAFEICAKFCYGMTVTLNAYN 81
F LHKFPL+SK ++KL S ++ ++ N I+I D PGGA FE+ KFCY ++ +N N
Sbjct: 7 FLLHKFPLVSKCGFIKKLASESSNDS-NIIRIPDFPGGAEGFELVIKFCYDISFEINTEN 65
Query: 82 VIATRCAAEYLGMHENIEKGNLLFKIDVFLSSSIFRSWKDSIILLQTTKSMSPLDEEQKV 141
+ CAAEYL M E NL+ I+V+L+ I +S S+ +LQ ++ + P+ E ++
Sbjct: 66 IAMLLCAAEYLEMTEEHSVENLVETIEVYLNEVILKSLSKSVKVLQKSQDLLPIAERVRL 125
Query: 142 VNRCIESIANKACVDVSKVDWSYTYNRKKLPEENGFESNQNGVRTRNVPKDWWVEDLCEL 201
V+RCI+SIA C + +SN++ V DWW +DL L
Sbjct: 126 VDRCIDSIAYAICQES--------------------QSNEDIV-------DWWADDLAVL 158
Query: 202 EVDMYKSVITNIKTKEIQSNDVIGEALKAYAYRKLPN--------------FSKGMIPCE 247
++DM++ V+ + + + +G LK YA + L F K E
Sbjct: 159 KIDMFRRVLVAMIARGFKRYS-LGPVLKLYAEKALRGLVRFLNFLTEQCDIFGKEAKKME 217
Query: 248 DVSKH--RLIVETIVKLLPSDKGSVSCRFLVKLLKAVIFVESEDRIRDVLVKRIGQQLEE 305
+H RLI+ETIV LLP ++ SVS FL LL+A I++E+ R L KR+G QL +
Sbjct: 218 AEQEHEKRLILETIVSLLPRERNSVSVSFLSILLRAAIYLETTVACRLDLEKRMGLQLRQ 277
Query: 306 ASVNDILIK--APDGEITMYDVGIVQKIVREFLMKDHNSEIELVGGGELEGIRKPGILSD 363
A ++D+LI + +G+ TM DV VQ+I+ +L E E+ G SD
Sbjct: 278 AVIDDLLIPYYSFNGDNTMLDVDTVQRILMNYL------EFEVEGNSA-------DFASD 324
Query: 364 ASKLMVAKLIDGYLAEIAKDPNLPLLNFVNLAELVSRISRPSHDGLYRAIDTYLKEHPGI 423
+LM + YLAEIA D N+ F+ AE + + SR +YRAID +LK HP I
Sbjct: 325 IGELM-----ETYLAEIASDRNINFAKFIGFAECIPKQSR-----MYRAIDIFLKTHPNI 374
Query: 424 SKGEKKSICQLMDCRKLSVDASLHAVQNERLPTRVIVQVLHFEQVRTAASSGTSTPDIAK 483
S+ EKK +C LMDC+KLS D HA QN+R E + + S +T +
Sbjct: 375 SEVEKKKVCSLMDCKKLSRDVYAHAAQNDRFQ----------ENLSNSDSPAPATAEKTL 424
Query: 484 GIKDLSIGSNGSSRSGTTNPEYDTDGAGTAEELKALRKELASLRQSNGVGNSFKDGDAKP 543
+LS N S+ N + + K L KE A S D
Sbjct: 425 SPPELSSYKNELSKLNRENQYLKLELLKVKMKFKELEKEKAFEVMSG--------SDCSS 476
Query: 544 NMDKAVIGKMKGLLKSKKSFIKLWASKGGHGENSGSDSSESIGSVNPEEVKSTPSRNRRN 603
++ A + K + +KSFI + K G + I ++ ++ ++RR+
Sbjct: 477 SVSTASVAKPR---LPRKSFINSVSQKLG----------KLINPFGLKQGQTKQPKSRRH 523
Query: 604 SVS 606
S+S
Sbjct: 524 SIS 526
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.315 0.133 0.372
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,394,489
Number of Sequences: 26719
Number of extensions: 583012
Number of successful extensions: 1597
Number of sequences better than 10.0: 50
Number of HSP's better than 10.0 without gapping: 36
Number of HSP's successfully gapped in prelim test: 14
Number of HSP's that attempted gapping in prelim test: 1425
Number of HSP's gapped (non-prelim): 59
length of query: 606
length of database: 11,318,596
effective HSP length: 105
effective length of query: 501
effective length of database: 8,513,101
effective search space: 4265063601
effective search space used: 4265063601
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 63 (28.9 bits)
Medicago: description of AC146551.5


