

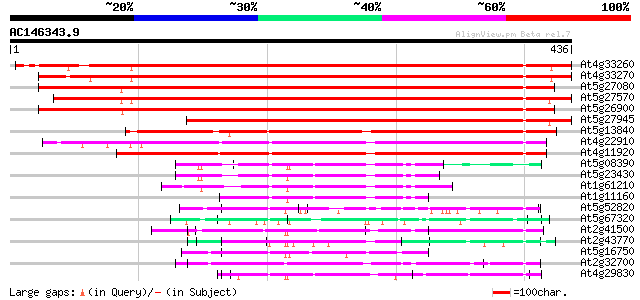
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146343.9 + phase: 0
(436 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g33260 WD-repeat protein -like protein 565 e-161
At4g33270 putative cdc20 protein (CDC20.1) 562 e-160
At5g27080 putative cdc20 protein 504 e-143
At5g27570 cdc20-like protein 502 e-142
At5g26900 WD-repeat protein - like 502 e-142
At5g27945 WD-repeat protein - like 420 e-118
At5g13840 cell cycle switch protein 322 2e-88
At4g22910 putative fizzy-related protein 312 3e-85
At4g11920 Srw1 like protein 311 3e-85
At5g08390 katanin p80 subunit - like protein 87 2e-17
At5g23430 unknown protein 80 2e-15
At1g61210 78 8e-15
At1g11160 hypothetical protein 77 1e-14
At5g52820 Notchless protein homolog 69 4e-12
At5g67320 unknown protein 66 4e-11
At2g41500 putative U4/U6 small nuclear ribonucleoprotein 65 1e-10
At2g43770 putative splicing factor 63 4e-10
At5g16750 WD40-repeat protein 60 2e-09
At2g32700 unknown protein 59 4e-09
At4g29830 unknown protein 59 5e-09
>At4g33260 WD-repeat protein -like protein
Length = 447
Score = 565 bits (1455), Expect = e-161
Identities = 291/453 (64%), Positives = 340/453 (74%), Gaps = 35/453 (7%)
Query: 5 PLLQRFLHLDDDNNSRWHQNIDRFIPNRSAIDWDHATTIL---------STTTTNVTKEN 55
PL + FL NS+ +N+DRFIPNRSA+++D+A L S T ++ +KE
Sbjct: 9 PLQEHFL---PRKNSK--ENLDRFIPNRSAMNFDYAHFALTEERKGKDQSATVSSPSKE- 62
Query: 56 LWLASNVYQKKLAEAADLP-TRILAFRNKPRKRNVISPP--------PPPRSKPMRYIPK 106
Y+K+LAE +L TRILAFRNKP+ + P P KP RYIP+
Sbjct: 63 ------AYRKQLAETMNLNHTRILAFRNKPQAPVELLPSNHSASLHQQPKSVKPRRYIPQ 116
Query: 107 TCEGTFDLPDLSDDFSLNLLDWGSRNVLSIALDHTIYFWNASDSSGSEFVTVDEEEGPVT 166
T E T D PD+ DDF LNLLDWGS NVL+IALDHT+Y W+AS S SE VT+DEE+GPVT
Sbjct: 117 TSERTLDAPDIVDDFYLNLLDWGSANVLAIALDHTVYLWDASTGSTSELVTIDEEKGPVT 176
Query: 167 SVCWAPDGRHLAVGLTNSHVQLWDTAANKQLRTLKGGHRARVGSLAWNGHVLTTGGMDGK 226
S+ WAPDGRH+AVGL NS VQLWD+A+N+QLRTLKGGH++RVGSLAWN H+LTTGGMDG
Sbjct: 177 SINWAPDGRHVAVGLNNSEVQLWDSASNRQLRTLKGGHQSRVGSLAWNNHILTTGGMDGL 236
Query: 227 IVNNDVRLRSQIINTYRGHRREVCGLKWSLDGKQLASGGNDNVVHIWDMSAVSSNSPTRW 286
I+NNDVR+RS I+ TYRGH +EVCGLKWS G+QLASGGNDNVVHIWD S SSNS T+W
Sbjct: 237 IINNDVRIRSPIVETYRGHTQEVCGLKWSGSGQQLASGGNDNVVHIWDRSVASSNSTTQW 296
Query: 287 LYRFDEHKAAVKALAWCPFQGNLLASGGGGGDCCVKLWNTGMGERMNSVDTGSQVCALLW 346
L+R +EH +AVKALAWCPFQ NLLA+GGGGGD +K WNT G +NSVDTGSQVC+LLW
Sbjct: 297 LHRLEEHTSAVKALAWCPFQANLLATGGGGGDRTIKFWNTHTGACLNSVDTGSQVCSLLW 356
Query: 347 SKNERELLSSHGLTQNQLTLWKYPSMLKIAELHGHTSRVLHMTQSPDGSTVASAAAAADQ 406
SKNERELLSSHG TQNQLTLWKYPSM+K+AEL GHTSRVL+M QSPDG TVAS AA D+
Sbjct: 357 SKNERELLSSHGFTQNQLTLWKYPSMVKMAELTGHTSRVLYMAQSPDGCTVAS--AAGDE 414
Query: 407 TLRFWEVFGTPPA---APPKRNKMPFADSNRIR 436
TLRFW VFG P A PK PF+ NRIR
Sbjct: 415 TLRFWNVFGVPETAKKAAPKAVAEPFSHVNRIR 447
>At4g33270 putative cdc20 protein (CDC20.1)
Length = 457
Score = 562 bits (1448), Expect = e-160
Identities = 284/430 (66%), Positives = 330/430 (76%), Gaps = 20/430 (4%)
Query: 23 QNIDRFIPNRSAIDWDHATTILSTTTTNVTKENLWLASN----VYQKKLAEAADLP-TRI 77
+N+DRFIPNRSA+++D+A L T K+ S+ Y+K+LAE +L TRI
Sbjct: 32 ENLDRFIPNRSAMNFDYAHFAL--TEGRKGKDQTAAVSSPSKEAYRKQLAETMNLNHTRI 89
Query: 78 LAFRNKPRKRNVISPP--------PPPRSKPMRYIPKTCEGTFDLPDLSDDFSLNLLDWG 129
LAFRNKP+ + P P KP RYIP+T E T D PD+ DDF LNLLDWG
Sbjct: 90 LAFRNKPQAPVELLPSNHSASLHQQPKSVKPRRYIPQTSERTLDAPDIVDDFYLNLLDWG 149
Query: 130 SRNVLSIALDHTIYFWNASDSSGSEFVTVDEEEGPVTSVCWAPDGRHLAVGLTNSHVQLW 189
S NVL+IALDHT+Y W+AS S SE VT+DEE+GPVTS+ WAPDGRH+AVGL NS VQLW
Sbjct: 150 SANVLAIALDHTVYLWDASTGSTSELVTIDEEKGPVTSINWAPDGRHVAVGLNNSEVQLW 209
Query: 190 DTAANKQLRTLKGGHRARVGSLAWNGHVLTTGGMDGKIVNNDVRLRSQIINTYRGHRREV 249
D+A+N+QLRTLKGGH++RVGSLAWN H+LTTGGMDG I+NNDVR+RS I+ TYRGH +EV
Sbjct: 210 DSASNRQLRTLKGGHQSRVGSLAWNNHILTTGGMDGLIINNDVRIRSPIVETYRGHTQEV 269
Query: 250 CGLKWSLDGKQLASGGNDNVVHIWDMSAVSSNSPTRWLYRFDEHKAAVKALAWCPFQGNL 309
CGLKWS G+QLASGGNDNVVHIWD S SSNS T+WL+R +EH +AVKALAWCPFQ NL
Sbjct: 270 CGLKWSGSGQQLASGGNDNVVHIWDRSVASSNSTTQWLHRLEEHTSAVKALAWCPFQANL 329
Query: 310 LASGGGGGDCCVKLWNTGMGERMNSVDTGSQVCALLWSKNERELLSSHGLTQNQLTLWKY 369
LA+GGGGGD +K WNT G +NSVDTGSQVC+LLWSKNERELLSSHG TQNQLTLWKY
Sbjct: 330 LATGGGGGDRTIKFWNTHTGACLNSVDTGSQVCSLLWSKNERELLSSHGFTQNQLTLWKY 389
Query: 370 PSMLKIAELHGHTSRVLHMTQSPDGSTVASAAAAADQTLRFWEVFGTPPA---APPKRNK 426
PSM+K+AEL GHTSRVL+M QSPDG TVAS AA D+TLRFW VFG P A PK
Sbjct: 390 PSMVKMAELTGHTSRVLYMAQSPDGCTVAS--AAGDETLRFWNVFGVPETAKKAAPKAVS 447
Query: 427 MPFADSNRIR 436
PF+ NRIR
Sbjct: 448 EPFSHVNRIR 457
>At5g27080 putative cdc20 protein
Length = 466
Score = 504 bits (1297), Expect = e-143
Identities = 257/408 (62%), Positives = 299/408 (72%), Gaps = 9/408 (2%)
Query: 23 QNIDRFIPNRSAIDWDHATTILSTTTTNVTKENLWLASNVYQKKLAEAADLP-TRILAFR 81
QN+DRFIPNRSA+D+D A L+ E + Y +LA + TRILAFR
Sbjct: 20 QNLDRFIPNRSAMDFDFANYALTQGRKRNVDEITSASRKAYMTQLAVVMNQNRTRILAFR 79
Query: 82 NKPR----KRNVISPPPPPRS-KPMRYIPKTCEGTFDLPDLSDDFSLNLLDWGSRNVLSI 136
NKP+ + SP P+S KP RYIP+ E D P L DDF LNLLDWGS NVL+I
Sbjct: 80 NKPKALLSSNHSDSPHQNPKSVKPRRYIPQNSERVLDAPGLMDDFYLNLLDWGSANVLAI 139
Query: 137 ALDHTIYFWNASDSSGSEFVTVDEEEGPVTSVCWAPDGRHLAVGLTNSHVQLWDTAANKQ 196
AL T+Y W+AS S SE VT+DE++GPVTS+ W DG LAVGL NS VQLWD +N+Q
Sbjct: 140 ALGDTVYLWDASSGSTSELVTIDEDKGPVTSINWTQDGLDLAVGLDNSEVQLWDFVSNRQ 199
Query: 197 LRTLKGGHRARVGSLAWNGHVLTTGGMDGKIVNNDVRLRSQIINTYRGHRREVCGLKWSL 256
+RTL GGH +RVGSLAWN H+LTTGGMDGKIVNNDVR+RS I+ TY GH EVCGLKWS
Sbjct: 200 VRTLIGGHESRVGSLAWNNHILTTGGMDGKIVNNDVRIRSSIVGTYLGHTEEVCGLKWSE 259
Query: 257 DGKQLASGGNDNVVHIWDMSAVSSNSPTR-WLYRFDEHKAAVKALAWCPFQGNLLASGGG 315
GK+LASGGN NVVHIWD +V+S+ PTR WL+RF+EH AAV+ALAWCPFQ LLA+GGG
Sbjct: 260 SGKKLASGGNYNVVHIWDHRSVASSKPTRQWLHRFEEHTAAVRALAWCPFQATLLATGGG 319
Query: 316 GGDCCVKLWNTGMGERMNSVDTGSQVCALLWSKNERELLSSHGLTQNQLTLWKYPSMLKI 375
GD +K WNT G +NSV+TGSQVC+LLWS+ ERELLSSHG TQNQLTLWKYPSM K+
Sbjct: 320 VGDGKIKFWNTHTGACLNSVETGSQVCSLLWSQRERELLSSHGFTQNQLTLWKYPSMSKM 379
Query: 376 AELHGHTSRVLHMTQSPDGSTVASAAAAADQTLRFWEVFGTPPAAPPK 423
AEL+GHTSRVL M QSP+G TVAS AA D+ LR W VFG PP K
Sbjct: 380 AELNGHTSRVLFMAQSPNGCTVAS--AAGDENLRLWNVFGEPPKTTKK 425
>At5g27570 cdc20-like protein
Length = 411
Score = 502 bits (1292), Expect = e-142
Identities = 257/413 (62%), Positives = 302/413 (72%), Gaps = 13/413 (3%)
Query: 35 IDWDHATTILSTTTTNVTKENLWLASNVYQKKLAEAADLP-TRILAFRNKPR---KRNVI 90
+D+D A L+ E + Y +LAEA + TRILAFRNKP+ N
Sbjct: 1 MDFDFANYALTQGRKRNVDEVTSASRKAYMTQLAEAMNQNRTRILAFRNKPKALLSSNHS 60
Query: 91 SPP--PPPRSKPMRYIPKTCEGTFDLPDLSDDFSLNLLDWGSRNVLSIALDHTIYFWNAS 148
PP P KP RYIP+ E D P ++DDF LNLLDWGS NVL+IAL T+Y W+AS
Sbjct: 61 DPPHQQPISVKPRRYIPQNSERVLDAPGIADDFYLNLLDWGSSNVLAIALGDTVYLWDAS 120
Query: 149 DSSGSEFVTVDEEEGPVTSVCWAPDGRHLAVGLTNSHVQLWDTAANKQLRTLKGGHRARV 208
S + VT+DEEEGPVTS+ W DG LA+GL NS VQLWD +N+Q+RTL+GGH +RV
Sbjct: 121 SGSTYKLVTIDEEEGPVTSINWTQDGLDLAIGLDNSEVQLWDCVSNRQVRTLRGGHESRV 180
Query: 209 GSLAWNGHVLTTGGMDGKIVNNDVRLRSQIINTYRGHRREVCGLKWSLDGKQLASGGNDN 268
GSLAWN H+LTTGGMDGKIVNNDVR+RS I+ TY GH EVCGLKWS GK+LASGGNDN
Sbjct: 181 GSLAWNNHILTTGGMDGKIVNNDVRIRSSIVETYLGHTEEVCGLKWSESGKKLASGGNDN 240
Query: 269 VVHIWDMSAVSSNSPTR-WLYRFDEHKAAVKALAWCPFQGNLLASGGGGGDCCVKLWNTG 327
VVHIWD +V+S++PTR WL+RF+EH AAV+ALAWCPFQ +LLA+GGG GD +K WNT
Sbjct: 241 VVHIWDHRSVASSNPTRQWLHRFEEHTAAVRALAWCPFQASLLATGGGVGDGKIKFWNTH 300
Query: 328 MGERMNSVDTGSQVCALLWSKNERELLSSHGLTQNQLTLWKYPSMLKIAELHGHTSRVLH 387
G +NSV+TGSQVC+LLWSK+ERELLSSHG TQNQLTLWKYPSM+K+AEL+GHTSRVL
Sbjct: 301 TGACLNSVETGSQVCSLLWSKSERELLSSHGFTQNQLTLWKYPSMVKMAELNGHTSRVLF 360
Query: 388 MTQSPDGSTVASAAAAADQTLRFWEVFGTPP----AAPPKRNKMPFADSNRIR 436
M QSPDG TVAS AA D+TLR W VFG PP A K+ PFA N IR
Sbjct: 361 MAQSPDGCTVAS--AAGDETLRLWNVFGEPPKTTKKAASKKYTDPFAHVNHIR 411
>At5g26900 WD-repeat protein - like
Length = 444
Score = 502 bits (1292), Expect = e-142
Identities = 253/407 (62%), Positives = 297/407 (72%), Gaps = 8/407 (1%)
Query: 23 QNIDRFIPNRSAIDWDHATTILSTTTTNVTKENLWLASNVYQKKLAEAADLP-TRILAFR 81
QN+DRFIPNRSA D+D A L+ + E + Y +LA + TRILAFR
Sbjct: 23 QNLDRFIPNRSAKDFDFANYALTQGSKRNLDEVTSASRKAYMTQLAVVMNQNRTRILAFR 82
Query: 82 NKPRK----RNVISPPPPPRS-KPMRYIPKTCEGTFDLPDLSDDFSLNLLDWGSRNVLSI 136
NKP+ + SP P+ KP RYIP+ E D P L DDFSLNLLDWGS NVL+I
Sbjct: 83 NKPKSLLSTNHSDSPHQNPKPVKPRRYIPQNSERVLDAPGLRDDFSLNLLDWGSANVLAI 142
Query: 137 ALDHTIYFWNASDSSGSEFVTVDEEEGPVTSVCWAPDGRHLAVGLTNSHVQLWDTAANKQ 196
AL T+Y W+AS S SE VT+DE++GPVTS+ W DG LAVGL NS VQLWD +N+Q
Sbjct: 143 ALGDTVYLWDASSGSTSELVTIDEDKGPVTSINWTQDGLDLAVGLDNSEVQLWDCVSNRQ 202
Query: 197 LRTLKGGHRARVGSLAWNGHVLTTGGMDGKIVNNDVRLRSQIINTYRGHRREVCGLKWSL 256
+RTL+GGH +RVGSLAW+ H+LTTGGMDGKIVNNDVR+RS I+ TY GH EVCGLKWS
Sbjct: 203 VRTLRGGHESRVGSLAWDNHILTTGGMDGKIVNNDVRIRSSIVETYLGHTEEVCGLKWSE 262
Query: 257 DGKQLASGGNDNVVHIWDMSAVSSNSPTRWLYRFDEHKAAVKALAWCPFQGNLLASGGGG 316
G + ASGGNDNVVHIWD S SS +WL+RF+EH AAV+ALAWCPFQ +LLA+GGG
Sbjct: 263 SGNKQASGGNDNVVHIWDRSLASSKQTRQWLHRFEEHTAAVRALAWCPFQASLLATGGGV 322
Query: 317 GDCCVKLWNTGMGERMNSVDTGSQVCALLWSKNERELLSSHGLTQNQLTLWKYPSMLKIA 376
GD +K WNT G +NSV+TGSQVC+LLWS++ERELLSSHG TQNQLTLWKYPSM K+A
Sbjct: 323 GDGKIKFWNTHTGACLNSVETGSQVCSLLWSQSERELLSSHGFTQNQLTLWKYPSMSKMA 382
Query: 377 ELHGHTSRVLHMTQSPDGSTVASAAAAADQTLRFWEVFGTPPAAPPK 423
EL+GHTSRVL M QSP+G TVAS AA D+ LR W VFG PP K
Sbjct: 383 ELNGHTSRVLFMAQSPNGCTVAS--AAGDENLRLWNVFGEPPKTTKK 427
>At5g27945 WD-repeat protein - like
Length = 428
Score = 420 bits (1080), Expect = e-118
Identities = 203/303 (66%), Positives = 235/303 (76%), Gaps = 6/303 (1%)
Query: 138 LDHTIYFWNASDSSGSEFVTVDEEEGPVTSVCWAPDGRHLAVGLTNSHVQLWDTAANKQL 197
L T+Y W+AS S+ VT+D+E GPVTS+ W DG LAVGL NS VQ+WD +N+ +
Sbjct: 128 LGDTVYLWDASSCYTSKLVTIDDENGPVTSINWTQDGLDLAVGLDNSEVQVWDCVSNRHV 187
Query: 198 RTLKGGHRARVGSLAWNGHVLTTGGMDGKIVNNDVRLRSQIINTYRGHRREVCGLKWSLD 257
RTL+GGH +RVGSLAWN H+LTTGGMDGKIVNNDVR+RS II TY GH EVCGLKWS
Sbjct: 188 RTLRGGHESRVGSLAWNNHILTTGGMDGKIVNNDVRIRSSIIGTYVGHTEEVCGLKWSES 247
Query: 258 GKQLASGGNDNVVHIWDMSAVSSNSPTRWLYRFDEHKAAVKALAWCPFQGNLLASGGGGG 317
GK+LASGGNDNVVHIWD S SSN +WL+RF+EH AAV+ALAWCPFQ +LLA+GGG G
Sbjct: 248 GKKLASGGNDNVVHIWDRSLASSNPTRQWLHRFEEHTAAVRALAWCPFQASLLATGGGVG 307
Query: 318 DCCVKLWNTGMGERMNSVDTGSQVCALLWSKNERELLSSHGLTQNQLTLWKYPSMLKIAE 377
D + WNT G +NSV+TGSQVC+LLWSK+ERELLS+HG TQNQLTLWKYPSM+K+AE
Sbjct: 308 DGKINFWNTHTGACLNSVETGSQVCSLLWSKSERELLSAHGFTQNQLTLWKYPSMVKMAE 367
Query: 378 LHGHTSRVLHMTQSPDGSTVASAAAAADQTLRFWEVFGTPP----AAPPKRNKMPFADSN 433
L+GHTSRVL M QSPDG TVAS AA D+TLR W VFG PP A K+ PFA N
Sbjct: 368 LNGHTSRVLFMAQSPDGCTVAS--AAGDETLRLWNVFGEPPKTTKKAASKKYTEPFAHVN 425
Query: 434 RIR 436
IR
Sbjct: 426 HIR 428
>At5g13840 cell cycle switch protein
Length = 481
Score = 322 bits (826), Expect = 2e-88
Identities = 156/338 (46%), Positives = 220/338 (64%), Gaps = 20/338 (5%)
Query: 91 SPPPPPRSKPMRYIPKTCEGTFDLPDLSDDFSLNLLDWGSRNVLSIALDHTIYFWNASDS 150
SPPP KP R +PKT D P L DDF LN++DW S+NVL++ L +Y W AS+S
Sbjct: 149 SPPP----KPPRKVPKTPHKVLDAPSLQDDFYLNVVDWSSQNVLAVGLGTCVYLWTASNS 204
Query: 151 SGSEFVTVDEEEGPVTSVC---WAPDGRHLAVGLTNSHVQLWDTAANKQLRTLKGGHRAR 207
++ + GP SVC W +G ++++G ++ VQ+WD K++RT+ GGH+ R
Sbjct: 205 KVTKLCDL----GPNDSVCSVQWTREGSYISIGTSHGQVQVWDGTQCKRVRTM-GGHQTR 259
Query: 208 VGSLAWNGHVLTTGGMDGKIVNNDVRLRSQIINTYRGHRREVCGLKWSLDGKQLASGGND 267
G LAWN +L++G D I+ +D+R++S ++ GH+ EVCGLKWS D ++LASGGND
Sbjct: 260 TGVLAWNSRILSSGSRDRNILQHDIRVQSDFVSKLVGHKSEVCGLKWSHDDRELASGGND 319
Query: 268 NVVHIWDMSAVSSNSPTRWLYRFDEHKAAVKALAWCPFQGNLLASGGGGGDCCVKLWNTG 327
N + +W+ N + + + EH AAVKA+ W P Q +LLASGGG D C++ WNT
Sbjct: 320 NQLLVWN------NHSQQPILKLTEHTAAVKAITWSPHQSSLLASGGGTADRCIRFWNTT 373
Query: 328 MGERMNSVDTGSQVCALLWSKNERELLSSHGLTQNQLTLWKYPSMLKIAELHGHTSRVLH 387
G ++NS+DTGSQVC L WSKN E++S+HG +QNQ+ LWKYPSM K+A L GH+ RVL+
Sbjct: 374 NGNQLNSIDTGSQVCNLAWSKNVNEIVSTHGYSQNQIMLWKYPSMSKVATLTGHSMRVLY 433
Query: 388 MTQSPDGSTVASAAAAADQTLRFWEVFGTPPAAPPKRN 425
+ SPDG T+ + A D+TLRFW VF + P ++
Sbjct: 434 LATSPDGQTIVT--GAGDETLRFWNVFPSVKMQTPVKD 469
>At4g22910 putative fizzy-related protein
Length = 483
Score = 312 bits (799), Expect = 3e-85
Identities = 174/414 (42%), Positives = 245/414 (59%), Gaps = 34/414 (8%)
Query: 26 DRFIPNRSAIDWDHATTILSTTTTNVTKEN------LWLASNVYQKKLAEAADLP----- 74
DRFIP+RS ++ A LS + + KE+ L + ++ + E D+
Sbjct: 62 DRFIPSRSGSNF--ALFDLSPSPSKDGKEDGAGSYATLLRAAMFGPETPEKRDITGFSSS 119
Query: 75 TRILAFRNKP-RKRNVISP-------PPPPRSKPM---RYIPKTCEGTFDLPDLSDDFSL 123
I F+ + R N SP P S P+ R +P++ D P L DDF L
Sbjct: 120 RNIFRFKTETHRSLNSFSPFGVDDDSPGVSHSGPVKAPRKVPRSPYKVLDAPALQDDFYL 179
Query: 124 NLLDWGSRNVLSIALDHTIYFWNASDSSGSEFVTVDEEEGPVTSVCWAPDGRHLAVGLTN 183
NL+DW ++NVL++ L + +Y WNA S ++ + E+ V SV WA G HLAVG +
Sbjct: 180 NLVDWSAQNVLAVGLGNCVYLWNACSSKVTKLCDLGAEDS-VCSVGWALRGTHLAVGTST 238
Query: 184 SHVQLWDTAANKQLRTLKGGHRARVGSLAWNGHVLTTGGMDGKIVNNDVRLRSQIINTYR 243
VQ+WD + K+ RT++G HR RVG+LAW VL++G D I+ D+R + ++
Sbjct: 239 GKVQIWDASRCKRTRTMEG-HRLRVGALAWGSSVLSSGSRDKSILQRDIRCQEDHVSKLA 297
Query: 244 GHRREVCGLKWSLDGKQLASGGNDNVVHIWDMSAVSSNSPTRWLYRFDEHKAAVKALAWC 303
GH+ EVCGLKWS D ++LASGGNDN + +W+ + T+ + ++ EH AAVKA+AW
Sbjct: 298 GHKSEVCGLKWSYDNRELASGGNDNRLFVWNQHS------TQPVLKYSEHTAAVKAIAWS 351
Query: 304 PFQGNLLASGGGGGDCCVKLWNTGMGERMNSVDTGSQVCALLWSKNERELLSSHGLTQNQ 363
P LLASGGG D C++ WNT ++S+DT SQVC L WSKN EL+S+HG +QNQ
Sbjct: 352 PHVHGLLASGGGTADRCIRFWNTTTNTHLSSIDTCSQVCNLAWSKNVNELVSTHGYSQNQ 411
Query: 364 LTLWKYPSMLKIAELHGHTSRVLHMTQSPDGSTVASAAAAADQTLRFWEVFGTP 417
+ +WKYP+M KIA L GHT RVL++ SPDG T+ + A D+TLRFW VF +P
Sbjct: 412 IIVWKYPTMSKIATLTGHTYRVLYLAVSPDGQTIVT--GAGDETLRFWNVFPSP 463
>At4g11920 Srw1 like protein
Length = 475
Score = 311 bits (798), Expect = 3e-85
Identities = 154/334 (46%), Positives = 215/334 (64%), Gaps = 10/334 (2%)
Query: 84 PRKRNVISPPPPPRSKPMRYIPKTCEGTFDLPDLSDDFSLNLLDWGSRNVLSIALDHTIY 143
P +V+S P K R I ++ D P L DDF LNL+DW ++NVL++ L + +Y
Sbjct: 132 PFDSDVVSGVSPSPVKSPRKILRSPYKVLDAPALQDDFYLNLVDWSAQNVLAVGLGNCVY 191
Query: 144 FWNASDSSGSEFVTVDEEEGPVTSVCWAPDGRHLAVGLTNSHVQLWDTAANKQLRTLKGG 203
WNA S ++ + +E V SV WA G HLA+G ++ VQ+WD K +RT++G
Sbjct: 192 LWNACSSKVTKLCDLGVDE-TVCSVGWALRGTHLAIGTSSGTVQIWDVLRCKNIRTMEG- 249
Query: 204 HRARVGSLAWNGHVLTTGGMDGKIVNNDVRLRSQIINTYRGHRREVCGLKWSLDGKQLAS 263
HR RVG+LAW+ VL++G D I+ D+R + ++ +GH+ E+CGLKWS D ++LAS
Sbjct: 250 HRLRVGALAWSSSVLSSGSRDKSILQRDIRTQEDHVSKLKGHKSEICGLKWSSDNRELAS 309
Query: 264 GGNDNVVHIWDMSAVSSNSPTRWLYRFDEHKAAVKALAWCPFQGNLLASGGGGGDCCVKL 323
GGNDN + +W+ + T+ + RF EH AAVKA+AW P LLASGGG D C++
Sbjct: 310 GGNDNKLFVWNQHS------TQPVLRFCEHAAAVKAIAWSPHHFGLLASGGGTADRCIRF 363
Query: 324 WNTGMGERMNSVDTGSQVCALLWSKNERELLSSHGLTQNQLTLWKYPSMLKIAELHGHTS 383
WNT +N VDT SQVC L+WSKN EL+S+HG +QNQ+ +WKYP+M K+A L GH+
Sbjct: 364 WNTTTNTHLNCVDTNSQVCNLVWSKNVNELVSTHGYSQNQIIVWKYPTMSKLATLTGHSY 423
Query: 384 RVLHMTQSPDGSTVASAAAAADQTLRFWEVFGTP 417
RVL++ SPDG T+ + A D+TLRFW VF +P
Sbjct: 424 RVLYLAVSPDGQTIVT--GAGDETLRFWNVFPSP 455
>At5g08390 katanin p80 subunit - like protein
Length = 823
Score = 87.0 bits (214), Expect = 2e-17
Identities = 70/220 (31%), Positives = 109/220 (48%), Gaps = 35/220 (15%)
Query: 130 SRNVLSIALDHTIYFW-----NA-----SDSSGSEFVTVDEEEGPVTSVCWAPDGRHLAV 179
SR +++ DH + W NA SSG + VT D EG V A
Sbjct: 122 SRVLVTGGEDHKVNLWAIGKPNAILSLYGHSSGIDSVTFDASEGLV------------AA 169
Query: 180 GLTNSHVQLWDTAANKQLRTLKGGHRARVGSLAWN--GHVLTTGGMDGKIVNNDVRLRSQ 237
G + ++LWD K +RTL G HR+ S+ ++ G +G +D + D+R +
Sbjct: 170 GAASGTIKLWDLEEAKVVRTLTG-HRSNCVSVNFHPFGEFFASGSLDTNLKIWDIRKKG- 227
Query: 238 IINTYRGHRREVCGLKWSLDGKQLASGGNDNVVHIWDMSAVSSNSPTRWLYRFDEHKAAV 297
I+TY+GH R V L+++ DG+ + SGG DNVV +WD++A + L+ F H+ +
Sbjct: 228 CIHTYKGHTRGVNVLRFTPDGRWIVSGGEDNVVKVWDLTA------GKLLHEFKSHEGKI 281
Query: 298 KALAWCPFQGNLLASGGGGGDCCVKLWNTGMGERMNSVDT 337
++L + P + LLA+ G D VK W+ E + S T
Sbjct: 282 QSLDFHPHE-FLLAT--GSADKTVKFWDLETFELIGSGGT 318
Score = 42.0 bits (97), Expect = 7e-04
Identities = 54/241 (22%), Positives = 89/241 (36%), Gaps = 58/241 (24%)
Query: 175 RHLAVGLTNSHVQLWDTAANKQLRTLKGGHRARVGSLAWNGH--VLTTGGMDGKIVNNDV 232
R L G + V LW + +L G H + + S+ ++ ++ G G I D+
Sbjct: 123 RVLVTGGEDHKVNLWAIGKPNAILSLYG-HSSGIDSVTFDASEGLVAAGAASGTIKLWDL 181
Query: 233 RLRSQIINTYRGHRREVCGLKWSLDGKQLASGGNDNVVHIWDMSAVSSNSPTRWLYRFDE 292
++++ T GHR + + G+ ASG D + IWD+ ++ +
Sbjct: 182 E-EAKVVRTLTGHRSNCVSVNFHPFGEFFASGSLDTNLKIWDIRKKGC------IHTYKG 234
Query: 293 HKAAVKALAWCPFQGNLLASGGGGGDCCVKLWNTGMGERMNSVDTGSQVCALLWSKNERE 352
H V L + P G + SGG D VK+W+ G+ ++ E
Sbjct: 235 HTRGVNVLRFTP-DGRWIVSGGE--DNVVKVWDLTAGKLLH------------------E 273
Query: 353 LLSSHGLTQNQLTLWKYPSMLKIAELHGHTSRVLHMTQSPDGSTVASAAAAADQTLRFWE 412
S G KI L H L T S AD+T++FW+
Sbjct: 274 FKSHEG---------------KIQSLDFHPHEFLLATGS------------ADKTVKFWD 306
Query: 413 V 413
+
Sbjct: 307 L 307
Score = 33.5 bits (75), Expect = 0.24
Identities = 32/125 (25%), Positives = 50/125 (39%), Gaps = 7/125 (5%)
Query: 290 FDEHKAAVKALAWCPFQGNLLASGGGGGDCCVKLWNTGMGERMNSV-DTGSQVCALLWSK 348
F H AAV L +L +GG D V LW G + S+ S + ++ +
Sbjct: 105 FVAHSAAVNCLKIGRKSSRVLVTGGE--DHKVNLWAIGKPNAILSLYGHSSGIDSVTFDA 162
Query: 349 NERELLSSHGLTQNQLTLWKYPSMLKIAELHGHTSRVLHMTQSPDGSTVASAAAAADQTL 408
+E L + G + LW + L GH S + + P G AS + D L
Sbjct: 163 SEG--LVAAGAASGTIKLWDLEEAKVVRTLTGHRSNCVSVNFHPFGEFFAS--GSLDTNL 218
Query: 409 RFWEV 413
+ W++
Sbjct: 219 KIWDI 223
>At5g23430 unknown protein
Length = 837
Score = 80.1 bits (196), Expect = 2e-15
Identities = 67/217 (30%), Positives = 106/217 (47%), Gaps = 35/217 (16%)
Query: 130 SRNVLSIALDHTIYFW-----NA-----SDSSGSEFVTVDEEEGPVTSVCWAPDGRHLAV 179
SR +++ DH + W NA SSG + VT D E V A
Sbjct: 29 SRVLVTGGEDHKVNLWAIGKPNAILSLYGHSSGIDSVTFDASEVLV------------AA 76
Query: 180 GLTNSHVQLWDTAANKQLRTLKGGHRARVGSLAWN--GHVLTTGGMDGKIVNNDVRLRSQ 237
G + ++LWD K +RTL G HR+ S+ ++ G +G +D + D+R +
Sbjct: 77 GAASGTIKLWDLEEAKIVRTLTG-HRSNCISVDFHPFGEFFASGSLDTNLKIWDIRKKG- 134
Query: 238 IINTYRGHRREVCGLKWSLDGKQLASGGNDNVVHIWDMSAVSSNSPTRWLYRFDEHKAAV 297
I+TY+GH R V L+++ DG+ + SGG DN+V +WD++A + L F H+ +
Sbjct: 135 CIHTYKGHTRGVNVLRFTPDGRWVVSGGEDNIVKVWDLTA------GKLLTEFKSHEGQI 188
Query: 298 KALAWCPFQGNLLASGGGGGDCCVKLWNTGMGERMNS 334
++L + P + LLA+ G D VK W+ E + S
Sbjct: 189 QSLDFHPHE-FLLAT--GSADRTVKFWDLETFELIGS 222
Score = 40.8 bits (94), Expect = 0.001
Identities = 52/241 (21%), Positives = 89/241 (36%), Gaps = 58/241 (24%)
Query: 175 RHLAVGLTNSHVQLWDTAANKQLRTLKGGHRARVGSLAWNGH--VLTTGGMDGKIVNNDV 232
R L G + V LW + +L G H + + S+ ++ ++ G G I D+
Sbjct: 30 RVLVTGGEDHKVNLWAIGKPNAILSLYG-HSSGIDSVTFDASEVLVAAGAASGTIKLWDL 88
Query: 233 RLRSQIINTYRGHRREVCGLKWSLDGKQLASGGNDNVVHIWDMSAVSSNSPTRWLYRFDE 292
++I+ T GHR + + G+ ASG D + IWD+ ++ +
Sbjct: 89 E-EAKIVRTLTGHRSNCISVDFHPFGEFFASGSLDTNLKIWDIRKKGC------IHTYKG 141
Query: 293 HKAAVKALAWCPFQGNLLASGGGGGDCCVKLWNTGMGERMNSVDTGSQVCALLWSKNERE 352
H V L + P G + SGG D VK+W+ G+ + E
Sbjct: 142 HTRGVNVLRFTP-DGRWVVSGGE--DNIVKVWDLTAGKLLT------------------E 180
Query: 353 LLSSHGLTQNQLTLWKYPSMLKIAELHGHTSRVLHMTQSPDGSTVASAAAAADQTLRFWE 412
S G Q+ + H H + A +AD+T++FW+
Sbjct: 181 FKSHEGQIQS-------------LDFHPHEFLL--------------ATGSADRTVKFWD 213
Query: 413 V 413
+
Sbjct: 214 L 214
Score = 35.4 bits (80), Expect = 0.062
Identities = 33/128 (25%), Positives = 52/128 (39%), Gaps = 7/128 (5%)
Query: 287 LYRFDEHKAAVKALAWCPFQGNLLASGGGGGDCCVKLWNTGMGERMNSV-DTGSQVCALL 345
L F H AAV L +L +GG D V LW G + S+ S + ++
Sbjct: 9 LQEFVAHSAAVNCLKIGRKSSRVLVTGGE--DHKVNLWAIGKPNAILSLYGHSSGIDSVT 66
Query: 346 WSKNERELLSSHGLTQNQLTLWKYPSMLKIAELHGHTSRVLHMTQSPDGSTVASAAAAAD 405
+ +E +L + G + LW + L GH S + + P G AS + D
Sbjct: 67 FDASE--VLVAAGAASGTIKLWDLEEAKIVRTLTGHRSNCISVDFHPFGEFFAS--GSLD 122
Query: 406 QTLRFWEV 413
L+ W++
Sbjct: 123 TNLKIWDI 130
>At1g61210
Length = 282
Score = 78.2 bits (191), Expect = 8e-15
Identities = 62/230 (26%), Positives = 108/230 (46%), Gaps = 28/230 (12%)
Query: 119 DDFSLNLLDWGSRNVLSIALDHTIYFWNA--SDSSGSEFVTVDEEEGPVTSVCWAPDGRH 176
DD+ +NL G L + D ++W + +S + V D E V +
Sbjct: 56 DDYKVNLWAIGKPTSL-MKNDAIAFYWQSLCGHTSAVDSVAFDSAEVLVLA--------- 105
Query: 177 LAVGLTNSHVQLWDTAANKQLRTLKGGHRARVGSLAWN--GHVLTTGGMDGKIVNNDVRL 234
G ++ ++LWD K +R G HR+ ++ ++ G L +G D + D+R
Sbjct: 106 ---GASSGVIKLWDVEEAKMVRAFTG-HRSNCSAVEFHPFGEFLASGSSDANLKIWDIRK 161
Query: 235 RSQIINTYRGHRREVCGLKWSLDGKQLASGGNDNVVHIWDMSAVSSNSPTRWLYRFDEHK 294
+ I TY+GH R + ++++ DG+ + SGG DNVV +WD++A + L+ F H+
Sbjct: 162 KG-CIQTYKGHSRGISTIRFTPDGRWVVSGGLDNVVKVWDLTA------GKLLHEFKFHE 214
Query: 295 AAVKALAWCPFQGNLLASGGGGGDCCVKLWNTGMGERMNSVDTGSQVCAL 344
+++L + P + LLA+ G D VK W+ E + S C +
Sbjct: 215 GPIRSLDFHPLE-FLLAT--GSADRTVKFWDLETFELIGSTRPEVLACGV 261
Score = 37.0 bits (84), Expect = 0.021
Identities = 35/170 (20%), Positives = 73/170 (42%), Gaps = 12/170 (7%)
Query: 244 GHRREVCGLKWSLDGKQLASGGNDNVVHIWDMSAVSSNSPTRWLYRFDEHKAAVKALAWC 303
GH V + + + +G + V+ +WD+ + + F H++ A+ +
Sbjct: 86 GHTSAVDSVAFDSAEVLVLAGASSGVIKLWDVEEA------KMVRAFTGHRSNCSAVEFH 139
Query: 304 PFQGNLLASGGGGGDCCVKLWNTGMGERMNSVDTGSQVCALLWSKNERELLSSHGLTQNQ 363
PF G LASG D +K+W+ + + S+ + + + + S GL N
Sbjct: 140 PF-GEFLASGSS--DANLKIWDIRKKGCIQTYKGHSRGISTIRFTPDGRWVVSGGL-DNV 195
Query: 364 LTLWKYPSMLKIAELHGHTSRVLHMTQSPDGSTVASAAAAADQTLRFWEV 413
+ +W + + E H + + P +A+ +A D+T++FW++
Sbjct: 196 VKVWDLTAGKLLHEFKFHEGPIRSLDFHPLEFLLATGSA--DRTVKFWDL 243
Score = 30.0 bits (66), Expect = 2.6
Identities = 33/143 (23%), Positives = 48/143 (33%), Gaps = 21/143 (14%)
Query: 284 TRWLYRFDEHKAAVKALAWCPFQGNLLASGGGGGDCCVKLWNTGMGERMNSVDTGSQVCA 343
T L F H A V L+ L +GG D V LW G + D A
Sbjct: 25 TESLGEFLAHSANVNCLSIGKKTSRLFITGGD--DYKVNLWAIGKPTSLMKNDA----IA 78
Query: 344 LLWSK-------------NERELLSSHGLTQNQLTLWKYPSMLKIAELHGHTSRVLHMTQ 390
W + E+L G + + LW + GH S +
Sbjct: 79 FYWQSLCGHTSAVDSVAFDSAEVLVLAGASSGVIKLWDVEEAKMVRAFTGHRSNCSAVEF 138
Query: 391 SPDGSTVASAAAAADQTLRFWEV 413
P G +AS ++D L+ W++
Sbjct: 139 HPFGEFLAS--GSSDANLKIWDI 159
>At1g11160 hypothetical protein
Length = 974
Score = 77.4 bits (189), Expect = 1e-14
Identities = 51/164 (31%), Positives = 88/164 (53%), Gaps = 13/164 (7%)
Query: 164 PVTSVCWAPDGRHLAVGLTNSHVQLWDTAANKQLRTLKGGHRARVGSLAWN--GHVLTTG 221
PV SV + + + G ++ ++LWD +K +R G HR+ ++ ++ G L +G
Sbjct: 9 PVDSVAFNSEEVLVLAGASSGVIKLWDLEESKMVRAFTG-HRSNCSAVEFHPFGEFLASG 67
Query: 222 GMDGKIVNNDVRLRSQIINTYRGHRREVCGLKWSLDGKQLASGGNDNVVHIWDMSAVSSN 281
D + D R + I TY+GH R + +++S DG+ + SGG DNVV +WD++A
Sbjct: 68 SSDTNLRVWDTRKKG-CIQTYKGHTRGISTIEFSPDGRWVVSGGLDNVVKVWDLTA---- 122
Query: 282 SPTRWLYRFDEHKAAVKALAWCPFQGNLLASGGGGGDCCVKLWN 325
+ L+ F H+ +++L + P + LLA+ G D VK W+
Sbjct: 123 --GKLLHEFKCHEGPIRSLDFHPLE-FLLAT--GSADRTVKFWD 161
Score = 39.3 bits (90), Expect = 0.004
Identities = 34/170 (20%), Positives = 77/170 (45%), Gaps = 12/170 (7%)
Query: 244 GHRREVCGLKWSLDGKQLASGGNDNVVHIWDMSAVSSNSPTRWLYRFDEHKAAVKALAWC 303
GH V + ++ + + +G + V+ +WD+ ++ + F H++ A+ +
Sbjct: 5 GHTSPVDSVAFNSEEVLVLAGASSGVIKLWDLEE------SKMVRAFTGHRSNCSAVEFH 58
Query: 304 PFQGNLLASGGGGGDCCVKLWNTGMGERMNSVDTGSQVCALLWSKNERELLSSHGLTQNQ 363
PF G LASG D +++W+T + + ++ + + + + S GL N
Sbjct: 59 PF-GEFLASGSS--DTNLRVWDTRKKGCIQTYKGHTRGISTIEFSPDGRWVVSGGL-DNV 114
Query: 364 LTLWKYPSMLKIAELHGHTSRVLHMTQSPDGSTVASAAAAADQTLRFWEV 413
+ +W + + E H + + P +A+ +A D+T++FW++
Sbjct: 115 VKVWDLTAGKLLHEFKCHEGPIRSLDFHPLEFLLATGSA--DRTVKFWDL 162
>At5g52820 Notchless protein homolog
Length = 473
Score = 69.3 bits (168), Expect = 4e-12
Identities = 75/293 (25%), Positives = 122/293 (41%), Gaps = 63/293 (21%)
Query: 165 VTSVCWAPDGRHLAVGLTNSHVQLWDTAANKQLRTLKGGHRARVGSLAW--NGHVLTTGG 222
V V ++PDG+ LA G ++ V+LWD L T K GH+ V ++AW +G L +G
Sbjct: 112 VLCVSFSPDGKQLASGSGDTTVRLWDLYTETPLFTCK-GHKNWVLTVAWSPDGKHLVSGS 170
Query: 223 MDGKIVNNDVRLRSQIINTYRGHRREVCGLKW-----SLDGKQLASGGNDNVVHIWDMSA 277
G+I + + + GH++ + G+ W S ++ + D IWD++
Sbjct: 171 KSGEICCWNPKKGELEGSPLTGHKKWITGISWEPVHLSSPCRRFVTSSKDGDARIWDITL 230
Query: 278 VSSNSPTRWLYRFDEHKAAVKALAWCPFQGNLLASGGGGGDCCVKLWNT----------G 327
S + H AV + W G G DC +K+W T G
Sbjct: 231 KKS------IICLSGHTLAVTCVKW----GGDGIIYTGSQDCTIKMWETTQGKLIRELKG 280
Query: 328 MGERMNSV--------------DTGSQVCALLWSKNERELLSSHGLTQNQ---------- 363
G +NS+ TG Q ++ +++ L + T+
Sbjct: 281 HGHWINSLALSTEYVLRTGAFDHTGRQYPP---NEEKQKALERYNKTKGDSPERLVSGSD 337
Query: 364 ---LTLWKYPSMLKIAE--LHGHTSRVLHMTQSPDGSTVASAAAAADQTLRFW 411
+ LW+ PS+ K + L GH V H+ SPDG +AS A+ D+++R W
Sbjct: 338 DFTMFLWE-PSVSKQPKKRLTGHQQLVNHVYFSPDGKWIAS--ASFDKSVRLW 387
Score = 68.2 bits (165), Expect = 9e-12
Identities = 74/255 (29%), Positives = 114/255 (44%), Gaps = 33/255 (12%)
Query: 165 VTSVCWAPDGRHLAVGLTNSHVQLWDTAANKQLRTLKGGHRARVGSLAWNG-HVLTTGGM 223
VT V W DG + G + +++W+T K +R LKG H + SLA + +VL TG
Sbjct: 244 VTCVKWGGDGI-IYTGSQDCTIKMWETTQGKLIRELKG-HGHWINSLALSTEYVLRTGAF 301
Query: 224 D--GKIV--NNDVRLRSQIINTYRGHRREVCGLKWSLDGKQLASGGNDNVVHIWDMSAVS 279
D G+ N + + + N +G E +L SG +D + +W+ S
Sbjct: 302 DHTGRQYPPNEEKQKALERYNKTKGDSPE-----------RLVSGSDDFTMFLWEPSV-- 348
Query: 280 SNSPTRWLYRFDEHKAAVKALAWCPFQGNLLASGGGGGDCCVKLWNTGMGERMNSVDTG- 338
S P + R H+ V + + P G +AS D V+LWN G+ + +V G
Sbjct: 349 SKQPKK---RLTGHQQLVNHVYFSP-DGKWIASASF--DKSVRLWN-GITGQFVTVFRGH 401
Query: 339 -SQVCALLWSKNERELLSSHGLTQNQLTLWKYPSMLKIAELHGHTSRVLHMTQSPDGSTV 397
V + WS + R LLS G + L +W+ + +L GH V + SPDG V
Sbjct: 402 VGPVYQVSWSADSRLLLS--GSKDSTLKIWEIRTKKLKQDLPGHADEVFAVDWSPDGEKV 459
Query: 398 ASAAAAADQTLRFWE 412
S D+ L+ W+
Sbjct: 460 VS--GGKDRVLKLWK 472
Score = 63.5 bits (153), Expect = 2e-10
Identities = 56/189 (29%), Positives = 76/189 (39%), Gaps = 23/189 (12%)
Query: 232 VRLRSQIINTYRGHRREVCGLKWSLDGKQLASGGNDNVVHIWDMSAVSSNSPTRWLYRFD 291
+R ++ T GH V + +S DGKQLASG D V +WD+ + L+
Sbjct: 95 IRPVNRCSQTIAGHAEAVLCVSFSPDGKQLASGSGDTTVRLWDLYTETP------LFTCK 148
Query: 292 EHKAAVKALAWCPFQGNLLASGGGGGDCCVKLWNTGMGERMNSVDTGSQ--VCALLW--- 346
HK V +AW P G L SG G+ C WN GE S TG + + + W
Sbjct: 149 GHKNWVLTVAWSP-DGKHLVSGSKSGEIC--CWNPKKGELEGSPLTGHKKWITGISWEPV 205
Query: 347 ---SKNERELLSSHGLTQNQLTLWKYPSMLKIAELHGHTSRVLHMTQSPDGSTVASAAAA 403
S R + SS +W I L GHT V + DG +
Sbjct: 206 HLSSPCRRFVTSS---KDGDARIWDITLKKSIICLSGHTLAVTCVKWGGDGIIY---TGS 259
Query: 404 ADQTLRFWE 412
D T++ WE
Sbjct: 260 QDCTIKMWE 268
Score = 43.9 bits (102), Expect = 2e-04
Identities = 29/95 (30%), Positives = 46/95 (47%), Gaps = 7/95 (7%)
Query: 133 VLSIALDHTIYFWNASDSSGSEFVTVDEEE-GPVTSVCWAPDGRHLAVGLTNSHVQLWDT 191
+ S + D ++ WN +FVTV GPV V W+ D R L G +S +++W+
Sbjct: 375 IASASFDKSVRLWNGITG---QFVTVFRGHVGPVYQVSWSADSRLLLSGSKDSTLKIWEI 431
Query: 192 AANKQLRTLKGGHRARVGSLAW--NGHVLTTGGMD 224
K + L GH V ++ W +G + +GG D
Sbjct: 432 RTKKLKQDLP-GHADEVFAVDWSPDGEKVVSGGKD 465
Score = 33.1 bits (74), Expect = 0.31
Identities = 17/47 (36%), Positives = 26/47 (55%), Gaps = 2/47 (4%)
Query: 380 GHTSRVLHMTQSPDGSTVASAAAAADQTLRFWEVFGTPPAAPPKRNK 426
GH VL ++ SPDG +AS + D T+R W+++ P K +K
Sbjct: 107 GHAEAVLCVSFSPDGKQLASGSG--DTTVRLWDLYTETPLFTCKGHK 151
>At5g67320 unknown protein
Length = 613
Score = 65.9 bits (159), Expect = 4e-11
Identities = 70/316 (22%), Positives = 120/316 (37%), Gaps = 71/316 (22%)
Query: 162 EGPVTSVC---WAPDGRHLAVGLTNSHVQLWD----------TAANKQ---LRTLKGGHR 205
EG + VC W+P LA G ++ ++W T N L+ KG
Sbjct: 262 EGHTSEVCACAWSPSASLLASGSGDATARIWSIPEGSFKAVHTGRNINALILKHAKGKSN 321
Query: 206 AR---VGSLAWNGH--VLTTGGMDGKIVNNDVRLRSQIINTYRGHRREVCGLKWSLDGKQ 260
+ V +L WNG +L TG DG+ L ++I+T H+ + LKW+ G
Sbjct: 322 EKSKDVTTLDWNGEGTLLATGSCDGQA--RIWTLNGELISTLSKHKGPIFSLKWNKKGDY 379
Query: 261 LASGGNDNVVHIWDMSA------------------------VSSNSPTRWLY-------- 288
L +G D +WD+ A +++S +Y
Sbjct: 380 LLTGSVDRTAVVWDVKAEEWKQQFEFHSGPTLDVDWRNNVSFATSSTDSMIYLCKIGETR 439
Query: 289 ---RFDEHKAAVKALAWCPFQGNLLASGGGGGDCCVKLWNTGMGERMNSV-DTGSQVCAL 344
F H+ V + W P G+LLAS D K+WN ++ + + ++ +
Sbjct: 440 PAKTFTGHQGEVNCVKWDP-TGSLLAS--CSDDSTAKIWNIKQSTFVHDLREHTKEIYTI 496
Query: 345 LWSKN-------ERELLSSHGLTQNQLTLWKYPSMLKIAELHGHTSRVLHMTQSPDGSTV 397
WS ++L + + + LW + +GH V + SP+G +
Sbjct: 497 RWSPTGPGTNNPNKQLTLASASFDSTVKLWDAELGKMLCSFNGHREPVYSLAFSPNGEYI 556
Query: 398 ASAAAAADQTLRFWEV 413
AS + D+++ W +
Sbjct: 557 AS--GSLDKSIHIWSI 570
Score = 65.5 bits (158), Expect = 6e-11
Identities = 62/287 (21%), Positives = 113/287 (38%), Gaps = 24/287 (8%)
Query: 126 LDWGSRNVLSI--ALDHTIYFWNASDSSGSEFVTVDEEEGPVTSVCWAPDGRHLAVGLTN 183
LDW L + D W + G T+ + +GP+ S+ W G +L G +
Sbjct: 330 LDWNGEGTLLATGSCDGQARIWTLN---GELISTLSKHKGPIFSLKWNKKGDYLLTGSVD 386
Query: 184 SHVQLWDTAANKQLRTLKGGHRARVGSLAWNGHV-LTTGGMDGKIVNNDVRLRSQIINTY 242
+WD A + + + H + W +V T D I + ++ T+
Sbjct: 387 RTAVVWDVKAEEWKQQFE-FHSGPTLDVDWRNNVSFATSSTDSMIYLCKIG-ETRPAKTF 444
Query: 243 RGHRREVCGLKWSLDGKQLASGGNDNVVHIWDMSAVSSNSPTRWLYRFDEHKAAVKALAW 302
GH+ EV +KW G LAS +D+ IW++ + +++ EH + + W
Sbjct: 445 TGHQGEVNCVKWDPTGSLLASCSDDSTAKIWNI------KQSTFVHDLREHTKEIYTIRW 498
Query: 303 CPF------QGNLLASGGGGGDCCVKLWNTGMGERMNSVDTGSQ-VCALLWSKNERELLS 355
P L D VKLW+ +G+ + S + + V +L +S N + S
Sbjct: 499 SPTGPGTNNPNKQLTLASASFDSTVKLWDAELGKMLCSFNGHREPVYSLAFSPNGEYIAS 558
Query: 356 SHGLTQNQLTLWKYPSMLKIAELHGHTSRVLHMTQSPDGSTVASAAA 402
G + +W KI + + + + + +G+ +A+ A
Sbjct: 559 --GSLDKSIHIWSIKEG-KIVKTYTGNGGIFEVCWNKEGNKIAACFA 602
Score = 45.1 bits (105), Expect = 8e-05
Identities = 53/215 (24%), Positives = 84/215 (38%), Gaps = 28/215 (13%)
Query: 217 VLTTGGMDGKIVNNDVRLRSQIINTYRGHRREVCGLKWSLDGKQLASGGNDNVVHIWDM- 275
V+T I N+DVR+ GH EVC WS LASG D IW +
Sbjct: 243 VMTPTSQTSHIPNSDVRI-------LEGHTSEVCACAWSPSASLLASGSGDATARIWSIP 295
Query: 276 ----SAVSSNSPTRWLY------RFDEHKAAVKALAWCPFQGNLLASGGGGGDCCVKLWN 325
AV + L + +E V L W +G LLA+G G ++W
Sbjct: 296 EGSFKAVHTGRNINALILKHAKGKSNEKSKDVTTLDW-NGEGTLLATGSCDGQ--ARIW- 351
Query: 326 TGMGERMNSVDT-GSQVCALLWSKNERELLSSHGLTQNQLTLWKYPSMLKIAELHGHTSR 384
T GE ++++ + +L W+K LL+ G +W + + H+
Sbjct: 352 TLNGELISTLSKHKGPIFSLKWNKKGDYLLT--GSVDRTAVVWDVKAEEWKQQFEFHSGP 409
Query: 385 VLHMTQSPDGSTVASAAAAADQTLRFWEVFGTPPA 419
L + + V+ A ++ D + ++ T PA
Sbjct: 410 TLDVDWR---NNVSFATSSTDSMIYLCKIGETRPA 441
Score = 37.7 bits (86), Expect = 0.013
Identities = 23/102 (22%), Positives = 45/102 (43%), Gaps = 6/102 (5%)
Query: 135 SIALDHTIYFWNASDSSGSEFVTVDEEEGPVTSVCWAPDGRHLAVGLTNSHVQLWDTAAN 194
S + D T+ W+A G + + PV S+ ++P+G ++A G + + +W
Sbjct: 516 SASFDSTVKLWDAE--LGKMLCSFNGHREPVYSLAFSPNGEYIASGSLDKSIHIWSIKEG 573
Query: 195 KQLRTLKGGHRARVGSLAWN--GHVLTTGGMDGKIVNNDVRL 234
K ++T G + + WN G+ + D + D R+
Sbjct: 574 KIVKTYTG--NGGIFEVCWNKEGNKIAACFADNSVCVLDFRM 613
>At2g41500 putative U4/U6 small nuclear ribonucleoprotein
Length = 554
Score = 64.7 bits (156), Expect = 1e-10
Identities = 63/273 (23%), Positives = 117/273 (42%), Gaps = 17/273 (6%)
Query: 145 WNASDSSGSEFVTVDEEEGPVTSVCWAPDGRHLAVGLTNSHVQLWDTAANKQLRTLKGGH 204
W + + V D +E T V ++P LA + +LW T L+T +G H
Sbjct: 282 WEMPQVTNTIAVLKDHKER-ATDVVFSPVDDCLATASADRTAKLWKTDGTL-LQTFEG-H 338
Query: 205 RARVGSLAWN--GHVLTTGGMDGKIVNNDVRLRSQIINTYRGHRREVCGLKWSLDGKQLA 262
R+ +A++ G L T D D+ ++++ GH R V G+ + DG A
Sbjct: 339 LDRLARVAFHPSGKYLGTTSYDKTWRLWDINTGAELL-LQEGHSRSVYGIAFQQDGALAA 397
Query: 263 SGGNDNVVHIWDMSAVSSNSPTRWLYRFDEHKAAVKALAWCPFQGNLLASGGGGGDCCVK 322
S G D++ +WD+ S + F H V ++ + P G LASGG C +
Sbjct: 398 SCGLDSLARVWDLRTGRS------ILVFQGHIKPVFSVNFSP-NGYHLASGGEDNQC--R 448
Query: 323 LWNTGMGERMNSVDTGSQVCALLWSKNERELLSSHGLTQNQLTLWKYPSMLKIAELHGHT 382
+W+ M + + + + + + + + + + ++ +W + L GH
Sbjct: 449 IWDLRMRKSLYIIPAHANLVSQVKYEPQEGYFLATASYDMKVNIWSGRDFSLVKSLAGHE 508
Query: 383 SRVLHMTQSPDGSTVASAAAAADQTLRFWEVFG 415
S+V + + D S +A+ + D+T++ W G
Sbjct: 509 SKVASLDITADSSCIAT--VSHDRTIKLWTSSG 539
Score = 60.1 bits (144), Expect = 2e-09
Identities = 51/189 (26%), Positives = 83/189 (42%), Gaps = 15/189 (7%)
Query: 139 DHTIYFWNASDSSGSEFVTVDEEEGPVTSVCWAPDGRHLAVGLTNSHVQLWDTAANKQLR 198
D T W + G+ T + + V + P G++L + +LWD +L
Sbjct: 319 DRTAKLWK---TDGTLLQTFEGHLDRLARVAFHPSGKYLGTTSYDKTWRLWDINTGAEL- 374
Query: 199 TLKGGHRARVGSLAW--NGHVLTTGGMDGKIVNNDVRLRSQIINTYRGHRREVCGLKWSL 256
L+ GH V +A+ +G + + G+D D+R I+ ++GH + V + +S
Sbjct: 375 LLQEGHSRSVYGIAFQQDGALAASCGLDSLARVWDLRTGRSIL-VFQGHIKPVFSVNFSP 433
Query: 257 DGKQLASGGNDNVVHIWDMSAVSSNSPTRWLYRFDEHKAAVKALAWCPFQGNLLASGGGG 316
+G LASGG DN IWD+ S LY H V + + P +G LA+
Sbjct: 434 NGYHLASGGEDNQCRIWDLRMRKS------LYIIPAHANLVSQVKYEPQEGYFLAT--AS 485
Query: 317 GDCCVKLWN 325
D V +W+
Sbjct: 486 YDMKVNIWS 494
Score = 49.3 bits (116), Expect = 4e-06
Identities = 46/181 (25%), Positives = 75/181 (41%), Gaps = 19/181 (10%)
Query: 111 TFDLPDLSDDFSLNLLDWGSRNVLSIA------------LDHTIYFWNASDSSGSEFVTV 158
T+ L D++ L L + SR+V IA LD W+ +G +
Sbjct: 362 TWRLWDINTGAELLLQEGHSRSVYGIAFQQDGALAASCGLDSLARVWDLR--TGRSILVF 419
Query: 159 DEEEGPVTSVCWAPDGRHLAVGLTNSHVQLWDTAANKQLRTLKGGHRARVGSLAWN---G 215
PV SV ++P+G HLA G ++ ++WD K L + H V + + G
Sbjct: 420 QGHIKPVFSVNFSPNGYHLASGGEDNQCRIWDLRMRKSLYIIPA-HANLVSQVKYEPQEG 478
Query: 216 HVLTTGGMDGKIVNNDVRLRSQIINTYRGHRREVCGLKWSLDGKQLASGGNDNVVHIWDM 275
+ L T D K VN ++ + GH +V L + D +A+ +D + +W
Sbjct: 479 YFLATASYDMK-VNIWSGRDFSLVKSLAGHESKVASLDITADSSCIATVSHDRTIKLWTS 537
Query: 276 S 276
S
Sbjct: 538 S 538
Score = 35.0 bits (79), Expect = 0.081
Identities = 41/172 (23%), Positives = 64/172 (36%), Gaps = 16/172 (9%)
Query: 244 GHRREVCGLKWSLDGKQLASGGNDNVVHIWDMSAVSSNSPTRWLYRFDEHKAAVKALAWC 303
G R + G +S DGK LA+ V +W+M V T + +HK + +
Sbjct: 253 GDDRPLTGCSFSRDGKILATCSLSGVTKLWEMPQV-----TNTIAVLKDHKERATDVVFS 307
Query: 304 PFQGNLLASGGGGGDCCVKLWNTGMGERMNSVDTGSQVCALLWSKNERELLSSHGLTQNQ 363
P L + D KLW T G + + + A + + L G T
Sbjct: 308 PVDDCLATA---SADRTAKLWKTD-GTLLQTFEGHLDRLARVAFHPSGKYL---GTTSYD 360
Query: 364 LT--LWKYPSMLKIAELHGHTSRVLHMTQSPDGSTVASAAAAADQTLRFWEV 413
T LW + ++ GH+ V + DG+ AS D R W++
Sbjct: 361 KTWRLWDINTGAELLLQEGHSRSVYGIAFQQDGALAAS--CGLDSLARVWDL 410
Score = 30.4 bits (67), Expect = 2.0
Identities = 18/52 (34%), Positives = 27/52 (51%), Gaps = 3/52 (5%)
Query: 366 LWKYPSMLK-IAELHGHTSRVLHMTQSPDGSTVASAAAAADQTLRFWEVFGT 416
LW+ P + IA L H R + SP +A+A+A D+T + W+ GT
Sbjct: 281 LWEMPQVTNTIAVLKDHKERATDVVFSPVDDCLATASA--DRTAKLWKTDGT 330
>At2g43770 putative splicing factor
Length = 343
Score = 62.8 bits (151), Expect = 4e-10
Identities = 54/227 (23%), Positives = 92/227 (39%), Gaps = 17/227 (7%)
Query: 200 LKGGHRARVGSLAWN--GHVLTTGGMDGKIVNNDVRLRSQIINTYRGHRREVCGLKWSLD 257
L GH + V ++ +N G ++ +G D +I V + +GH+ + L W+ D
Sbjct: 48 LLSGHPSAVYTMKFNPAGTLIASGSHDREIFLWRVHGDCKNFMVLKGHKNAILDLHWTSD 107
Query: 258 GKQLASGGNDNVVHIWDMSAVSSNSPTRWLYRFDEHKAAVKALAWCPFQGNLLASGGGGG 317
G Q+ S D V WD+ + + + EH + V + CP + G
Sbjct: 108 GSQIVSASPDKTVRAWDVET------GKQIKKMAEHSSFVNSC--CPTRRGPPLIISGSD 159
Query: 318 DCCVKLWNTGMGERMNSVDTGSQVCALLWSKNERELLSSHGLTQNQLTLWKYPSMLKIAE 377
D KLW+ + + Q+ A+ +S ++ + G N + +W
Sbjct: 160 DGTAKLWDMRQRGAIQTFPDKYQITAVSFSDAADKIFT--GGVDNDVKVWDLRKGEATMT 217
Query: 378 LHGHTSRVLHMTQSPDGSTVASAAAAADQTLRFWEVFGTPPAAPPKR 424
L GH + M+ SPDGS + + D L W++ P AP R
Sbjct: 218 LEGHQDTITGMSLSPDGSYLLT--NGMDNKLCVWDM---RPYAPQNR 259
Score = 62.0 bits (149), Expect = 6e-10
Identities = 39/148 (26%), Positives = 73/148 (48%), Gaps = 14/148 (9%)
Query: 165 VTSVCWAPDGRHLAVGLTNSHVQLWDTAANKQLRTLKGGHRARVG-SLAWNGHVLTTGGM 223
+T+V ++ + G ++ V++WD + TL+G G SL+ +G L T GM
Sbjct: 183 ITAVSFSDAADKIFTGGVDNDVKVWDLRKGEATMTLEGHQDTITGMSLSPDGSYLLTNGM 242
Query: 224 DGKIVNNDVRL---RSQIINTYRGHR----REVCGLKWSLDGKQLASGGNDNVVHIWDMS 276
D K+ D+R +++ + + GH+ + + WS DG ++ +G +D +VHIWD +
Sbjct: 243 DNKLCVWDMRPYAPQNRCVKIFEGHQHNFEKNLLKCSWSPDGTKVTAGSSDRMVHIWDTT 302
Query: 277 AVSSNSPTRWLYRFDEHKAAVKALAWCP 304
+ R +Y+ H +V + P
Sbjct: 303 S------RRTIYKLPGHTGSVNECVFHP 324
Score = 50.4 bits (119), Expect = 2e-06
Identities = 44/231 (19%), Positives = 80/231 (34%), Gaps = 46/231 (19%)
Query: 139 DHTIYFWNASDSSGSEFVTVDEEEGPVTSVCWAPDGRHLAVGLTNSHVQLWDTAANKQLR 198
D I+ W F+ + + + + W DG + + V+ WD KQ++
Sbjct: 74 DREIFLWRVHGDC-KNFMVLKGHKNAILDLHWTSDGSQIVSASPDKTVRAWDVETGKQIK 132
Query: 199 TL-----------------------KGGHRARVGSLAWNGHVLT---------------- 219
+ A++ + G + T
Sbjct: 133 KMAEHSSFVNSCCPTRRGPPLIISGSDDGTAKLWDMRQRGAIQTFPDKYQITAVSFSDAA 192
Query: 220 ----TGGMDGKIVNNDVRLRSQIINTYRGHRREVCGLKWSLDGKQLASGGNDNVVHIWDM 275
TGG+D + D+R + + T GH+ + G+ S DG L + G DN + +WDM
Sbjct: 193 DKIFTGGVDNDVKVWDLR-KGEATMTLEGHQDTITGMSLSPDGSYLLTNGMDNKLCVWDM 251
Query: 276 SAVSSNSPTRWLYRFDEHKAAVKALAWCPFQGNLLASGGGGGDCCVKLWNT 326
+ + ++ +H K L C + + G D V +W+T
Sbjct: 252 RPYAPQNRCVKIFEGHQHNFE-KNLLKCSWSPDGTKVTAGSSDRMVHIWDT 301
Score = 43.1 bits (100), Expect = 3e-04
Identities = 62/279 (22%), Positives = 109/279 (38%), Gaps = 27/279 (9%)
Query: 146 NASDSSGSEFVTVDEEEGPVTSVCWAPDGRHLAVGLTNSHVQLWDTAAN-KQLRTLKGGH 204
N + S + + + V ++ + P G +A G + + LW + K LKG H
Sbjct: 37 NRTSSLEAPIMLLSGHPSAVYTMKFNPAGTLIASGSHDREIFLWRVHGDCKNFMVLKG-H 95
Query: 205 RARVGSLAW--NGHVLTTGGMDGKIVNNDVRLRSQIINTYRGHRREVCGLKWSLDGKQL- 261
+ + L W +G + + D + DV Q I H V + G L
Sbjct: 96 KNAILDLHWTSDGSQIVSASPDKTVRAWDVETGKQ-IKKMAEHSSFVNSCCPTRRGPPLI 154
Query: 262 ASGGNDNVVHIWDMSAVSSNSPTRWLYRFDEHKAAVKALAWCPFQGNLLASGGGGGDCCV 321
SG +D +WDM R + K + A+++ + GG D V
Sbjct: 155 ISGSDDGTAKLWDMR-------QRGAIQTFPDKYQITAVSFSDAADKIFT---GGVDNDV 204
Query: 322 KLWNTGMGERMNSVDTGSQVCALLWSKNERELLSSHGLTQNQLTLW------KYPSMLKI 375
K+W+ GE +++ + + L ++G+ N+L +W +KI
Sbjct: 205 KVWDLRKGEATMTLEGHQDTITGMSLSPDGSYLLTNGM-DNKLCVWDMRPYAPQNRCVKI 263
Query: 376 AELHGHT--SRVLHMTQSPDGSTVASAAAAADQTLRFWE 412
E H H +L + SPDG+ V A ++D+ + W+
Sbjct: 264 FEGHQHNFEKNLLKCSWSPDGTKV--TAGSSDRMVHIWD 300
>At5g16750 WD40-repeat protein
Length = 876
Score = 60.5 bits (145), Expect = 2e-09
Identities = 53/196 (27%), Positives = 95/196 (48%), Gaps = 13/196 (6%)
Query: 134 LSIALDHTIYFWNASDSSGSEFVTVDEEEGPVTSVCWAPDGRHLAVGLTNSHVQLWDTAA 193
++ A I +++DSS T++ E +T++ +PD + L + +++WD
Sbjct: 34 IACACGDVINIVDSTDSSVKS--TIEGESDTLTALALSPDDKLLFSAGHSRQIRVWDLET 91
Query: 194 NKQLRTLKGGHRARVGSLAW--NGHVLTTGGMDGKIVNNDVRLRSQIINTYRGHRREVCG 251
K +R+ K GH V +A +G +L T G D K++ DV + +RGH+ V
Sbjct: 92 LKCIRSWK-GHEGPVMGMACHASGGLLATAGADRKVLVWDVD-GGFCTHYFRGHKGVVSS 149
Query: 252 LKWSLDGKQ--LASGGNDNVVHIWDMSAVSSNSPTRWLYRFDEHKAAVKALAWCPFQGNL 309
+ + D + L SG +D V +WD++A N+ + L ++H +AV ++A +
Sbjct: 150 ILFHPDSNKNILISGSDDATVRVWDLNA--KNTEKKCLAIMEKHFSAVTSIA---LSEDG 204
Query: 310 LASGGGGGDCCVKLWN 325
L G D V LW+
Sbjct: 205 LTLFSAGRDKVVNLWD 220
Score = 45.8 bits (107), Expect = 5e-05
Identities = 41/164 (25%), Positives = 73/164 (44%), Gaps = 15/164 (9%)
Query: 165 VTSVCWAPDGRHLAVGLTNSHVQLWDTAANKQLRTLKGGHRARVGSLAWNGH---VLTTG 221
+ SV A + + G + +W + TLKG H+ R+ S+ ++ V+T
Sbjct: 502 INSVAVARNDSLVCTGSEDRTASIWRLPDLVHVVTLKG-HKRRIFSVEFSTVDQCVMTAS 560
Query: 222 GMDGKIVNNDVRLRSQIINTYRGHRREVCGLKWSLDGKQLASGGNDNVVHIWDMSAVSSN 281
G K V + T+ GH V + DG Q S G D ++ +W+++
Sbjct: 561 G--DKTVKIWAISDGSCLKTFEGHTSSVLRASFITDGTQFVSCGADGLLKLWNVNT---- 614
Query: 282 SPTRWLYRFDEHKAAVKALAWCPFQGNLLASGGGGGDCCVKLWN 325
+ + +D+H+ V ALA + ++A+GGG D + LW+
Sbjct: 615 --SECIATYDQHEDKVWALA-VGKKTEMIATGGG--DAVINLWH 653
Score = 41.2 bits (95), Expect = 0.001
Identities = 57/252 (22%), Positives = 105/252 (41%), Gaps = 28/252 (11%)
Query: 122 SLNLLDWGSRNVLSI--ALDHTIYFWNASDSSGSEFVTVDEEEGPVTSVCWAPDGRHLAV 179
SL+ S NVL + + D T+ WNA+ S T G + +V +A V
Sbjct: 406 SLDTCVSSSGNVLIVTGSKDKTVRLWNATSKSCIGVGT--GHNGDILAVAFAKKSFSFFV 463
Query: 180 -GLTNSHVQLW---------DTAANKQLRTLKGGHRARVGSLAW--NGHVLTTGGMDGKI 227
G + +++W + N + R++ H + S+A N ++ TG D
Sbjct: 464 SGSGDRTLKVWSLDGISEDSEEPINLKTRSVVAAHDKDINSVAVARNDSLVCTGSEDRTA 523
Query: 228 VNNDVRLRSQI-INTYRGHRREVCGLKWSLDGKQLASGGNDNVVHIWDMSAVSSNSPTRW 286
+ RL + + T +GH+R + +++S + + + D V IW A+S S
Sbjct: 524 --SIWRLPDLVHVVTLKGHKRRIFSVEFSTVDQCVMTASGDKTVKIW---AISDGS---C 575
Query: 287 LYRFDEHKAAVKALAWCPFQGNLLASGGGGGDCCVKLWNTGMGERMNSVDTGSQVCALLW 346
L F+ H ++V ++ ++ G G +KLWN E + + D L
Sbjct: 576 LKTFEGHTSSVLRASFITDGTQFVSCGADG---LLKLWNVNTSECIATYDQHEDKVWALA 632
Query: 347 SKNERELLSSHG 358
+ E++++ G
Sbjct: 633 VGKKTEMIATGG 644
Score = 38.9 bits (89), Expect = 0.006
Identities = 36/143 (25%), Positives = 62/143 (43%), Gaps = 6/143 (4%)
Query: 139 DHTIYFWNASDSSGSEFVTVDEEEGPVTSVCWAPDGRHLAVGLTNSHVQLWDTAANKQLR 198
D T W D VT+ + + SV ++ + + + V++W + L+
Sbjct: 520 DRTASIWRLPDLV--HVVTLKGHKRRIFSVEFSTVDQCVMTASGDKTVKIWAISDGSCLK 577
Query: 199 TLKGGHRARV--GSLAWNGHVLTTGGMDGKIVNNDVRLRSQIINTYRGHRREVCGLKWSL 256
T +G H + V S +G + G DG + +V S+ I TY H +V L
Sbjct: 578 TFEG-HTSSVLRASFITDGTQFVSCGADGLLKLWNVNT-SECIATYDQHEDKVWALAVGK 635
Query: 257 DGKQLASGGNDNVVHIWDMSAVS 279
+ +A+GG D V+++W S S
Sbjct: 636 KTEMIATGGGDAVINLWHDSTAS 658
Score = 37.4 bits (85), Expect = 0.016
Identities = 39/182 (21%), Positives = 72/182 (39%), Gaps = 22/182 (12%)
Query: 244 GHRREVCGLKWSLDGKQ---LASGGNDNVVHIWDMSAVSSNSPTRWLYRFDEHKAAVKAL 300
GH+ V L + + +G D V +W+ ++ S H + A+
Sbjct: 399 GHKEVVLSLDTCVSSSGNVLIVTGSKDKTVRLWNATSKSCIGVGTG------HNGDILAV 452
Query: 301 AWCPFQGNLLASGGGGGDCCVKLWNT-GMGERMN---SVDTGSQVCALLWSKNE-----R 351
A+ + SG G D +K+W+ G+ E ++ T S V A N
Sbjct: 453 AFAKKSFSFFVSGSG--DRTLKVWSLDGISEDSEEPINLKTRSVVAAHDKDINSVAVARN 510
Query: 352 ELLSSHGLTQNQLTLWKYPSMLKIAELHGHTSRVLHMTQSPDGSTVASAAAAADQTLRFW 411
+ L G ++W+ P ++ + L GH R+ + S V + A+ D+T++ W
Sbjct: 511 DSLVCTGSEDRTASIWRLPDLVHVVTLKGHKRRIFSVEFSTVDQCVMT--ASGDKTVKIW 568
Query: 412 EV 413
+
Sbjct: 569 AI 570
Score = 28.5 bits (62), Expect = 7.6
Identities = 21/83 (25%), Positives = 39/83 (46%), Gaps = 3/83 (3%)
Query: 333 NSVDTGSQVCALLWSKNERELLSSHGLTQNQLTLWKYPSMLKIAELHGHTSRVLHMTQSP 392
++++ S L + +LL S G ++ Q+ +W ++ I GH V+ M
Sbjct: 54 STIEGESDTLTALALSPDDKLLFSAGHSR-QIRVWDLETLKCIRSWKGHEGPVMGMACHA 112
Query: 393 DGSTVASAAAAADQTLRFWEVFG 415
G +A+ A AD+ + W+V G
Sbjct: 113 SGGLLAT--AGADRKVLVWDVDG 133
>At2g32700 unknown protein
Length = 787
Score = 59.3 bits (142), Expect = 4e-09
Identities = 61/275 (22%), Positives = 119/275 (43%), Gaps = 19/275 (6%)
Query: 139 DHTIYFWNASDSSGSEFVTVDEEEGPVTSVCWAPDGRHLAVGLTNSHVQLWDTAANKQLR 198
D ++ WN T +E +T V + P+ LA + +++WD +
Sbjct: 531 DKKVFIWNMETLQVES--TPEEHAHIITDVRFRPNSTQLATSSFDKTIKIWDASDPGYFL 588
Query: 199 TLKGGHRARVGSLAWNGHVLTTGGMDGKIVNNDVRLRSQIINTYRGHRREVCGLKWSLDG 258
GH A V S+ + H T + NND+R + R + +++
Sbjct: 589 RTISGHAAPVMSIDF--HPKKTELLCSCDSNNDIRFWDINASCVRAVKGASTQVRFQPRT 646
Query: 259 KQLASGGNDNVVHIWDMSAVSSNSPTRWLYRFDEHKAAVKALAWCPFQGNLLASGGGGGD 318
Q + ++N V I+D+ + + + F H + V ++ W P G L+AS +
Sbjct: 647 GQFLAAASENTVSIFDI-----ENNNKRVNIFKGHSSNVHSVCWSP-NGELVAS---VSE 697
Query: 319 CCVKLWNTGMGERMNSV-DTGSQVCALLWSKNERELLSSHGLTQNQLTLWKYPSMLKIAE 377
VKLW+ G+ ++ + ++G++ ++++ + +LL G + LW K
Sbjct: 698 DAVKLWSLSSGDCIHELSNSGNKFHSVVFHPSYPDLLVIGG--YQAIELWNTMEN-KCMT 754
Query: 378 LHGHTSRVLHMTQSPDGSTVASAAAAADQTLRFWE 412
+ GH + + QSP VAS A+ D++++ W+
Sbjct: 755 VAGHECVISALAQSPSTGVVAS--ASHDKSVKIWK 787
Score = 52.0 bits (123), Expect = 6e-07
Identities = 57/241 (23%), Positives = 101/241 (41%), Gaps = 19/241 (7%)
Query: 130 SRNVLSIALDHTIYFWNASDSSGSEFVTVDEEEGPVTSVCWAPDGRHLAVGL-TNSHVQL 188
S + + + D TI W+ASD G T+ PV S+ + P L +N+ ++
Sbjct: 564 STQLATSSFDKTIKIWDASDP-GYFLRTISGHAAPVMSIDFHPKKTELLCSCDSNNDIRF 622
Query: 189 WDTAANKQLRTLKGGHRARVGSLAWNGHVLTTGGMDGKIVNNDVRLRSQIINTYRGHRRE 248
WD A+ +R +KG +V G L + + D+ ++ +N ++GH
Sbjct: 623 WDINASC-VRAVKGAS-TQVRFQPRTGQFLAAAS-ENTVSIFDIENNNKRVNIFKGHSSN 679
Query: 249 VCGLKWSLDGKQLASGGNDNVVHIWDMSAVSSNSPTRWLYRFDEHKAAVKALAWCPFQGN 308
V + WS +G+ +AS D V +W +S+ ++ ++ + P +
Sbjct: 680 VHSVCWSPNGELVASVSED-AVKLWSLSSGDC------IHELSNSGNKFHSVVFHPSYPD 732
Query: 309 LLASGGGGGDCCVKLWNTGMGERMNSVDTGSQVCALLWSKNERELLS-SHGLTQNQLTLW 367
LL GG ++LWNT + M + AL S + + S SH + +W
Sbjct: 733 LLVIGGYQ---AIELWNTMENKCMTVAGHECVISALAQSPSTGVVASASH---DKSVKIW 786
Query: 368 K 368
K
Sbjct: 787 K 787
Score = 34.3 bits (77), Expect = 0.14
Identities = 24/87 (27%), Positives = 38/87 (43%), Gaps = 9/87 (10%)
Query: 239 INTYRGHRREVCGLKWSLDGKQLASGGNDNVVHIWDMSAVSSNSPTRWLYRFDEHKAAVK 298
++ R +V +S DGK LAS G+D V IW+M + S +EH +
Sbjct: 503 VSCIRKSASKVICCSFSYDGKLLASAGHDKKVFIWNMETLQVESTP------EEHAHIIT 556
Query: 299 ALAWCPFQGNLLASGGGGGDCCVKLWN 325
+ + P L S D +K+W+
Sbjct: 557 DVRFRPNSTQLATS---SFDKTIKIWD 580
>At4g29830 unknown protein
Length = 321
Score = 58.9 bits (141), Expect = 5e-09
Identities = 42/152 (27%), Positives = 75/152 (48%), Gaps = 11/152 (7%)
Query: 165 VTSVCWAPDGRHLAVGLTNSHVQLWDTAANKQLRTLKGGHRARVGSLAW---NGHVLTTG 221
V SV W+P+G+ LA G + + ++D +K L L+ GH V SL + + VL +G
Sbjct: 161 VLSVAWSPNGKRLACGSMDGTICVFDVDRSKLLHQLE-GHNMPVRSLVFSPVDPRVLFSG 219
Query: 222 GMDGKIVNNDVRLRSQIINTYRGHRREVCGLKWSLDGKQLASGGNDNVVHIWDMSAVSSN 281
DG + +D ++ ++ + GH V + S DG +A+G +D V +WD+ ++
Sbjct: 220 SDDGHVNMHDAEGKT-LLGSMSGHTSWVLSVDASPDGGAIATGSSDRTVRLWDLKMRAA- 277
Query: 282 SPTRWLYRFDEHKAAVKALAWCPFQGNLLASG 313
+ H V ++A+ P G + +G
Sbjct: 278 -----IQTMSNHNDQVWSVAFRPPGGTGVRAG 304
Score = 55.8 bits (133), Expect = 4e-08
Identities = 69/268 (25%), Positives = 118/268 (43%), Gaps = 25/268 (9%)
Query: 162 EGPVTSVCWAPDGRH----LAVGLTNSHVQLWDTAANKQLRTLKGGHRARVGSLAWN--G 215
E V + W P L G + V+LW +RT GH V +LA + G
Sbjct: 14 EDSVWAATWVPATEDRPALLLTGSLDETVKLWRPDELDLVRT-NTGHSLGVAALAAHPSG 72
Query: 216 HVLTTGGMDGKIVNNDVRLRSQIINTYRGHRREVCGLKWSLDGKQLA-SGGNDNVVHIWD 274
+ + +D + DV + I EV G+++ G LA +GG+ V +WD
Sbjct: 73 IIAASSSIDSFVRVFDVDTNATIA-VLEAPPSEVWGMQFEPKGTILAVAGGSSASVKLWD 131
Query: 275 MSAVSSNSPTRWLYRFDEHKAAVK--------ALAWCPFQGNLLASGGGGGDCCVKLWNT 326
++ S T + R D K + K ++AW P G LA G G CV ++
Sbjct: 132 TASWRLIS-TLSIPRPDAPKPSDKTSSKKFVLSVAWSP-NGKRLACGSMDGTICV--FDV 187
Query: 327 GMGERMNSVDTGSQ-VCALLWSKNERELLSSHGLTQNQLTLWKYPSMLKIAELHGHTSRV 385
+ ++ ++ + V +L++S + +L S G + + + + GHTS V
Sbjct: 188 DRSKLLHQLEGHNMPVRSLVFSPVDPRVLFS-GSDDGHVNMHDAEGKTLLGSMSGHTSWV 246
Query: 386 LHMTQSPDGSTVASAAAAADQTLRFWEV 413
L + SPDG +A+ ++D+T+R W++
Sbjct: 247 LSVDASPDGGAIAT--GSSDRTVRLWDL 272
Score = 42.4 bits (98), Expect = 5e-04
Identities = 50/234 (21%), Positives = 89/234 (37%), Gaps = 39/234 (16%)
Query: 172 PDGRHLAV-GLTNSHVQLWDTAANKQLRTLKGGHRARVGSLAWNGHVLTTGGMDGKIVNN 230
P G LAV G +++ V+LWDTA+ + + TL I
Sbjct: 112 PKGTILAVAGGSSASVKLWDTASWRLISTLS-------------------------IPRP 146
Query: 231 DVRLRSQIINTYRGHRREVCGLKWSLDGKQLASGGNDNVVHIWDMSAVSSNSPTRWLYRF 290
D S ++ ++ V + WS +GK+LA G D + ++D+ ++ L++
Sbjct: 147 DAPKPSDKTSS----KKFVLSVAWSPNGKRLACGSMDGTICVFDVDR------SKLLHQL 196
Query: 291 DEHKAAVKALAWCPFQGNLLASGGGGGDCCVKLWNTGMGERMNSVDTGSQVCALLWSKNE 350
+ H V++L + P +L SG G + + G+ + +G L +
Sbjct: 197 EGHNMPVRSLVFSPVDPRVLFSGSDDGHVNM---HDAEGKTLLGSMSGHTSWVLSVDASP 253
Query: 351 RELLSSHGLTQNQLTLWKYPSMLKIAELHGHTSRVLHMTQSPDGSTVASAAAAA 404
+ G + + LW I + H +V + P G T A A
Sbjct: 254 DGGAIATGSSDRTVRLWDLKMRAAIQTMSNHNDQVWSVAFRPPGGTGVRAGRLA 307
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.133 0.424
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,117,783
Number of Sequences: 26719
Number of extensions: 502657
Number of successful extensions: 3103
Number of sequences better than 10.0: 195
Number of HSP's better than 10.0 without gapping: 113
Number of HSP's successfully gapped in prelim test: 84
Number of HSP's that attempted gapping in prelim test: 2209
Number of HSP's gapped (non-prelim): 602
length of query: 436
length of database: 11,318,596
effective HSP length: 102
effective length of query: 334
effective length of database: 8,593,258
effective search space: 2870148172
effective search space used: 2870148172
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC146343.9


