

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146548.14 - phase: 0
(180 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
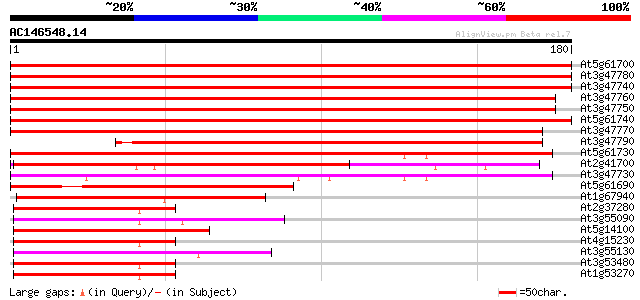
At5g61700 ABC family transporter - like protein 289 6e-79
At3g47780 ABC-type transport protein-like protein 287 2e-78
At3g47740 ABC-type transport protein-like protein 283 5e-77
At3g47760 ABC-type transport protein-like protein 276 3e-75
At3g47750 ABC-type transport protein-like protein 276 3e-75
At5g61740 ABC family transporter - like protein 276 6e-75
At3g47770 ABC-type transport protein-like protein 271 1e-73
At3g47790 ABC-type transport protein-like protein 182 8e-47
At5g61730 ABC transport protein - like 130 5e-31
At2g41700 ATP-binding cassette transporter ABCA1 127 4e-30
At3g47730 ABC-type transport protein-like protein 116 7e-27
At5g61690 ABC transport protein - like 101 2e-22
At1g67940 ABC transporter like protein 51 3e-07
At2g37280 putative ABC transporter 48 2e-06
At3g55090 ABC transporter - like protein 48 3e-06
At5g14100 unknown protein 47 4e-06
At4g15230 ABC transporter like protein 47 4e-06
At3g55130 ABC transporter - like protein 47 6e-06
At3g53480 ABC transporter - like protein 47 6e-06
At1g53270 putative ABC transporter emb|AAD22683.1 47 6e-06
>At5g61700 ABC family transporter - like protein
Length = 888
Score = 289 bits (739), Expect = 6e-79
Identities = 141/180 (78%), Positives = 162/180 (89%)
Query: 1 MKRRLSVAISLIGDPKVVYMDEPSTGLDPASRKCLWNVIKLAKQDRAIILTTHSMEEADA 60
MKRRLSVAISLIG+PKVVY+DEPSTGLDPASRK LWNVIK AKQ+ AIILTTHSMEEA+
Sbjct: 709 MKRRLSVAISLIGNPKVVYLDEPSTGLDPASRKNLWNVIKRAKQNTAIILTTHSMEEAEF 768
Query: 61 LCDRLGIFVDGSLQCVGYPKELKGRYGGTYVFAMTTSSDHEKDVENMVQRLTPNANKIYH 120
LCDRLGIFVDG LQC+G KELK RYGG+YVF MTTSS HE++VE +V+ ++PNA KIYH
Sbjct: 769 LCDRLGIFVDGGLQCIGNSKELKSRYGGSYVFTMTTSSKHEEEVERLVESVSPNAKKIYH 828
Query: 121 LSGTQKFELPKEEIKMANVFRAIEAAKRNFTVFAWGLSDTTLEDVFIKVAREAQAFDTLT 180
L+GTQKFELPK+E+++A VFRA+E AK NFTVFAWGL+DTTLEDVFIKVAR AQAF +L+
Sbjct: 829 LAGTQKFELPKQEVRIAEVFRAVEKAKANFTVFAWGLADTTLEDVFIKVARTAQAFISLS 888
>At3g47780 ABC-type transport protein-like protein
Length = 900
Score = 287 bits (735), Expect = 2e-78
Identities = 137/180 (76%), Positives = 161/180 (89%)
Query: 1 MKRRLSVAISLIGDPKVVYMDEPSTGLDPASRKCLWNVIKLAKQDRAIILTTHSMEEADA 60
MKRRLSVAISLIG+PKVVYMDEPSTGLDPASRK LW VIK AKQ+ AIILTTHSMEEA+
Sbjct: 721 MKRRLSVAISLIGNPKVVYMDEPSTGLDPASRKNLWTVIKRAKQNTAIILTTHSMEEAEF 780
Query: 61 LCDRLGIFVDGSLQCVGYPKELKGRYGGTYVFAMTTSSDHEKDVENMVQRLTPNANKIYH 120
LCDRLGIFVDG LQC+G PKELKGRYGG+YVF MTTSS+HE++VE +++ ++PNA KIYH
Sbjct: 781 LCDRLGIFVDGGLQCIGNPKELKGRYGGSYVFTMTTSSEHEQNVEKLIKDVSPNAKKIYH 840
Query: 121 LSGTQKFELPKEEIKMANVFRAIEAAKRNFTVFAWGLSDTTLEDVFIKVAREAQAFDTLT 180
++GTQKFELPKEE++++ VF+A+E AK NFTVFAWGL+DTTLEDVFIKV R QAF+ +
Sbjct: 841 IAGTQKFELPKEEVRISEVFQAVEKAKSNFTVFAWGLADTTLEDVFIKVVRNGQAFNVFS 900
>At3g47740 ABC-type transport protein-like protein
Length = 925
Score = 283 bits (723), Expect = 5e-77
Identities = 136/180 (75%), Positives = 158/180 (87%)
Query: 1 MKRRLSVAISLIGDPKVVYMDEPSTGLDPASRKCLWNVIKLAKQDRAIILTTHSMEEADA 60
MKRRLSVAISLIG+PKVVYMDEPSTGLDPASRK LW VIK AK+ AIILTTHSMEEA+
Sbjct: 746 MKRRLSVAISLIGNPKVVYMDEPSTGLDPASRKNLWTVIKNAKRHTAIILTTHSMEEAEF 805
Query: 61 LCDRLGIFVDGSLQCVGYPKELKGRYGGTYVFAMTTSSDHEKDVENMVQRLTPNANKIYH 120
LCDRLGIFVDG LQC+G PKELKGRYGG+YV MTTSS+HEKDVE +VQ ++PN KIYH
Sbjct: 806 LCDRLGIFVDGRLQCIGNPKELKGRYGGSYVLTMTTSSEHEKDVEMLVQEVSPNVKKIYH 865
Query: 121 LSGTQKFELPKEEIKMANVFRAIEAAKRNFTVFAWGLSDTTLEDVFIKVAREAQAFDTLT 180
++GTQKFE+PK+E++++ VF+ +E AK NF VFAWGL+DTTLEDVFIKVAR AQAF+ +
Sbjct: 866 IAGTQKFEIPKDEVRISEVFQVVEKAKSNFKVFAWGLADTTLEDVFIKVARTAQAFNVFS 925
>At3g47760 ABC-type transport protein-like protein
Length = 664
Score = 276 bits (707), Expect = 3e-75
Identities = 135/175 (77%), Positives = 152/175 (86%)
Query: 1 MKRRLSVAISLIGDPKVVYMDEPSTGLDPASRKCLWNVIKLAKQDRAIILTTHSMEEADA 60
MKRRLSVAISLIG PKVVYMDEPSTGLDPASR+ LW IK AK AIILTTHSMEEA+
Sbjct: 485 MKRRLSVAISLIGSPKVVYMDEPSTGLDPASRRSLWTAIKRAKNHTAIILTTHSMEEAEF 544
Query: 61 LCDRLGIFVDGSLQCVGYPKELKGRYGGTYVFAMTTSSDHEKDVENMVQRLTPNANKIYH 120
LCDRLGIFVDG LQCVG PKELK RYGG+YV MTT S+HEKDVE +VQ ++PNA KIYH
Sbjct: 545 LCDRLGIFVDGRLQCVGNPKELKARYGGSYVLTMTTPSEHEKDVEMLVQDVSPNAKKIYH 604
Query: 121 LSGTQKFELPKEEIKMANVFRAIEAAKRNFTVFAWGLSDTTLEDVFIKVAREAQA 175
++GTQKFE+PKEE++++ VF+A+E AK NF VFAWGL+DTTLEDVFIKVAR AQA
Sbjct: 605 IAGTQKFEIPKEEVRISEVFQAVEKAKDNFRVFAWGLADTTLEDVFIKVARTAQA 659
>At3g47750 ABC-type transport protein-like protein
Length = 895
Score = 276 bits (707), Expect = 3e-75
Identities = 134/175 (76%), Positives = 154/175 (87%)
Query: 1 MKRRLSVAISLIGDPKVVYMDEPSTGLDPASRKCLWNVIKLAKQDRAIILTTHSMEEADA 60
MKRRLSVAISLIG PKVVYMDEPSTGLDPASR+ LW IK AK+ AIILTTHSMEEA+
Sbjct: 716 MKRRLSVAISLIGSPKVVYMDEPSTGLDPASRRSLWTAIKGAKKHTAIILTTHSMEEAEF 775
Query: 61 LCDRLGIFVDGSLQCVGYPKELKGRYGGTYVFAMTTSSDHEKDVENMVQRLTPNANKIYH 120
LCDRLGIFVDG LQCVG PKELK RYGG+YV MTTSS+HEKDVE ++Q ++PNA KIYH
Sbjct: 776 LCDRLGIFVDGRLQCVGNPKELKARYGGSYVLTMTTSSEHEKDVEMLIQDVSPNAKKIYH 835
Query: 121 LSGTQKFELPKEEIKMANVFRAIEAAKRNFTVFAWGLSDTTLEDVFIKVAREAQA 175
++GTQKFE+PK+E+++A +F+A+E AK NF VFAWGL+DTTLEDVFIKVAR AQA
Sbjct: 836 IAGTQKFEIPKDEVRIAELFQAVEKAKGNFRVFAWGLADTTLEDVFIKVARTAQA 890
>At5g61740 ABC family transporter - like protein
Length = 848
Score = 276 bits (705), Expect = 6e-75
Identities = 133/180 (73%), Positives = 157/180 (86%)
Query: 1 MKRRLSVAISLIGDPKVVYMDEPSTGLDPASRKCLWNVIKLAKQDRAIILTTHSMEEADA 60
MKRRLSVAISLIG+PKVVYMDEPSTGLDPASRK LW VI+ AKQ+ AIILTTHSMEEA+
Sbjct: 669 MKRRLSVAISLIGNPKVVYMDEPSTGLDPASRKDLWTVIQRAKQNTAIILTTHSMEEAEF 728
Query: 61 LCDRLGIFVDGSLQCVGYPKELKGRYGGTYVFAMTTSSDHEKDVENMVQRLTPNANKIYH 120
LCDRLGIFVDG LQCVG PKELKGRYGG+YVF MTTS +HE+ VE MV+ ++PN+ ++YH
Sbjct: 729 LCDRLGIFVDGGLQCVGNPKELKGRYGGSYVFTMTTSVEHEEKVERMVKHISPNSKRVYH 788
Query: 121 LSGTQKFELPKEEIKMANVFRAIEAAKRNFTVFAWGLSDTTLEDVFIKVAREAQAFDTLT 180
L+GTQKFE+PK+E+ +A+VF +E K FTVFAWGL+DTTLEDVF KVA AQAF++L+
Sbjct: 789 LAGTQKFEIPKQEVMIADVFFMVEKVKSKFTVFAWGLADTTLEDVFFKVATTAQAFNSLS 848
>At3g47770 ABC-type transport protein-like protein
Length = 722
Score = 271 bits (693), Expect = 1e-73
Identities = 131/171 (76%), Positives = 150/171 (87%)
Query: 1 MKRRLSVAISLIGDPKVVYMDEPSTGLDPASRKCLWNVIKLAKQDRAIILTTHSMEEADA 60
MKRRLSVAISLIG PKVVYMDEPSTGLDPASR LW VIK AK+ AIILTTHSMEEA+
Sbjct: 549 MKRRLSVAISLIGSPKVVYMDEPSTGLDPASRINLWTVIKRAKKHAAIILTTHSMEEAEF 608
Query: 61 LCDRLGIFVDGSLQCVGYPKELKGRYGGTYVFAMTTSSDHEKDVENMVQRLTPNANKIYH 120
LCDRLGIFVDG LQC+G PKELKGRYGG+YV +TTS +HEKDVE +VQ ++ NA KIYH
Sbjct: 609 LCDRLGIFVDGRLQCIGNPKELKGRYGGSYVLTITTSPEHEKDVETLVQEVSSNARKIYH 668
Query: 121 LSGTQKFELPKEEIKMANVFRAIEAAKRNFTVFAWGLSDTTLEDVFIKVAR 171
++GTQKFE PKEE++++ VF+A+E AKRNFTVFAWG +DTTLEDVFIKVA+
Sbjct: 669 IAGTQKFEFPKEEVRISEVFQAVENAKRNFTVFAWGFADTTLEDVFIKVAK 719
>At3g47790 ABC-type transport protein-like protein
Length = 727
Score = 182 bits (462), Expect = 8e-47
Identities = 87/137 (63%), Positives = 107/137 (77%), Gaps = 3/137 (2%)
Query: 35 LWNVIKLAKQDRAIILTTHSMEEADALCDRLGIFVDGSLQCVGYPKELKGRYGGTYVFAM 94
LW K + R + TTHSMEEA+ LCDR+GIFVDGSLQC+G PKELK RYGG+YV +
Sbjct: 592 LW---KAQESQRFCLNTTHSMEEAEILCDRIGIFVDGSLQCIGNPKELKSRYGGSYVLTV 648
Query: 95 TTSSDHEKDVENMVQRLTPNANKIYHLSGTQKFELPKEEIKMANVFRAIEAAKRNFTVFA 154
TTS +HEK+VE +V ++ NA KIY +GTQKFELPK+E+K+ VF+A+E AK F V A
Sbjct: 649 TTSEEHEKEVEQLVHNISTNAKKIYRTAGTQKFELPKQEVKIGEVFKALEKAKTMFPVVA 708
Query: 155 WGLSDTTLEDVFIKVAR 171
WGL+DTTLEDVFIKVA+
Sbjct: 709 WGLADTTLEDVFIKVAQ 725
>At5g61730 ABC transport protein - like
Length = 940
Score = 130 bits (326), Expect = 5e-31
Identities = 71/178 (39%), Positives = 109/178 (60%), Gaps = 4/178 (2%)
Query: 1 MKRRLSVAISLIGDPKVVYMDEPSTGLDPASRKCLWNVIKLAKQDRAIILTTHSMEEADA 60
MKRRLSVAI+LIGDPK+V++DEP+TG+DP +R+ +W++I+ +K+ RAIILTTHSMEEAD
Sbjct: 661 MKRRLSVAIALIGDPKLVFLDEPTTGMDPITRRHVWDIIQESKKGRAIILTTHSMEEADI 720
Query: 61 LCDRLGIFVDGSLQCVGYPKELKGRYGGTYVFAMTTSSDHEKDVENMVQRLTPNANKIYH 120
L DR+GI G L+C+G LK R+G +V ++ + + ++R K+
Sbjct: 721 LSDRIGIMAKGRLRCIGTSIRLKSRFGTGFVATVSFIENKKDGAPEPLKRFFKERLKVEP 780
Query: 121 LSGTQ---KFELPKE-EIKMANVFRAIEAAKRNFTVFAWGLSDTTLEDVFIKVAREAQ 174
+ F +P + E + F ++ + F + L TLE+VF+ +AR A+
Sbjct: 781 TEENKAFMTFVIPHDKEQLLKGFFAELQDRESEFGIADIQLGLATLEEVFLNIARRAE 838
>At2g41700 ATP-binding cassette transporter ABCA1
Length = 1884
Score = 127 bits (318), Expect = 4e-30
Identities = 71/186 (38%), Positives = 108/186 (57%), Gaps = 16/186 (8%)
Query: 1 MKRRLSVAISLIGDPKVVYMDEPSTGLDPASRKCLWNVIKLAKQDRAIILTTHSMEEADA 60
MKR+LS+ I+LIG+ KV+ +DEP++G+DP S + W +IK K+ R I+LTTHSM+EA+
Sbjct: 652 MKRKLSLGIALIGNSKVIILDEPTSGMDPYSMRLTWQLIKKIKKGRIILLTTHSMDEAEE 711
Query: 61 LCDRLGIFVDGSLQCVGYPKELKGRYGGTYVFAMTTSSDHEKDVENMVQRLTPNANKIYH 120
L DR+GI +GSL+C G LK YG Y + +S ++V R P+A +
Sbjct: 712 LGDRIGIMANGSLKCCGSSIFLKHHYGVGYTLTLVKTSPTVSVAAHIVHRHIPSATCVSE 771
Query: 121 LSGTQKFELPKEEIK-MANVFRAIEAAKRNFT---------------VFAWGLSDTTLED 164
+ F+LP + N+FR IE+ +N + ++G+S TTLE+
Sbjct: 772 VGNEISFKLPLASLPCFENMFREIESCMKNSVDRSKISEIEDSDYPGIQSYGISVTTLEE 831
Query: 165 VFIKVA 170
VF++VA
Sbjct: 832 VFLRVA 837
Score = 108 bits (271), Expect = 1e-24
Identities = 54/111 (48%), Positives = 77/111 (68%), Gaps = 3/111 (2%)
Query: 2 KRRLSVAISLIGDPKVVYMDEPSTGLDPASRKCLWNVI-KLAKQD--RAIILTTHSMEEA 58
KR+LSVAI++IGDP +V +DEPSTG+DP +++ +W+VI +L+ + A+ILTTHSM EA
Sbjct: 1597 KRKLSVAIAMIGDPPIVILDEPSTGMDPVAKRFMWDVISRLSTRSGKTAVILTTHSMNEA 1656
Query: 59 DALCDRLGIFVDGSLQCVGYPKELKGRYGGTYVFAMTTSSDHEKDVENMVQ 109
ALC R+GI V G L+C+G P+ LK RYG + + ++EN Q
Sbjct: 1657 QALCTRIGIMVGGRLRCIGSPQHLKTRYGNHLELEVKPNEVSNVELENFCQ 1707
>At3g47730 ABC-type transport protein-like protein
Length = 1011
Score = 116 bits (290), Expect = 7e-27
Identities = 73/212 (34%), Positives = 114/212 (53%), Gaps = 38/212 (17%)
Query: 1 MKRRLSVAISLIGDPKVVYMDEP----------------------------STGLDPASR 32
MKRRLSVA+SLIGDPK+V++DEP +TG+DP +R
Sbjct: 669 MKRRLSVAVSLIGDPKLVFLDEPVCNIYSVDVYLFKSLFLFSCVLVIHGLQTTGMDPITR 728
Query: 33 KCLWNVIKLAKQDRAIILTTHSMEEADALCDRLGIFVDGSLQCVGYPKELKGRYGGTYV- 91
+ +W++I+ K+ RAIILTTHSMEEAD L DR+GI G L+C+G LK R+G ++
Sbjct: 729 RHVWDIIQETKKGRAIILTTHSMEEADILSDRIGIMAKGRLRCIGTSIRLKSRFGTGFIA 788
Query: 92 -FAMTTSSDHE----KDVENMVQRLTPNANKIYHLSGTQ---KFELPKE-EIKMANVFRA 142
+ S++H D V++ + K+ + + F +P + E + + F
Sbjct: 789 NISFVESNNHNGEAGSDSREPVKKFFKDHLKVKPIEENKAFMTFVIPHDKENLLTSFFAE 848
Query: 143 IEAAKRNFTVFAWGLSDTTLEDVFIKVAREAQ 174
++ + F + L TLE+VF+ +AR+A+
Sbjct: 849 LQDREEEFGISDIQLGLATLEEVFLNIARKAE 880
>At5g61690 ABC transport protein - like
Length = 954
Score = 101 bits (251), Expect = 2e-22
Identities = 50/91 (54%), Positives = 68/91 (73%), Gaps = 6/91 (6%)
Query: 1 MKRRLSVAISLIGDPKVVYMDEPSTGLDPASRKCLWNVIKLAKQDRAIILTTHSMEEADA 60
MKRRLSVA++LIGDPK+ +TG+DP +R+ +W++I+ +K+ RAIILTTHSMEEAD
Sbjct: 647 MKRRLSVAVALIGDPKL------TTGMDPITRRHVWDIIQESKKGRAIILTTHSMEEADI 700
Query: 61 LCDRLGIFVDGSLQCVGYPKELKGRYGGTYV 91
L DR+GI G L+C+G LK R+G +V
Sbjct: 701 LSDRIGIMAKGRLRCIGTSIRLKSRFGTGFV 731
>At1g67940 ABC transporter like protein
Length = 263
Score = 51.2 bits (121), Expect = 3e-07
Identities = 27/82 (32%), Positives = 52/82 (62%), Gaps = 2/82 (2%)
Query: 3 RRLSVAISLIGDPKVVYMDEPSTGLDPASRKCLWNVIKLAKQDRAI--ILTTHSMEEADA 60
+R+++A +L +P+V+ +DEP++ LDP S + + +VI K+ R I ++ +HS+++
Sbjct: 166 QRVALARTLANEPEVLLLDEPTSALDPISTENIEDVIVKLKKQRGITTVIVSHSIKQIQK 225
Query: 61 LCDRLGIFVDGSLQCVGYPKEL 82
+ D + + VDG + V P EL
Sbjct: 226 VADIVCLVVDGEIVEVLKPSEL 247
>At2g37280 putative ABC transporter
Length = 1413
Score = 48.1 bits (113), Expect = 2e-06
Identities = 20/53 (37%), Positives = 36/53 (67%), Gaps = 1/53 (1%)
Query: 2 KRRLSVAISLIGDPKVVYMDEPSTGLDPASRKCLWNVIK-LAKQDRAIILTTH 53
++RL+VA+ L+ +P +++MDEP+TGLD + + +K +A+ R I+ T H
Sbjct: 970 RKRLTVAVELVANPSIIFMDEPTTGLDARAAAIVMRAVKNVAETGRTIVCTIH 1022
Score = 34.3 bits (77), Expect = 0.037
Identities = 47/197 (23%), Positives = 79/197 (39%), Gaps = 47/197 (23%)
Query: 2 KRRLSVAISLIGDPKVVYMDEPSTGLDPASR----KCLWNVIKLAKQDRAIILTTHSMEE 57
K+RL+ A ++G K ++MDE + GLD ++ K L V + + L + E
Sbjct: 316 KKRLTTAEMIVGPTKALFMDEITNGLDSSTAFQIIKSLQQVAHITNATVFVSLLQPAPES 375
Query: 58 ADALCDRLGIFVDGSL--------------QCVGYPKELKG-----------RYGGTY-- 90
D L D + + +G + +C E KG + G Y
Sbjct: 376 YD-LFDDIVLMAEGKIVYHGPRDDVLKFFEECGFQCPERKGVADFLQEVISKKDQGQYWL 434
Query: 91 -------VFAMTTSSDHEKDVENMVQRLTPNANKIYHLSGTQK-------FELPKEEIKM 136
++ T S KD+E + +++ +K Y +S T K + LPK E+
Sbjct: 435 HQNLPHSFVSVDTLSKRFKDLE-IGRKIEEALSKPYDISKTHKDALSFNVYSLPKWELFR 493
Query: 137 ANVFRAIEAAKRNFTVF 153
A + R KRN+ V+
Sbjct: 494 ACISREFLLMKRNYFVY 510
>At3g55090 ABC transporter - like protein
Length = 720
Score = 47.8 bits (112), Expect = 3e-06
Identities = 30/89 (33%), Positives = 47/89 (52%), Gaps = 2/89 (2%)
Query: 2 KRRLSVAISLIGDPKVVYMDEPSTGLDPASRKCLWNVIK-LAKQDRAIILTTHS-MEEAD 59
+RR+S+ I +I DP V+++DEP++GLD S + V+K +A+ II++ H
Sbjct: 221 RRRVSIGIDIIHDPIVLFLDEPTSGLDSTSAFMVVKVLKRIAESGSIIIMSIHQPSHRVL 280
Query: 60 ALCDRLGIFVDGSLQCVGYPKELKGRYGG 88
+L DRL G G P L + G
Sbjct: 281 SLLDRLIFLSRGHTVFSGSPASLPSFFAG 309
>At5g14100 unknown protein
Length = 278
Score = 47.4 bits (111), Expect = 4e-06
Identities = 22/63 (34%), Positives = 40/63 (62%)
Query: 2 KRRLSVAISLIGDPKVVYMDEPSTGLDPASRKCLWNVIKLAKQDRAIILTTHSMEEADAL 61
KRRL++AI L+ P ++ +DEP GLD +R + ++K K++ +++ +H + E AL
Sbjct: 199 KRRLALAIQLVQTPDLLILDEPLAGLDWKARADVAKLLKHLKKELTLLVVSHDLRELAAL 258
Query: 62 CDR 64
D+
Sbjct: 259 VDQ 261
>At4g15230 ABC transporter like protein
Length = 979
Score = 47.4 bits (111), Expect = 4e-06
Identities = 18/53 (33%), Positives = 36/53 (66%), Gaps = 1/53 (1%)
Query: 2 KRRLSVAISLIGDPKVVYMDEPSTGLDPASRKCLWNVIK-LAKQDRAIILTTH 53
++RL++A+ L+ +P +++MDEP+TGLD + + +K +A+ R ++ T H
Sbjct: 582 RKRLTIAVELVANPSIIFMDEPTTGLDARAAAIVMRAVKNVAETGRTVVCTIH 634
Score = 28.5 bits (62), Expect = 2.0
Identities = 19/66 (28%), Positives = 31/66 (46%), Gaps = 4/66 (6%)
Query: 2 KRRLSVAISLIGDPKVVYMDEPSTGLDPASR----KCLWNVIKLAKQDRAIILTTHSMEE 57
KRRL+ ++G ++MDE S GLD ++ CL + +A+ I L + E
Sbjct: 49 KRRLTTGELVVGPATTLFMDEISNGLDSSTTFQIVSCLQQLAHIAEATILISLLQPAPET 108
Query: 58 ADALCD 63
+ D
Sbjct: 109 FELFDD 114
>At3g55130 ABC transporter - like protein
Length = 725
Score = 47.0 bits (110), Expect = 6e-06
Identities = 29/85 (34%), Positives = 45/85 (52%), Gaps = 2/85 (2%)
Query: 2 KRRLSVAISLIGDPKVVYMDEPSTGLDPASRKCLWNVIKLAKQDRAIILTTHSMEEAD-- 59
+RR+S+ I +I DP V+++DEP++GLD + + V+K Q +I++ + A
Sbjct: 230 RRRVSIGIDIIHDPIVLFLDEPTSGLDSTNAFMVVQVLKRIAQSGSIVIMSIHQPSARIV 289
Query: 60 ALCDRLGIFVDGSLQCVGYPKELKG 84
L DRL I G G P L G
Sbjct: 290 ELLDRLIILSRGKSVFNGSPASLPG 314
>At3g53480 ABC transporter - like protein
Length = 1450
Score = 47.0 bits (110), Expect = 6e-06
Identities = 19/53 (35%), Positives = 35/53 (65%), Gaps = 1/53 (1%)
Query: 2 KRRLSVAISLIGDPKVVYMDEPSTGLDPASRKCLWNVIK-LAKQDRAIILTTH 53
++RL++A+ L+ +P +++MDEP+TGLD + + +K +A R I+ T H
Sbjct: 1007 RKRLTIAVELVANPSIIFMDEPTTGLDARAAAIVMRAVKNVADTGRTIVCTIH 1059
Score = 30.0 bits (66), Expect = 0.70
Identities = 12/30 (40%), Positives = 21/30 (70%)
Query: 2 KRRLSVAISLIGDPKVVYMDEPSTGLDPAS 31
K+RL+ A ++G K ++MDE + GLD ++
Sbjct: 351 KKRLTTAEMIVGPTKALFMDEITNGLDSST 380
>At1g53270 putative ABC transporter emb|AAD22683.1
Length = 590
Score = 47.0 bits (110), Expect = 6e-06
Identities = 22/54 (40%), Positives = 36/54 (65%), Gaps = 2/54 (3%)
Query: 2 KRRLSVAISLIGDPKVVYMDEPSTGLDPASRKCLWNVIK--LAKQDRAIILTTH 53
+RR+S+ + L+ DP V+ +DEP++GLD AS + ++K KQ + I+LT H
Sbjct: 178 RRRVSIGVELVHDPNVILIDEPTSGLDSASALQVVTLLKDMTIKQGKTIVLTIH 231
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.134 0.389
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,843,639
Number of Sequences: 26719
Number of extensions: 150559
Number of successful extensions: 511
Number of sequences better than 10.0: 107
Number of HSP's better than 10.0 without gapping: 85
Number of HSP's successfully gapped in prelim test: 22
Number of HSP's that attempted gapping in prelim test: 378
Number of HSP's gapped (non-prelim): 147
length of query: 180
length of database: 11,318,596
effective HSP length: 93
effective length of query: 87
effective length of database: 8,833,729
effective search space: 768534423
effective search space used: 768534423
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 57 (26.6 bits)
Medicago: description of AC146548.14


