

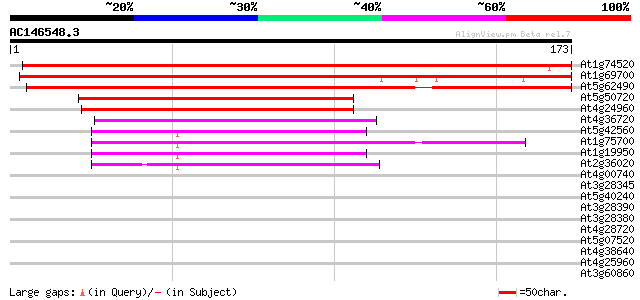
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146548.3 + phase: 0
(173 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g74520 AtHVA22a 268 1e-72
At1g69700 AtHVA22c 211 1e-55
At5g62490 AtHVA22b-like protein 199 6e-52
At5g50720 unknown protein 120 3e-28
At4g24960 abscisic acid-induced like protein 112 1e-25
At4g36720 putative protein 67 4e-12
At5g42560 unknown protein 64 4e-11
At1g75700 hypothetical protein 62 2e-10
At1g19950 unknown protein 62 2e-10
At2g36020 unknown protein 56 9e-09
At4g00740 unknown protein 30 0.50
At3g28345 P-glycoprotein, putative, 3'partial 29 1.5
At5g40240 unknown protein 28 2.5
At3g28390 P-glycoprotein, putative 28 2.5
At3g28380 P-glycoprotein, putative 28 2.5
At4g28720 unknown protein 27 4.2
At5g07520 glycine-rich protein atGRP 27 5.5
At4g38640 unknown protein 27 7.2
At4g25960 P-glycoprotein-2 (pgp2) 27 7.2
At3g60860 guanine nucleotide exchange factor - like protein 27 7.2
>At1g74520 AtHVA22a
Length = 177
Score = 268 bits (684), Expect = 1e-72
Identities = 124/174 (71%), Positives = 147/174 (84%), Gaps = 5/174 (2%)
Query: 5 GSGAGSFLMVLLRNFDVLAGPVISLVYPLYASVRAIESKSPVDDQQWLTYWVLYSLITLF 64
GSGAG+FL VLLRNFDVLAGPV+SLVYPLYASV+AIE++S DD+QWLTYWVLYSL+TL
Sbjct: 2 GSGAGNFLKVLLRNFDVLAGPVVSLVYPLYASVQAIETQSHADDKQWLTYWVLYSLLTLI 61
Query: 65 ELTFAKILEWIPIWPYAKLILTCWLVLPYFTGAAYVYEHYVRPFLANPQTINIWYVPRKK 124
ELTFAK++EW+PIW Y KLILTCWLV+PYF+GAAYVYEH+VRP NP++INIWYVP+K
Sbjct: 62 ELTFAKLIEWLPIWSYMKLILTCWLVIPYFSGAAYVYEHFVRPVFVNPRSINIWYVPKKM 121
Query: 125 DVFTKQDDIITAAEKYIKENGTEAFENLIHRADKSKWGSSH-----HTMYDETY 173
D+F K DD++TAAEKYI ENG +AFE ++ RADKSK + H TMY E Y
Sbjct: 122 DIFRKPDDVLTAAEKYIAENGPDAFEKILSRADKSKRYNKHEYESYETMYGEGY 175
>At1g69700 AtHVA22c
Length = 184
Score = 211 bits (538), Expect = 1e-55
Identities = 103/180 (57%), Positives = 138/180 (76%), Gaps = 10/180 (5%)
Query: 4 SGSGAGSFLMVLLRNFDVLAGPVISLVYPLYASVRAIESKSPVDDQQWLTYWVLYSLITL 63
S SG + L VL++NFDVLA P+++LVYPLYASV+AIE++S +D+QWLTYWVLY+LI+L
Sbjct: 3 SNSGDDNVLQVLIKNFDVLALPLVTLVYPLYASVKAIETRSLPEDEQWLTYWVLYALISL 62
Query: 64 FELTFAKILEWIPIWPYAKLILTCWLVLPYFTGAAYVYEHYVRPFLANPQ--TINIWYVP 121
FELTF+K LEW PIWPY KL CWLVLP F GA ++Y+H++RPF +PQ T IWYVP
Sbjct: 63 FELTFSKPLEWFPIWPYMKLFGICWLVLPQFNGAEHIYKHFIRPFYRDPQRATTKIWYVP 122
Query: 122 RKK-DVFTKQ--DDIITAAEKYIKENGTEAFENLIHRAD-----KSKWGSSHHTMYDETY 173
KK + F K+ DDI+TAAEKY++++GTEAFE +I + D +S G ++H ++D+ Y
Sbjct: 123 HKKFNFFPKRDDDDILTAAEKYMEQHGTEAFERMIVKKDSYERGRSSRGINNHMIFDDDY 182
>At5g62490 AtHVA22b-like protein
Length = 167
Score = 199 bits (506), Expect = 6e-52
Identities = 98/168 (58%), Positives = 121/168 (71%), Gaps = 5/168 (2%)
Query: 6 SGAGSFLMVLLRNFDVLAGPVISLVYPLYASVRAIESKSPVDDQQWLTYWVLYSLITLFE 65
SG GS + V+ +NFDV+AGPVISLVYPLYASVRAIES+S DD+QWLTYW LYSLI LFE
Sbjct: 3 SGIGSLVKVIFKNFDVIAGPVISLVYPLYASVRAIESRSHGDDKQWLTYWALYSLIKLFE 62
Query: 66 LTFAKILEWIPIWPYAKLILTCWLVLPYFTGAAYVYEHYVRPFLANPQTINIWYVPRKKD 125
LTF ++LEWIP++PYAKL LT WLVLP GAAY+YEHYVR FL +P T+N+WYVP KKD
Sbjct: 63 LTFFRLLEWIPLYPYAKLALTSWLVLPGMNGAAYLYEHYVRSFLLSPHTVNVWYVPAKKD 122
Query: 126 VFTKQDDIITAAEKYIKENGTEAFENLIHRADKSKWGSSHHTMYDETY 173
DD+ A K+ N + A + I + + H+ +D+ Y
Sbjct: 123 -----DDLGATAGKFTPVNDSGAPQEKIVSSVDTSAKYVGHSAFDDAY 165
>At5g50720 unknown protein
Length = 116
Score = 120 bits (302), Expect = 3e-28
Identities = 55/85 (64%), Positives = 64/85 (74%)
Query: 22 LAGPVISLVYPLYASVRAIESKSPVDDQQWLTYWVLYSLITLFELTFAKILEWIPIWPYA 81
LAGPV+ L+YPLYASV AIES S VDD+QWL YW+LYS +TL EL +LEWIPIW A
Sbjct: 14 LAGPVVMLLYPLYASVIAIESPSKVDDEQWLAYWILYSFLTLSELILQSLLEWIPIWYTA 73
Query: 82 KLILTCWLVLPYFTGAAYVYEHYVR 106
KL+ WLVLP F GAA++Y VR
Sbjct: 74 KLVFVAWLVLPQFRGAAFIYNKVVR 98
>At4g24960 abscisic acid-induced like protein
Length = 135
Score = 112 bits (280), Expect = 1e-25
Identities = 47/84 (55%), Positives = 62/84 (72%)
Query: 23 AGPVISLVYPLYASVRAIESKSPVDDQQWLTYWVLYSLITLFELTFAKILEWIPIWPYAK 82
AGP++ L+YPLYASV A+ES + VDD+QWL YW++YS ++L EL ++EWIPIW K
Sbjct: 15 AGPIVMLLYPLYASVIAMESTTKVDDEQWLAYWIIYSFLSLTELILQSLIEWIPIWYTVK 74
Query: 83 LILTCWLVLPYFTGAAYVYEHYVR 106
L+ WLVLP F GAA++Y VR
Sbjct: 75 LVFVAWLVLPQFQGAAFIYNRVVR 98
>At4g36720 putative protein
Length = 227
Score = 67.4 bits (163), Expect = 4e-12
Identities = 30/87 (34%), Positives = 47/87 (53%)
Query: 27 ISLVYPLYASVRAIESKSPVDDQQWLTYWVLYSLITLFELTFAKILEWIPIWPYAKLILT 86
I + P+Y++ +AIES + Q+ L YW Y +L E+ KI+ W P++ + K
Sbjct: 44 IGIGLPVYSTFKAIESGDENEQQKMLIYWAAYGSFSLVEVFTDKIISWFPLYYHVKFAFL 103
Query: 87 CWLVLPYFTGAAYVYEHYVRPFLANPQ 113
WL LP G+ +Y + +RPFL Q
Sbjct: 104 VWLQLPTVEGSKQIYNNQIRPFLLRHQ 130
>At5g42560 unknown protein
Length = 296
Score = 63.9 bits (154), Expect = 4e-11
Identities = 29/87 (33%), Positives = 45/87 (51%), Gaps = 2/87 (2%)
Query: 26 VISLVYPLYASVRAIESKSPVDDQQ--WLTYWVLYSLITLFELTFAKILEWIPIWPYAKL 83
V+ YP Y + +E P +Q W YW+L + +T+FE + W+P++ AKL
Sbjct: 13 VLGYAYPAYECYKTVEKNRPEIEQLRFWCQYWILVACLTVFERVGDAFVSWVPMYSEAKL 72
Query: 84 ILTCWLVLPYFTGAAYVYEHYVRPFLA 110
+L P G YVYE + RP+L+
Sbjct: 73 AFFIYLWYPKTRGTTYVYESFFRPYLS 99
>At1g75700 hypothetical protein
Length = 166
Score = 62.0 bits (149), Expect = 2e-10
Identities = 36/136 (26%), Positives = 64/136 (46%), Gaps = 4/136 (2%)
Query: 26 VISLVYPLYASVRAIESKSPVDDQQ--WLTYWVLYSLITLFELTFAKILEWIPIWPYAKL 83
V YP Y + +E P Q W YW++ + +T+FE ++ W+P++ AKL
Sbjct: 2 VFGYAYPAYECFKTVELNKPEIQQLQFWCQYWIIVAALTIFERIGDALVSWLPMYSEAKL 61
Query: 84 ILTCWLVLPYFTGAAYVYEHYVRPFLANPQTINIWYVPRKKDVFTKQDDIITAAEKYIKE 143
+L P G YVY+ + RP++A + + + K +D + +K I +
Sbjct: 62 AFFIYLWFPKTKGTTYVYDSFFRPYIAKHENEIDRNLVKVKT--RAKDMAMIYLQKAINQ 119
Query: 144 NGTEAFENLIHRADKS 159
T+ FE L + ++S
Sbjct: 120 GQTKFFEILQYITEQS 135
>At1g19950 unknown protein
Length = 315
Score = 62.0 bits (149), Expect = 2e-10
Identities = 28/87 (32%), Positives = 44/87 (50%), Gaps = 2/87 (2%)
Query: 26 VISLVYPLYASVRAIESKSPVDDQQ--WLTYWVLYSLITLFELTFAKILEWIPIWPYAKL 83
V YP Y +A+E P Q W YW+L + +T+FE + W+P++ AKL
Sbjct: 13 VFGYAYPAYECYKAVEKNKPEMQQLRFWCQYWILVAALTIFERVGDALASWVPLYCEAKL 72
Query: 84 ILTCWLVLPYFTGAAYVYEHYVRPFLA 110
+L P G YVY+ + +P++A
Sbjct: 73 AFFIYLWFPKTRGTTYVYDSFFQPYVA 99
>At2g36020 unknown protein
Length = 258
Score = 56.2 bits (134), Expect = 9e-09
Identities = 26/92 (28%), Positives = 50/92 (54%), Gaps = 4/92 (4%)
Query: 26 VISLVYPLYASVRAIESKSPVDDQQ---WLTYWVLYSLITLFELTFAKILEWIPIWPYAK 82
++ YP + + +E K+ VD ++ W YW+L +LI+ FE + W+P++ K
Sbjct: 13 ILGYTYPAFECFKTVE-KNKVDIEELRFWCQYWILLALISSFERVGDFFISWLPLYGEMK 71
Query: 83 LILTCWLVLPYFTGAAYVYEHYVRPFLANPQT 114
++ +L P G +VYE ++P++A +T
Sbjct: 72 VVFFVYLWYPKTKGTRHVYETLLKPYMAQHET 103
>At4g00740 unknown protein
Length = 600
Score = 30.4 bits (67), Expect = 0.50
Identities = 28/126 (22%), Positives = 50/126 (39%), Gaps = 22/126 (17%)
Query: 68 FAKILEWIPIWPY----AKLILTCWLVLPYFTGAAYVYEHYVRPFLANPQTINIWYVPRK 123
FA L P+W A+ LT ++ Y G VY + PF P+T + +V
Sbjct: 457 FAATLASDPVWVMNVIPARKPLTLDVI--YDRGLIGVYHDWCEPFSTYPRTYDFIHVSGI 514
Query: 124 KDVFTKQD---------DIITAAEKYIKENG-------TEAFENLIHRADKSKWGSSHHT 167
+ + +QD D++ ++ ++ G E + + A +W SS H
Sbjct: 515 ESLIKRQDSSKSRCSLVDLMVEMDRILRPEGKVVIRDSPEVLDKVARMAHAVRWSSSIHE 574
Query: 168 MYDETY 173
E++
Sbjct: 575 KEPESH 580
>At3g28345 P-glycoprotein, putative, 3'partial
Length = 610
Score = 28.9 bits (63), Expect = 1.5
Identities = 12/27 (44%), Positives = 19/27 (69%)
Query: 1 MGGSGSGAGSFLMVLLRNFDVLAGPVI 27
+GGSGSG + + +L R +D LAG ++
Sbjct: 393 VGGSGSGKSTVISLLQRFYDPLAGEIL 419
>At5g40240 unknown protein
Length = 368
Score = 28.1 bits (61), Expect = 2.5
Identities = 28/121 (23%), Positives = 53/121 (43%), Gaps = 21/121 (17%)
Query: 1 MGGSGSGAGSFLMVLLRNFDVLAGPVISLVYP---LYASVRAIESKSPVDDQQWLTYWVL 57
+G S +G+ ++VL + VLA + V P L+ + +IES + + + L
Sbjct: 148 IGAILSISGALVVVLYKGPQVLASASFTTVLPTVTLHQQLTSIESSWIIGGLLLASQYFL 207
Query: 58 YSL-----------------ITLFELTFAKILEWIPIWPYAKLILTCWLVLPYFTGAAYV 100
S+ + F FA ++ +P+ +A+ LT W++ P + AA +
Sbjct: 208 ISVWYILQTRVMEVYPEEITVVFFYNLFATLIS-VPVCLFAESNLTSWVLKPDISLAAII 266
Query: 101 Y 101
Y
Sbjct: 267 Y 267
>At3g28390 P-glycoprotein, putative
Length = 1225
Score = 28.1 bits (61), Expect = 2.5
Identities = 11/27 (40%), Positives = 19/27 (69%)
Query: 1 MGGSGSGAGSFLMVLLRNFDVLAGPVI 27
+GGSGSG + + +L R +D +AG ++
Sbjct: 381 VGGSGSGKSTVISLLQRFYDPIAGEIL 407
>At3g28380 P-glycoprotein, putative
Length = 1240
Score = 28.1 bits (61), Expect = 2.5
Identities = 11/27 (40%), Positives = 19/27 (69%)
Query: 1 MGGSGSGAGSFLMVLLRNFDVLAGPVI 27
+GGSGSG + + +L R +D +AG ++
Sbjct: 393 VGGSGSGKSTVISLLQRFYDPIAGEIL 419
>At4g28720 unknown protein
Length = 426
Score = 27.3 bits (59), Expect = 4.2
Identities = 13/29 (44%), Positives = 18/29 (61%), Gaps = 2/29 (6%)
Query: 64 FELTFAKILEWIPIWPYAKLILT-CWLVL 91
FEL K+L W P+W K++L W+VL
Sbjct: 239 FELAM-KMLRWFPLWLVDKILLVLSWMVL 266
>At5g07520 glycine-rich protein atGRP
Length = 228
Score = 26.9 bits (58), Expect = 5.5
Identities = 12/33 (36%), Positives = 21/33 (63%)
Query: 10 SFLMVLLRNFDVLAGPVISLVYPLYASVRAIES 42
SFL+ LL F V+ V+S+V+ ++A + + S
Sbjct: 3 SFLIFLLEAFQVVIATVVSIVFLVFAGLTLVGS 35
>At4g38640 unknown protein
Length = 556
Score = 26.6 bits (57), Expect = 7.2
Identities = 14/60 (23%), Positives = 27/60 (44%), Gaps = 7/60 (11%)
Query: 47 DDQQWLTYWVLYSLITLFELTFAKILEWIPIWPYAKLILTCWLVLPYFTGAAYVYEHYVR 106
D W + V+Y +++ + F K L W L++T L +P+ + +HY +
Sbjct: 117 DGYYWASSSVMYGMVSSSDPVFEKDLIW-------TLVVTLILSVPFCFSVLLLLKHYTK 169
>At4g25960 P-glycoprotein-2 (pgp2)
Length = 1233
Score = 26.6 bits (57), Expect = 7.2
Identities = 12/27 (44%), Positives = 18/27 (66%)
Query: 1 MGGSGSGAGSFLMVLLRNFDVLAGPVI 27
+G SGSG S + ++LR +D AG V+
Sbjct: 1024 VGQSGSGKSSVISLILRFYDPTAGKVM 1050
>At3g60860 guanine nucleotide exchange factor - like protein
Length = 1793
Score = 26.6 bits (57), Expect = 7.2
Identities = 10/49 (20%), Positives = 24/49 (48%)
Query: 123 KKDVFTKQDDIITAAEKYIKENGTEAFENLIHRADKSKWGSSHHTMYDE 171
K ++ K+DD+ ++Y N + +++ + +WG S+ D+
Sbjct: 796 KHEIKMKEDDLRLQQKQYANSNRMLGLDGILNIVIRKQWGDSYAETSDD 844
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.323 0.139 0.446
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,153,628
Number of Sequences: 26719
Number of extensions: 171414
Number of successful extensions: 369
Number of sequences better than 10.0: 27
Number of HSP's better than 10.0 without gapping: 22
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 341
Number of HSP's gapped (non-prelim): 30
length of query: 173
length of database: 11,318,596
effective HSP length: 92
effective length of query: 81
effective length of database: 8,860,448
effective search space: 717696288
effective search space used: 717696288
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 56 (26.2 bits)
Medicago: description of AC146548.3


