

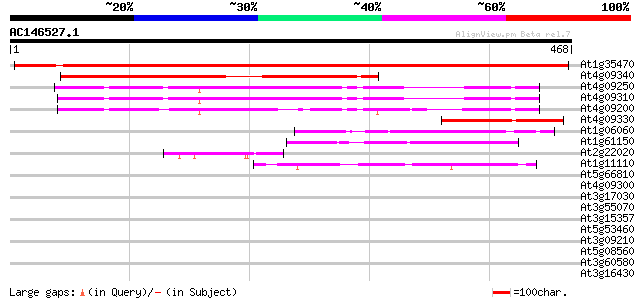
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146527.1 - phase: 0
(468 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g35470 unknown protein 619 e-177
At4g09340 putative protein 317 7e-87
At4g09250 putative protein 250 1e-66
At4g09310 putative protein 249 2e-66
At4g09200 putative protein 225 5e-59
At4g09330 putative protein 125 4e-29
At1g06060 unknown protein 79 5e-15
At1g61150 unknown protein 57 2e-08
At2g22020 hypothetical protein 50 2e-06
At1g11110 unknown protein 46 5e-05
At5g66810 putative protein 37 0.018
At4g09300 putative protein 37 0.030
At3g17030 hypothetical protein 37 0.030
At3g55070 unknown protein 33 0.34
At3g15357 unknown protein 31 1.3
At5g53460 NADH-dependent glutamate synthase 30 2.2
At3g09210 unknown protein 30 2.2
At5g08560 WD-repeat protein-like 30 2.8
At3g60580 zinc finger protein - like 30 2.8
At3g16430 putative lectin 30 2.8
>At1g35470 unknown protein
Length = 467
Score = 619 bits (1596), Expect = e-177
Identities = 300/464 (64%), Positives = 378/464 (80%), Gaps = 7/464 (1%)
Query: 5 NGKNTIEKPDQDQNQDLGFHFLELSRRKAKLDSEDFEVDDEEKEESPSELNTINSSGGFV 64
N N + + QDL +FL+ R AK D+++ +E EE P+ELNTINS+GGF+
Sbjct: 9 NSANGDTTNNGENGQDLNLNFLDKIRLSAKRDAKE-----DEGEELPTELNTINSAGGFL 63
Query: 65 VVATDKLSVKYTSVNLHGHDVGVIQANKVAPMKRIVYYFEIFVKDAGVKGQVSIGFTTES 124
VV+ DKLSVKYT+ NLHGHDVGV+QANK AP+K + YYFEIFVKD+G+KGQ++IGFT ES
Sbjct: 64 VVSPDKLSVKYTNTNLHGHDVGVVQANKPAPIKCLTYYFEIFVKDSGIKGQIAIGFTKES 123
Query: 125 FKMRRQPGWEANSCGYHGDDGFLYRGQGKGEAFGPTYTTGDIVGAGINYAAQEFFFTKNG 184
FKMRRQPGWE NSCGYHGDDG+LYRGQGKGE FGP +T D VG GINYA+QEFFFTKNG
Sbjct: 124 FKMRRQPGWEVNSCGYHGDDGYLYRGQGKGEPFGPKFTKDDAVGGGINYASQEFFFTKNG 183
Query: 185 QVVGTVFKEMKGPLFPTVAVHSQNEEVHVNFGQKPFTFDLKEYEAQERMKQQIKIEEVPV 244
+VG + K+++G LFPTVAVHSQNEEV VNFG+K F FD+K YEA ER KQQ+ IE++ +
Sbjct: 184 TIVGKIPKDIRGHLFPTVAVHSQNEEVLVNFGKKKFAFDIKGYEASERNKQQLAIEKISI 243
Query: 245 SPNASYGIVRSYLLHYGYEDTLNSFDEASRSTIPPINIVQENGIDDQETTYALNHRKTLR 304
PN YG+V++YLLHYGYE+TL++F+ A+++T+PPI+I QEN ID+ +++YAL RK LR
Sbjct: 244 PPNIGYGLVKTYLLHYGYEETLDAFNLATKNTVPPIHIDQENAIDEDDSSYALKQRKNLR 303
Query: 305 QLIRDGEIDAAFGKLREWYPQIAEDNTSAMCFLLHCQKFIELVRVGALEEAVKYGRIELS 364
QL+R+GEID A +L++ YPQI +D+ S +CFLLHCQKFIELVRVG LEE V YGR+EL+
Sbjct: 304 QLVRNGEIDTALAELQKLYPQIVQDDKSVVCFLLHCQKFIELVRVGKLEEGVNYGRLELA 363
Query: 365 SFFGLSLFEDIVQDCVALLAYERPLESAVGYLLKDSQREVVADAVNAMILSTNPNIK-VT 423
F GL+ F+DIV+DC ALLAYE+P ES+V Y L+DSQRE+VADAVNA ILSTNPN K V
Sbjct: 364 KFVGLTGFQDIVEDCFALLAYEKPEESSVWYFLEDSQRELVADAVNAAILSTNPNKKDVQ 423
Query: 424 KNC-LHSNLERLLRQLTACCLERRSLSGEQGEAFQLQRVLSSGK 466
++C L S+LE+LLRQLT CCLERRSL+G+QGE F+L+ VL++ +
Sbjct: 424 RSCHLQSHLEKLLRQLTVCCLERRSLNGDQGETFRLRHVLNNNR 467
>At4g09340 putative protein
Length = 237
Score = 317 bits (813), Expect = 7e-87
Identities = 157/265 (59%), Positives = 192/265 (72%), Gaps = 31/265 (11%)
Query: 43 DDEEKEESPSELNTINSSGGFVVVATDKLSVKYTSVNLHGHDVGVIQANKVAPMKRIVYY 102
D+EE+EESP+ELNTI GFVV++ DKLS+KYT V+ HGHDVGV+QANK AP KR YY
Sbjct: 3 DNEEEEESPTELNTITGLSGFVVISPDKLSLKYTGVSQHGHDVGVVQANKPAPCKRFAYY 62
Query: 103 FEIFVKDAGVKGQVSIGFTTESFKMRRQPGWEANSCGYHGDDGFLYRGQGKGEAFGPTYT 162
FEI+VKDAGVKG+VSIGFTT SF++ R PGWE N+CGYHG+DG +Y G+ +GEAFGPTYT
Sbjct: 63 FEIYVKDAGVKGRVSIGFTTNSFRIARHPGWELNTCGYHGEDGLIYLGKRQGEAFGPTYT 122
Query: 163 TGDIVGAGINYAAQEFFFTKNGQVVGTVFKEMKGPLFPTVAVHSQNEEVHVNFGQKPFTF 222
TGD VG GINY +QEFFFT V VNFGQ+ F F
Sbjct: 123 TGDTVGGGINYDSQEFFFT-----------------------------VTVNFGQEKFAF 153
Query: 223 DLKEYEAQERMKQQIKIEEVPVSPNASYGIVRSYLLHYGYEDTLNSFDEASRSTIPPINI 282
D K YE+ ER KQQI IE++ + N S G+V++YLLHYGYE+T ++ D A+ ST+PPIN
Sbjct: 154 DFKGYESSERNKQQIAIEKISIPSNMSLGLVKNYLLHYGYEETHHALDLATNSTLPPING 213
Query: 283 VQENGIDDQETTYALNHRKTLRQLI 307
QENGIDD T+YAL+ RK LRQ++
Sbjct: 214 AQENGIDD--TSYALHERKILRQVL 236
>At4g09250 putative protein
Length = 427
Score = 250 bits (639), Expect = 1e-66
Identities = 155/411 (37%), Positives = 223/411 (53%), Gaps = 72/411 (17%)
Query: 38 EDFEVDDEEKEESPSELNTINSSGGFVVVATDKLSVKYTSVNLHGHDVGVIQANKVAPMK 97
E E ++EE+E P+ L T+ S F V+ D LS +YT + HD V+Q AP
Sbjct: 77 EAMEENEEEEELRPTALYTVGGSDEFTFVSPDGLSGQYTGPD---HDGVVLQTENPAPTN 133
Query: 98 RIVYYFEIFVKDAGVKGQVSIGFTTESFKMRRQPGWEANSCGYHGDDGFLYRGQGKGEAF 157
+ YYFE+ + +AG KG+++IGF+ E G+ SC Y G+ G + G G+
Sbjct: 134 CLAYYFEVHIMNAGEKGEIAIGFSKEHIYSE---GYTVRSCAYIGNSGLICSQNGDGDRT 190
Query: 158 ----GPTYTTGDIVGAGINYAAQEFFFTKNGQVVGTVFKEMKGPLFPTVAVHSQNEEVHV 213
TYTTGD+VG GI+ +QEFFFTKNG +VGT+ ++ + P++PT+ +HSQNE V V
Sbjct: 191 VAKASDTYTTGDLVGCGIDSVSQEFFFTKNGTIVGTIPRQFRRPVYPTIVLHSQNEAVTV 250
Query: 214 NF-GQKPFTFDLKEYEAQERMKQQIKIEEVPVSPNASYGIVRSYLLHYGYEDTLNSFD-E 271
NF GQ F+FD + +E R+K++ +IE++ +S + S+G+V++YLL YGYED+ +F+
Sbjct: 251 NFGGQIAFSFDFEHFEESLRVKKEREIEKISMSRSISHGLVKTYLLRYGYEDSFRAFNLA 310
Query: 272 ASRSTIPPINIVQENGIDDQETTYALNHRKTLRQLIRDGEIDAAFGKLREWYPQIAEDNT 331
ASR T+ I QEN D+ Y L+ RK LR+LI EID A L++ YPQ+ ED
Sbjct: 311 ASRHTV----IAQENSFDE----YELHQRKKLRELIMTAEIDDAIAALKDRYPQLIED-- 360
Query: 332 SAMCFLLHCQKFIELVRVGALEEAVKYGRIELSSFFGLSLFEDIVQDCVALLAYERPLES 391
C AL E ES
Sbjct: 361 -----------------------------------------------CSALFECETIEES 373
Query: 392 AVGYLLKDSQREVVADAVNAMILSTNPNIKVTKNCLHSNLERLLRQLTACC 442
YLL ++Q+ +VA AV+ ILSTNP T++ ++LER+LRQ A C
Sbjct: 374 GAAYLLGETQKNIVAAAVSEAILSTNP---ATRDQQSASLERVLRQFEATC 421
>At4g09310 putative protein
Length = 349
Score = 249 bits (636), Expect = 2e-66
Identities = 154/408 (37%), Positives = 222/408 (53%), Gaps = 72/408 (17%)
Query: 41 EVDDEEKEESPSELNTINSSGGFVVVATDKLSVKYTSVNLHGHDVGVIQANKVAPMKRIV 100
E ++EE+E P+ L T+ S F V+ D LS +YT + HD V+Q AP +
Sbjct: 2 EENEEEEELRPTALYTVGGSDEFTFVSPDGLSGQYTGPD---HDGVVLQTENPAPTNCLA 58
Query: 101 YYFEIFVKDAGVKGQVSIGFTTESFKMRRQPGWEANSCGYHGDDGFLYRGQGKGEAF--- 157
YYFE+ + +AG KG+++IGF+ E G+ SC Y G+ G + G G+
Sbjct: 59 YYFEVHIMNAGEKGEIAIGFSKEHIYSE---GYTVRSCAYIGNSGLICSQNGDGDRTVAK 115
Query: 158 -GPTYTTGDIVGAGINYAAQEFFFTKNGQVVGTVFKEMKGPLFPTVAVHSQNEEVHVNF- 215
TYTTGD+VG GI+ +QEFFFTKNG +VGT+ ++ + P++PT+ +HSQNE V VNF
Sbjct: 116 ASDTYTTGDLVGCGIDSVSQEFFFTKNGTIVGTIPRQFRRPVYPTIVLHSQNEAVTVNFG 175
Query: 216 GQKPFTFDLKEYEAQERMKQQIKIEEVPVSPNASYGIVRSYLLHYGYEDTLNSFD-EASR 274
GQ F+FD + +E R+K++ +IE++ +S + S+G+V++YLL YGYED+ +F+ ASR
Sbjct: 176 GQIAFSFDFEHFEESLRVKKEREIEKISMSRSISHGLVKTYLLRYGYEDSFRAFNLAASR 235
Query: 275 STIPPINIVQENGIDDQETTYALNHRKTLRQLIRDGEIDAAFGKLREWYPQIAEDNTSAM 334
T+ I QEN D+ Y L+ RK LR+LI EID A L++ YPQ+ ED
Sbjct: 236 HTV----IAQENSFDE----YELHQRKKLRELIMTAEIDDAIAALKDRYPQLIED----- 282
Query: 335 CFLLHCQKFIELVRVGALEEAVKYGRIELSSFFGLSLFEDIVQDCVALLAYERPLESAVG 394
C AL E ES
Sbjct: 283 --------------------------------------------CSALFECETIEESGAA 298
Query: 395 YLLKDSQREVVADAVNAMILSTNPNIKVTKNCLHSNLERLLRQLTACC 442
YLL ++Q+ +VA AV+ ILSTNP T++ ++LER+LRQ A C
Sbjct: 299 YLLGETQKNIVAAAVSEAILSTNP---ATRDQQSASLERVLRQFEATC 343
>At4g09200 putative protein
Length = 377
Score = 225 bits (573), Expect = 5e-59
Identities = 159/442 (35%), Positives = 219/442 (48%), Gaps = 112/442 (25%)
Query: 41 EVDDEEKEESPSELNTINSSGGFVVVATDKLSVKYTSVNLHGHDVGVIQANKVAPMKRIV 100
E ++EE+E P+ L T+ S F V+ D LS +YT + HD V+Q AP +
Sbjct: 2 EENEEEEELRPTALYTVGGSDEFTFVSPDGLSGQYTGPD---HDGVVLQTENPAPTNCLA 58
Query: 101 YYFEIFVKDAGVKGQVSIGFTTESFKMRRQPGWEANSCGYHGDDGFLYRGQGKGEAF--- 157
YYFE+ + +AG KG+++IGF+ E + SC Y G+ G + G G+
Sbjct: 59 YYFEVHIMNAGEKGEIAIGFSKEHI-------YSEGSCAYIGNSGLICSQNGDGDRTVAK 111
Query: 158 -GPTYTTGDIVGAGINYAAQEFFFTKNGQVVGTVFKEMKGPLFPTVAVHSQNEEVHVNF- 215
TYTTGD+VG GI+ +QEFFFTKNG +VGT+ ++ + P++PT+ +HSQNE V VNF
Sbjct: 112 ASDTYTTGDLVGCGIDSVSQEFFFTKNGTIVGTIPRQFRRPVYPTIVLHSQNEAVTVNFG 171
Query: 216 GQKPFTFDLKEYEAQERMKQQIKIEEVPVSPNASYGIVRSYLLHYGYEDTLNSFD-EASR 274
GQ F+FD E+P S +V++YLL YGYED+ +F+ ASR
Sbjct: 172 GQIAFSFDF----------------ELPFS-----RLVKTYLLRYGYEDSFRAFNLAASR 210
Query: 275 STIPPINIVQENGIDDQETTYALNHRKTLRQ----------------------------- 305
T+ I QEN D+ Y L+ RK LR+
Sbjct: 211 HTV----IAQENSFDE----YELHQRKKLREVLFTAMVQNQPKASYIATSFLFHIELIHA 262
Query: 306 -----LIRDGEIDAAFGKLREWYPQIAEDNTSAMCFLLHCQKFIELVRVGALEEAVKYGR 360
LI EID A L++ YPQ+ E + A FLL CQK +ELVR
Sbjct: 263 FCLLKLIMTAEIDDAIAALKDRYPQLIEGGSEAY-FLLICQKIVELVR------------ 309
Query: 361 IELSSFFGLSLFEDIVQDCVALLAYERPLESAVGYLLKDSQREVVADAVNAMILSTNPNI 420
DC AL E ES YLL ++Q+ +VA AV+ ILSTNP
Sbjct: 310 -----------------DCSALFECETIEESGAAYLLGETQKNIVAAAVSEAILSTNP-- 350
Query: 421 KVTKNCLHSNLERLLRQLTACC 442
T++ ++LER+LRQ A C
Sbjct: 351 -ATRDQQSASLERVLRQFEATC 371
>At4g09330 putative protein
Length = 111
Score = 125 bits (315), Expect = 4e-29
Identities = 66/102 (64%), Positives = 78/102 (75%), Gaps = 3/102 (2%)
Query: 361 IELSSFFGLSLFEDIVQDCVALLAYERPLESAVGYLLKDSQREVVADAVNAMILSTNPNI 420
+EL+ F GL+ F+DI++DC ALL YERP ES VGY L+++QREVVAD VNA ILST P
Sbjct: 1 MELAKFIGLTTFQDILEDCFALLVYERPEESNVGYFLEETQREVVADTVNAAILSTKPK- 59
Query: 421 KVTKNCLHSNLERLLRQLTACCLERRSLSGEQGEAFQLQRVL 462
KN HS+LE LLRQLTACCLE RSL+ QGEAF L R+L
Sbjct: 60 --GKNQSHSHLETLLRQLTACCLELRSLNDGQGEAFSLNRLL 99
>At1g06060 unknown protein
Length = 213
Score = 79.0 bits (193), Expect = 5e-15
Identities = 63/217 (29%), Positives = 111/217 (51%), Gaps = 22/217 (10%)
Query: 238 KIEEVPVSPNASYGIVRSYLLHYGYEDTLNSFDEASRSTIPPINIVQENGIDDQETTYAL 297
+ E V+ N + IV SYLLH + +T +S ++ P I+ ++N +
Sbjct: 5 QFEHTVVADNDIHSIVMSYLLHNCFNETADSLASSTGVKQPAID--RDN----------M 52
Query: 298 NHRKTLRQLIRDGEIDAAFGKLREWYPQIAEDNTSAMCFLLHCQKFIELVRVGALEEAVK 357
RK + I + + AF +L E Q + + F L C F+EL+ G EA+K
Sbjct: 53 ERRKQIIHFILERKALKAF-ELTEQLAQDLLEKNKDLQFDLLCLHFVELICAGNCTEALK 111
Query: 358 YGRIELSSFFGLSLFEDIVQDCVALLAYERPLESAVGYLLKDSQREVVADAVNAMILSTN 417
+G+ L+ F + + + ++D +ALLAYE P +S + +LL R+ VAD +N IL
Sbjct: 112 FGKTRLAPFGKVKKYVEKLEDVMALLAYEDPEKSPMFHLLSSEYRQQVADNLNRTIL--- 168
Query: 418 PNIKVTKNCLHSNLERLLRQLTACCLERRSLSGEQGE 454
+ T + ++ +ER+++Q+T + R+ L+ E G+
Sbjct: 169 ---EHTNHPSYTPMERIIQQVT---VVRQYLTEENGK 199
>At1g61150 unknown protein
Length = 243
Score = 57.0 bits (136), Expect = 2e-08
Identities = 50/194 (25%), Positives = 91/194 (46%), Gaps = 15/194 (7%)
Query: 232 RMKQQIKIEEVPVSPNASYGIVRSYLLHYGYEDTLNSFDEASRSTIPPINIVQENGIDDQ 291
R + + K+ V + +V ++L+ GY + F S T P I++
Sbjct: 27 REEWEKKLNAVKLRKEDMNTLVMNFLVTEGYVEAAEKFQRES-GTKPEIDLA-------- 77
Query: 292 ETTYALNHRKTLRQLIRDGEIDAAFGKLREWYPQIAEDNTSAMCFLLHCQKFIELVRVGA 351
+ R +++ +++G ++ A K+ + P+I + N + F L Q+ IEL+R G
Sbjct: 78 ----TITDRMAVKKAVQNGNVEDAIEKVNDLNPEILDTNPE-LFFHLQQQRLIELIRQGK 132
Query: 352 LEEAVKYGRIELSSFFGLS-LFEDIVQDCVALLAYERPLESAVGYLLKDSQREVVADAVN 410
EEA+++ + EL+ + F + ++ VALL ++ V LL S R A VN
Sbjct: 133 TEEALEFAQEELAPRGEENQAFLEELEKTVALLVFDDASTCPVKELLDLSHRLKTASEVN 192
Query: 411 AMILSTNPNIKVTK 424
A IL++ + K K
Sbjct: 193 AAILTSQSHEKDPK 206
>At2g22020 hypothetical protein
Length = 795
Score = 50.4 bits (119), Expect = 2e-06
Identities = 30/118 (25%), Positives = 57/118 (47%), Gaps = 19/118 (16%)
Query: 129 RQPGWEANSCGY-------HGDDGFLYRGQG------KGEAFGPTYTTGDIVGAGINYAA 175
+Q GW +C + DD + + G+ + E +G ++ GD++G I+
Sbjct: 161 QQLGWATLACPFTDQKGVGDADDSYAFDGRRVSKWNKEAEPYGQSWVAGDVIGCCIDLNC 220
Query: 176 QEFFFTKNGQVVGTVFKEMK--GP---LFPTVAVHSQNEEVHVNFGQKPFTFDLKEYE 228
E +F +NG +G F ++ GP +P +++ SQ E +NFG PF + + ++
Sbjct: 221 DEIYFYRNGVSLGAAFTGIRKLGPGFGYYPAISL-SQGERCELNFGAYPFKYPVDGFQ 277
>At1g11110 unknown protein
Length = 277
Score = 45.8 bits (107), Expect = 5e-05
Identities = 57/247 (23%), Positives = 106/247 (42%), Gaps = 44/247 (17%)
Query: 204 VHSQNEEVHVNFGQKPFTFDLKEYEAQERMKQQIK-------IEEVPVSPNASYGIVRSY 256
V ++NEE ++ + +E+ QE+ K+ I + + +V ++
Sbjct: 31 VEAENEEAQIS--------ENEEFVIQEQPKKLIMSDVWEQYLTTAEIRKEDMNRLVMNF 82
Query: 257 LLHYGYEDTLNSFDEASRSTIPPINIVQENGIDDQETTYALNHRKTLRQLIRDGEIDAAF 316
L+ GY + + F + S + Q +++ R +++ I G+++ A
Sbjct: 83 LVVEGYLEAVEKFQKESGTK--------------QVGVASISDRLAVKRDIESGDLEDAV 128
Query: 317 GKLREWYPQIAEDNTSAMCFLLHCQKFIELVRVG-ALEEAVKYGRIELSSFF--GLSLFE 373
KL P+I + N F L+ Q+FIE +R+G ++E + EL L+ E
Sbjct: 129 EKLNAINPEILKTN-----FSLNQQRFIERIRIGVTIKETFNFAEKELKPLVEQNLAFLE 183
Query: 374 DIVQDCVALLAYERP-LESAVGYLLKDSQREVVADAVNAMILSTNPNIKVTKNCLHSNLE 432
++ + L + P + A LL +S+ A VNA IL++ +K K L
Sbjct: 184 ELEKTMAILRFRDLPDIPEAERELLDNSRWFKTAAEVNAAILTSQTGLKCPK------LL 237
Query: 433 RLLRQLT 439
LL+ LT
Sbjct: 238 DLLKMLT 244
>At5g66810 putative protein
Length = 752
Score = 37.4 bits (85), Expect = 0.018
Identities = 46/190 (24%), Positives = 75/190 (39%), Gaps = 36/190 (18%)
Query: 230 QERMKQQIKIEEVPVSPNASYGIVRSYLLHYGYEDTLNSFDEASRSTIPPINIVQENGID 289
+E K++ ++ PV+ A ++ ++ + D A+ PP + +
Sbjct: 48 EEPRKRKATMDSTPVNWEALDALIIDFVSSENLVE-----DAAAAVNSPPSPLSSPSSSS 102
Query: 290 DQETTYALNHRKTL----RQLIRDGEIDAAFGKLREWYPQIAEDNTSAMCFLLHCQKFIE 345
+ + H + + R I G+I+ A LR P + +D+ + F L QKFIE
Sbjct: 103 SPSISSSSYHSRLIIRRIRSSIESGDIETAIDILRSHAPFVLDDHR--ILFRLQKQKFIE 160
Query: 346 LVRVGALEEAVKYGRIELSSFFGLSLFEDIVQDCV---ALLAYERPLESAVGYLL----- 397
L+R G E A+ D ++ CV AL AY E LL
Sbjct: 161 LLRKGTHEAAI-----------------DCLRTCVAPCALDAYPEAYEEFKHVLLALIYD 203
Query: 398 KDSQREVVAD 407
KD Q VA+
Sbjct: 204 KDDQTSPVAN 213
>At4g09300 putative protein
Length = 226
Score = 36.6 bits (83), Expect = 0.030
Identities = 32/127 (25%), Positives = 55/127 (43%), Gaps = 16/127 (12%)
Query: 238 KIEEVPVSPNASYGIVRSYLLHYGYEDTLNSFDEASRSTIPPINIVQENGIDDQETTYAL 297
K+ +V + +V + L+ GY + F E S I + +E ++
Sbjct: 21 KLSDVEILIEDMNRLVMNLLVAEGYREAAEKFKEES--------ITMQTA---EEDLASM 69
Query: 298 NHRKTLRQLIRDGEIDAAFGKLREWYPQIAEDNTSAMCFLLHCQKFIELVRVGALEEAVK 357
N R + + I ++ A KL P+I + + F LH Q IEL+R EEAV
Sbjct: 70 NERLEVIKAIESRNLEDAIEKLNALNPEIIKTS-----FHLHQQMLIELIREKKTEEAVA 124
Query: 358 YGRIELS 364
+ + +L+
Sbjct: 125 FAQEKLA 131
>At3g17030 hypothetical protein
Length = 651
Score = 36.6 bits (83), Expect = 0.030
Identities = 24/68 (35%), Positives = 36/68 (52%), Gaps = 2/68 (2%)
Query: 1 MKETNGKNTIEKPDQDQNQDLGFHFLELSRRKAKLDSEDFEVDDEEKEESPSELNTINSS 60
M +TNG + IE DQ++ +D FL+ +R + ED E + EE + P E T S
Sbjct: 1 MGDTNGASLIEIGDQEEVEDPFLAFLDYAR--TVISPEDDEDEKEESKRGPGEAMTEASG 58
Query: 61 GGFVVVAT 68
G+ VA+
Sbjct: 59 PGWGWVAS 66
>At3g55070 unknown protein
Length = 418
Score = 33.1 bits (74), Expect = 0.34
Identities = 42/160 (26%), Positives = 71/160 (44%), Gaps = 20/160 (12%)
Query: 252 IVRSYLLHYGYEDTLNSFDEASRSTIPPINIVQENGIDDQETTYALNHRKTLRQLIRDGE 311
I+ Y+L Y +T E+S NI+ ID K + +++ E
Sbjct: 148 ILVDYMLRMSYFETATKLSESS-------NIMDLVDID------IFREAKKVIDALKNRE 194
Query: 312 IDAAFGKLREWYPQIAEDNTSAMCFLLHCQKFIELVRVGALE---EAVKYGRIELSSFFG 368
+ +A + ++ + S F L Q+FIELVRV E +A++Y R L+S+
Sbjct: 195 VASALTWCADNKTRLKKSK-SKFEFQLRLQEFIELVRVDTAESYKKAIQYARKHLASWGT 253
Query: 369 LSLFEDIVQDCVALLAYERPLE-SAVGYLLKDSQREVVAD 407
+ E +Q +A LA++ E S L + Q +V+ D
Sbjct: 254 THMKE--LQHVLATLAFKSTTECSKYKVLFELRQWDVLVD 291
>At3g15357 unknown protein
Length = 143
Score = 31.2 bits (69), Expect = 1.3
Identities = 16/49 (32%), Positives = 26/49 (52%), Gaps = 1/49 (2%)
Query: 1 MKETNGKNTIEKPDQDQNQDLGFHFLELSRRKAKLDSEDFEVDDEEKEE 49
M+ +GK ++ D D +D F E+ + D ED + DDEE+E+
Sbjct: 88 MEAAHGKRVVDNKDADSEEDTDFDEDEIDDVDFE-DDEDSDEDDEEEED 135
>At5g53460 NADH-dependent glutamate synthase
Length = 2216
Score = 30.4 bits (67), Expect = 2.2
Identities = 23/89 (25%), Positives = 43/89 (47%), Gaps = 5/89 (5%)
Query: 11 EKPDQDQNQDLGFH--FLELSRRKAKLDSEDFEVDDEEKEESPSELNTINSSGGFVVVAT 68
E+ D+ + ++L F EL A E+ + E PS+++ +GGF+ A
Sbjct: 1659 EEADETEEKELEEKDAFAELKNMAAASSKEEMSGNGVAAEARPSKVDNAVKNGGFI--AY 1716
Query: 69 DKLSVKYTSVNLHGHDVG-VIQANKVAPM 96
++ VKY N+ +D V++ +K P+
Sbjct: 1717 EREGVKYRDPNVRLNDWNEVMEESKPGPL 1745
>At3g09210 unknown protein
Length = 333
Score = 30.4 bits (67), Expect = 2.2
Identities = 24/72 (33%), Positives = 31/72 (42%), Gaps = 3/72 (4%)
Query: 2 KETNGKNTIEKPDQDQNQDLGFHFLELSRRKAKLDSEDFEVDDEEKEES---PSELNTIN 58
K N K I KP + DL F + + K DSE E D E+E S EL ++
Sbjct: 185 KVGNTKRQINKPRPVDDSDLEAIFKQAKEAQEKADSEFEEADRAEEEASILASQELLALS 244
Query: 59 SSGGFVVVATDK 70
+S VA K
Sbjct: 245 NSDVIETVAESK 256
>At5g08560 WD-repeat protein-like
Length = 589
Score = 30.0 bits (66), Expect = 2.8
Identities = 25/82 (30%), Positives = 42/82 (50%), Gaps = 8/82 (9%)
Query: 284 QENGIDDQETTYALNHRKTLRQLIRDGEIDAAFGKL-REWYPQIAEDNTSAMCFLLHCQK 342
+E+GI +T K Q ++DG+ D + L R +P E A FLL QK
Sbjct: 90 EESGISLHNSTI-----KLFLQQVKDGKWDQSVKTLHRIGFPD--EKAVKAASFLLLEQK 142
Query: 343 FIELVRVGALEEAVKYGRIELS 364
F+E ++V + +A++ R E++
Sbjct: 143 FLEFLKVEKIADALRTLRNEMA 164
>At3g60580 zinc finger protein - like
Length = 288
Score = 30.0 bits (66), Expect = 2.8
Identities = 23/94 (24%), Positives = 38/94 (39%), Gaps = 6/94 (6%)
Query: 14 DQDQNQDLGFHFLELSRRKAKLDSEDFEV----DDEEKEESPSELNTINSSGGFVVVATD 69
D +DL F + LSR K K + + EV + EE+ E +++N + G +
Sbjct: 120 DTTTEEDLAFCLMMLSRDKWKKNKSNKEVVEEIETEEESEGYNKINRATTKGRYKCETCG 179
Query: 70 KLSVKYTSVNLH--GHDVGVIQANKVAPMKRIVY 101
K+ Y ++ H H + NK Y
Sbjct: 180 KVFKSYQALGGHRASHKKNRVSNNKTEQRSETEY 213
>At3g16430 putative lectin
Length = 296
Score = 30.0 bits (66), Expect = 2.8
Identities = 19/55 (34%), Positives = 25/55 (44%), Gaps = 5/55 (9%)
Query: 168 GAGINYAAQEFFFTKNGQVVGTVFKEMKGPLFPT---VAVHSQNEEVHVNFGQKP 219
G GI Y +F + KNGQ T + +KG PT V H + V + KP
Sbjct: 34 GMGIQYI--QFDYVKNGQTEQTPLRGIKGSTIPTDPFVINHPEEHLVSIEIWYKP 86
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.317 0.135 0.388
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,642,516
Number of Sequences: 26719
Number of extensions: 474512
Number of successful extensions: 1796
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 12
Number of HSP's successfully gapped in prelim test: 18
Number of HSP's that attempted gapping in prelim test: 1749
Number of HSP's gapped (non-prelim): 35
length of query: 468
length of database: 11,318,596
effective HSP length: 103
effective length of query: 365
effective length of database: 8,566,539
effective search space: 3126786735
effective search space used: 3126786735
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 62 (28.5 bits)
Medicago: description of AC146527.1


