

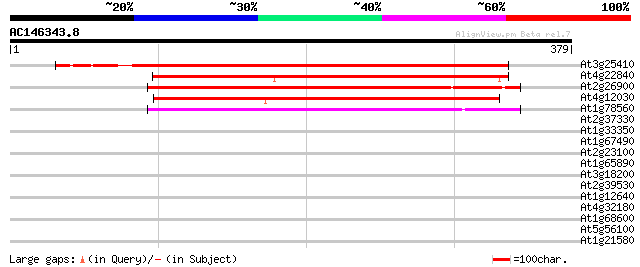
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146343.8 + phase: 0 /pseudo
(379 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g25410 unknown protein 409 e-114
At4g22840 unknown protein 201 5e-52
At2g26900 Na+ dependent ileal bile acid transporter like protein 194 9e-50
At4g12030 Sodium Bile acid symporter (SBF) like protein 187 1e-47
At1g78560 Unknown protein (T30F21.11) 154 8e-38
At2g37330 unknown protein 35 0.052
At1g33350 unknown protein 33 0.26
At1g67490 alpha-glucosidase I 31 1.3
At2g23100 hypothetical protein 30 2.9
At1g65890 unknown protein 29 3.7
At3g18200 integral membrane protein, putative 29 4.9
At2g39530 unknown protein 29 4.9
At1g12640 unknown protein 29 4.9
At4g32180 unknown protein 28 6.4
At1g68600 unknown protein 28 6.4
At5g56100 unknown protein (At5g56100) 28 8.3
At1g21580 hypothetical protein 28 8.3
>At3g25410 unknown protein
Length = 431
Score = 409 bits (1052), Expect = e-114
Identities = 217/307 (70%), Positives = 252/307 (81%), Gaps = 12/307 (3%)
Query: 32 FSLQRVSKGCSISSFACSTSKLHGGRVGFNHREGKTSFLKFGVDESVVGGGGNGDVGER- 90
F L+R + G + ACST+ G RVG + R+G S L F G DV E+
Sbjct: 57 FRLRRRNSGL-VPVVACSTTPFMG-RVGLHWRDGNMSLLSFC---------GGTDVTEKA 105
Query: 91 DWSQILSALLPFVVAVTAVAALSQPSTFTWVSKELYAPALGGIMLSIGIGLSMEDFALAF 150
D SQ SALLPFVVA+TAVAALS P +FTWVSK+LYAPALGGIMLSIGI LS++DFALAF
Sbjct: 106 DSSQFWSALLPFVVALTAVAALSYPPSFTWVSKDLYAPALGGIMLSIGIQLSVDDFALAF 165
Query: 151 KRPLPLSIGFIAQYVLKPVLGVLIAKAFGLPRMFYAGFVLTACVSGAQLSSYASFISKGD 210
KRP+PLS+GF+AQYVLKP+LGVL+A AFG+PR FYAGF+LT CV+GAQLSSYAS +SK D
Sbjct: 166 KRPVPLSVGFVAQYVLKPLLGVLVANAFGMPRTFYAGFILTCCVAGAQLSSYASSLSKAD 225
Query: 211 VALCIVLTSYTTIASVIVTPLLTGLLIGSVVPVDAVAMSKSILQVVLAPVTLGLLLNTYA 270
VA+ I+LTS TTIASVI TPLL+GLLIGSVVPVDAVAMSKSILQVVL P+TLGL+LNTYA
Sbjct: 226 VAMSILLTSSTTIASVIFTPLLSGLLIGSVVPVDAVAMSKSILQVVLVPITLGLVLNTYA 285
Query: 271 KPVVSILRPVMPFVAMICTSLCIGSPLAINRSQILSGEGLRLVAPVLIFHAAAFTLGYWF 330
KPVV++L+PVMPFVAM+CTSLCIGSPL+INRSQILS EGL L+ P++ FHA AF LGYWF
Sbjct: 286 KPVVTLLQPVMPFVAMVCTSLCIGSPLSINRSQILSAEGLGLIVPIVTFHAVAFALGYWF 345
Query: 331 SNLPSLR 337
S +P LR
Sbjct: 346 SKIPGLR 352
>At4g22840 unknown protein
Length = 409
Score = 201 bits (511), Expect = 5e-52
Identities = 106/251 (42%), Positives = 162/251 (64%), Gaps = 10/251 (3%)
Query: 97 SALLPFVVAVTAVAALSQPSTFTWVSKELYAPALGGIMLSIGIGLSMEDFALAFKRPLPL 156
+++LP VV + + AL P +FTW + + PALG +M ++GI + +DF AFKRP +
Sbjct: 98 NSILPHVVLASTILALIYPPSFTWFTSRYFVPALGFLMFAVGINSNEKDFLEAFKRPKAI 157
Query: 157 SIGFIAQYVLKPVLGVLIAKA----FGLPRMFYAGFVLTACVSGAQLSSYASFISKGDVA 212
+G++ QY++KPVLG + A F LP AG +L +CVSGAQLS+YA+F++ +A
Sbjct: 158 LLGYVGQYLVKPVLGFIFGLAAVSLFQLPTPIGAGIMLVSCVSGAQLSNYATFLTDPALA 217
Query: 213 -LCIVLTSYTTIASVIVTPLLTGLLIGSVVPVDAVAMSKSILQVVLAPVTLGLLLNTYAK 271
L IV+TS +T +V+VTP+L+ LLIG +PVD M SILQVV+AP+ GLLLN
Sbjct: 218 PLSIVMTSLSTATAVLVTPMLSLLLIGKKLPVDVKGMISSILQVVIAPIAAGLLLNKLFP 277
Query: 272 PVVSILRPVMPFVAMICTSLCIGSPLAINRSQILSGEGLRLVAPVLIFHAAAFTLGYW-- 329
V + +RP +P ++++ T+ C+G+PLA+N + ++S G ++ V +FH +AF GY+
Sbjct: 278 KVSNAIRPFLPILSVLDTACCVGAPLALNINSVMSPFGATILLLVTMFHLSAFLAGYFLT 337
Query: 330 ---FSNLPSLR 337
F N P +
Sbjct: 338 GSVFRNAPDAK 348
>At2g26900 Na+ dependent ileal bile acid transporter like protein
Length = 409
Score = 194 bits (492), Expect = 9e-50
Identities = 100/252 (39%), Positives = 154/252 (60%), Gaps = 2/252 (0%)
Query: 94 QILSALLPFVVAVTAVAALSQPSTFTWVSKELYAPALGGIMLSIGIGLSMEDFALAFKRP 153
++L+ L P V + + + +PS TW+ +L+ LG +MLS+G+ L+ EDF + P
Sbjct: 99 ELLTTLFPLWVILGTLVGIFKPSLVTWLETDLFTLGLGFLMLSMGLTLTFEDFRRCLRNP 158
Query: 154 LPLSIGFIAQYVLKPVLGVLIAKAFGLPRMFYAGFVLTACVSGAQLSSYASFISKGDVAL 213
+ +GF+AQY++KP+LG LIA L G +L +C G Q S+ A++ISKG+VAL
Sbjct: 159 WTVGVGFLAQYMIKPILGFLIAMTLKLSAPLATGLILVSCCPGGQASNVATYISKGNVAL 218
Query: 214 CIVLTSYTTIASVIVTPLLTGLLIGSVVPVDAVAMSKSILQVVLAPVTLGLLLNTYAKPV 273
+++T+ +TI ++I+TPLLT LL G +VPVDA ++ S QVVL P +G+L N +
Sbjct: 219 SVLMTTCSTIGAIIMTPLLTKLLAGQLVPVDAAGLALSTFQVVLVPTIIGVLANEFFPKF 278
Query: 274 VSILRPVMPFVAMICTSLCIGSPLAINRSQILSGEGLRLVAPVLIFHAAAFTLGYWFSNL 333
S + V P + +I T+L SP+ + +L +G +L+ PV + HAAAF +GYW S
Sbjct: 279 TSKIITVTPLIGVILTTLLCASPIG-QVADVLKTQGAQLILPVALLHAAAFAIGYWISKF 337
Query: 334 PSLRFLTSITIN 345
S TS TI+
Sbjct: 338 -SFGESTSRTIS 348
>At4g12030 Sodium Bile acid symporter (SBF) like protein
Length = 407
Score = 187 bits (474), Expect = 1e-47
Identities = 99/239 (41%), Positives = 147/239 (61%), Gaps = 5/239 (2%)
Query: 98 ALLPFVVAVTAVAALSQPSTFTWVSKELYAPALGGIMLSIGIGLSMEDFALAFKRPLPLS 157
+ +P + ++ + AL P +FTW + P LG +M ++GI + DF A KRP +
Sbjct: 100 SFIPHGILLSTILALVYPPSFTWFKPRYFVPGLGFMMFAVGINSNERDFLEALKRPDAIF 159
Query: 158 IGFIAQYVLKPVLG----VLIAKAFGLPRMFYAGFVLTACVSGAQLSSYASFISKGDVA- 212
G+I QY++KP+LG V+ F LP AG +L +CVSGAQLS+Y +F++ +A
Sbjct: 160 AGYIGQYLIKPLLGYIFGVIAVSLFNLPTSIGAGIMLVSCVSGAQLSNYTTFLTDPSLAA 219
Query: 213 LCIVLTSYTTIASVIVTPLLTGLLIGSVVPVDAVAMSKSILQVVLAPVTLGLLLNTYAKP 272
L IV+TS +T +V+VTP+L+ LLIG +PVD M SILQVV+ P+ GLLLN
Sbjct: 220 LSIVMTSISTATAVLVTPMLSLLLIGKKLPVDVFGMISSILQVVITPIAAGLLLNRLFPR 279
Query: 273 VVSILRPVMPFVAMICTSLCIGSPLAINRSQILSGEGLRLVAPVLIFHAAAFTLGYWFS 331
+ + ++P +P + +I S CIG+PLA+N ILS G ++ V+ FH AF GY+F+
Sbjct: 280 LSNAIKPFLPALTVIDMSCCIGAPLALNIDSILSPFGATILFLVITFHLLAFVAGYFFT 338
>At1g78560 Unknown protein (T30F21.11)
Length = 401
Score = 154 bits (389), Expect = 8e-38
Identities = 86/252 (34%), Positives = 138/252 (54%), Gaps = 1/252 (0%)
Query: 94 QILSALLPFVVAVTAVAALSQPSTFTWVSKELYAPALGGIMLSIGIGLSMEDFALAFKRP 153
+ +S P V++ + L +PSTF WV+ L ML +G+ L+++D A P
Sbjct: 95 EAVSTAFPIWVSLGCLLGLMRPSTFNWVTPNWTIVGLTITMLGMGMTLTLDDLRGALSMP 154
Query: 154 LPLSIGFIAQYVLKPVLGVLIAKAFGLPRMFYAGFVLTACVSGAQLSSYASFISKGDVAL 213
L GF+ QY + P+ ++K LP + AG +L C G S+ ++I++G+VAL
Sbjct: 155 KELFAGFLLQYSVMPLSAFFVSKLLNLPPHYAAGLILVGCCPGGTASNIVTYIARGNVAL 214
Query: 214 CIVLTSYTTIASVIVTPLLTGLLIGSVVPVDAVAMSKSILQVVLAPVTLGLLLNTYAKPV 273
+++T+ +T+++VI+TPLLT L + VDA+ + S LQVVL PV G LN Y K +
Sbjct: 215 SVLMTAASTVSAVIMTPLLTAKLAKQYITVDALGLLMSTLQVVLLPVLAGAFLNQYFKKL 274
Query: 274 VSILRPVMPFVAMICTSLCIGSPLAINRSQILSGEGLRLVAPVLIFHAAAFTLGYWFSNL 333
V + PVMP +A+ ++ G + N S IL G ++V + H + F GY FS +
Sbjct: 275 VKFVSPVMPPIAVGTVAILCGYAIGQNASAILM-SGKQVVLASCLLHISGFLFGYLFSRI 333
Query: 334 PSLRFLTSITIN 345
+ +S TI+
Sbjct: 334 LGIDVASSRTIS 345
>At2g37330 unknown protein
Length = 273
Score = 35.4 bits (80), Expect = 0.052
Identities = 32/147 (21%), Positives = 65/147 (43%), Gaps = 17/147 (11%)
Query: 135 LSIGIGLSMEDFALAFKRPLPLSIGFIAQYVLKPVLGVLIAKAFGLPRM----------F 184
LSI G S+ F L P F +Y++ P+ G+L+ A + +
Sbjct: 111 LSILAGTSITMFLLVLLNVFP----FTPRYMI-PIAGMLVGNAMTVTGVTMKQLRDDIKM 165
Query: 185 YAGFVLTACVSGA--QLSSYASFISKGDVALCIVLTSYTTIASVIVTPLLTGLLIGSVVP 242
V TA GA + ++ ++L VL S T+ + + +TG+++G P
Sbjct: 166 QLNLVETALALGATPRQATLQQVKRALVISLSPVLDSCKTVGLISLPGAMTGMIMGGASP 225
Query: 243 VDAVAMSKSILQVVLAPVTLGLLLNTY 269
++A+ + ++ +++ T+ + +TY
Sbjct: 226 LEAIQLQIVVMNMMVGAATVSSITSTY 252
>At1g33350 unknown protein
Length = 538
Score = 33.1 bits (74), Expect = 0.26
Identities = 31/96 (32%), Positives = 42/96 (43%), Gaps = 8/96 (8%)
Query: 118 FTWVSKELYAPALGGIMLSIGIGLS--MEDFALAFKRPLPLSIGFIAQYVLKPVLGVLIA 175
F++ + LYA L S+ + S F L R +P FI VLK ++
Sbjct: 83 FSFPNTHLYAAVLTAYSSSLPLHASSAFSFFRLMVNRSVPRPNHFIYPLVLKST--PYLS 140
Query: 176 KAFGLP----RMFYAGFVLTACVSGAQLSSYASFIS 207
AF P +F +GF L V A L SYAS +S
Sbjct: 141 SAFSTPLVHTHLFKSGFHLYVVVQTALLHSYASSVS 176
>At1g67490 alpha-glucosidase I
Length = 852
Score = 30.8 bits (68), Expect = 1.3
Identities = 11/20 (55%), Positives = 16/20 (80%)
Query: 354 GEQDNFIMHWNAELHTCRPS 373
G +DNF+++W AEL+T PS
Sbjct: 415 GSRDNFLLYWPAELYTAVPS 434
>At2g23100 hypothetical protein
Length = 1036
Score = 29.6 bits (65), Expect = 2.9
Identities = 22/61 (36%), Positives = 28/61 (45%), Gaps = 4/61 (6%)
Query: 16 KHKPLSFQPLLRTRSSFSLQRVSKGCSISS-FACSTSKLHGGRVGFNHREGKTSFLKFGV 74
+H PL PL+ +S SLQ G S+ FACS + H GF + F F V
Sbjct: 440 QHHPLHKHPLILNKSDPSLQLYVDGYSVEGMFACSACRRHS--TGFVY-SCNVEFCDFQV 496
Query: 75 D 75
D
Sbjct: 497 D 497
>At1g65890 unknown protein
Length = 578
Score = 29.3 bits (64), Expect = 3.7
Identities = 29/88 (32%), Positives = 38/88 (42%), Gaps = 6/88 (6%)
Query: 239 SVVPVDAVAMSKSILQVVLAPVTLGLLLNTY--AKPVVSILRPVMPFVAMICTSLCIGSP 296
SVV + AM + V +A L + NT A + +ILR P + I S P
Sbjct: 68 SVVAPNTPAMYEMHFAVPMAGAVLNPI-NTRLDATSIAAILRHAKPKILFIYRSF---EP 123
Query: 297 LAINRSQILSGEGLRLVAPVLIFHAAAF 324
LA Q+LS E L PV+ H F
Sbjct: 124 LAREILQLLSSEDSNLNLPVIFIHEIDF 151
>At3g18200 integral membrane protein, putative
Length = 360
Score = 28.9 bits (63), Expect = 4.9
Identities = 29/107 (27%), Positives = 51/107 (47%), Gaps = 11/107 (10%)
Query: 83 GNGDVGERDWSQILSALLPFVVAVTAVAALSQPSTFTWVSKELYAPALGGIMLSIGIGLS 142
G G V E+ +++ AL+ +S+ + VSK +Y P ++ + IG
Sbjct: 2 GKGVVSEK--VKLVVALITLQFCFAGFHIVSRVALNIGVSKVVY-PVYRNLLALLLIG-- 56
Query: 143 MEDFALAFK---RPLPLSIGFIAQYVLKPVLGVLIAKAFGLPRMFYA 186
FA F+ RP PL+I +AQ+ ++G+ + F L ++YA
Sbjct: 57 --PFAYFFEKKERP-PLTISLLAQFFFLALIGITANQGFYLLGLYYA 100
>At2g39530 unknown protein
Length = 178
Score = 28.9 bits (63), Expect = 4.9
Identities = 22/104 (21%), Positives = 43/104 (41%), Gaps = 2/104 (1%)
Query: 125 LYAPALGGIMLSIGIGLSMEDFALAFKRPLPLSIGFIAQYVLKPVLGVLIAKAFGLPRMF 184
L A +G + + + L++ FA PL F V+ +L A FG+ +
Sbjct: 63 LSAAVIGLVYAVVQLFLTISQFATGKTHPLTYQFDFYGDKVISYLLATGSAAGFGVSKDL 122
Query: 185 YAGFVLTACVSGAQLSSYASFISKGDVALCIVLTSYTTIASVIV 228
++ A + F SKG + ++L ++ ++A + V
Sbjct: 123 KDTYI--ALIEFDSTDPVDKFFSKGYASASLLLFAFVSLAVLSV 164
>At1g12640 unknown protein
Length = 462
Score = 28.9 bits (63), Expect = 4.9
Identities = 22/80 (27%), Positives = 37/80 (45%), Gaps = 9/80 (11%)
Query: 95 ILSALLPFVVAV-TAVAALSQPSTFTWVSKELYAPALGGIMLSIGIGLSMEDFALAFKRP 153
+L LL FV + + A PS + K LYA A G + + G S L
Sbjct: 16 VLRFLLCFVATIPVSFACRIVPSR---LGKHLYAAASGAFLSYLSFGFSSNLHFL----- 67
Query: 154 LPLSIGFIAQYVLKPVLGVL 173
+P++IG+ + + +P G++
Sbjct: 68 VPMTIGYASMAIYRPKCGII 87
>At4g32180 unknown protein
Length = 870
Score = 28.5 bits (62), Expect = 6.4
Identities = 22/69 (31%), Positives = 33/69 (46%), Gaps = 10/69 (14%)
Query: 144 EDFALAFKRPLPLSIGFIAQYVLKPVLGVLIAKAFGLPRMFYAGFVLTACVSGAQLSSYA 203
ED +L+ R + +IG I+ L A FGL R+F+ GF + S+A
Sbjct: 314 EDISLSLLRMISYNIGQISY---------LNALRFGLKRIFFGGFFIRGHAYTMDTISFA 364
Query: 204 -SFISKGDV 211
F SKG++
Sbjct: 365 VHFWSKGEM 373
>At1g68600 unknown protein
Length = 537
Score = 28.5 bits (62), Expect = 6.4
Identities = 26/94 (27%), Positives = 38/94 (39%), Gaps = 12/94 (12%)
Query: 135 LSIGIGLSMEDFALAFKRPLPLSIGFIAQYVLKPVL------GVLIAKAF--GLPRMFYA 186
+ +GI L++ F + K PL + F +L VL G + K F L M
Sbjct: 61 IKMGIALALCSFVIFLKEPLQDASKFAVWAILTVVLIFEYYVGATLVKGFNRALGTMLAG 120
Query: 187 GFVLTACVSGAQLSSYASFISKGDVALCIVLTSY 220
G L AQLS A + + +CI L +
Sbjct: 121 GLALGV----AQLSVLAGEFEEVIIVICIFLAGF 150
>At5g56100 unknown protein (At5g56100)
Length = 150
Score = 28.1 bits (61), Expect = 8.3
Identities = 22/76 (28%), Positives = 37/76 (47%), Gaps = 3/76 (3%)
Query: 222 TIASVIVTPLLTGLLIGSVVPVDAVAMSKSILQVVLAP---VTLGLLLNTYAKPVVSILR 278
T+A+V V L GL+ S V + + S L ++ AP VT+ +L++ V+
Sbjct: 29 TVAAVAVVGPLFGLMSFSFVATVTLFLIASPLLLIFAPAFMVTVAVLVSAMVGVGVAAAM 88
Query: 279 PVMPFVAMICTSLCIG 294
+M A++C IG
Sbjct: 89 WMMGIAALVCCGREIG 104
>At1g21580 hypothetical protein
Length = 1696
Score = 28.1 bits (61), Expect = 8.3
Identities = 23/77 (29%), Positives = 38/77 (48%), Gaps = 9/77 (11%)
Query: 12 PMKQKHKPLSFQ------PLLRTRSSFSLQRVSKGCSISSFACSTSKLHGGRVGFNHREG 65
P++Q+H+PLS Q + + S+ QR+S+ + SS C+ S+ VGFN +
Sbjct: 94 PLQQQHQPLSLQQQQQHPQYIPQQVSYEPQRISQPLT-SSIQCTESQ--SDWVGFNEKRL 150
Query: 66 KTSFLKFGVDESVVGGG 82
+ + G S V G
Sbjct: 151 DSWTVDSGPGRSRVDDG 167
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.324 0.138 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,911,778
Number of Sequences: 26719
Number of extensions: 322336
Number of successful extensions: 1112
Number of sequences better than 10.0: 17
Number of HSP's better than 10.0 without gapping: 7
Number of HSP's successfully gapped in prelim test: 11
Number of HSP's that attempted gapping in prelim test: 1096
Number of HSP's gapped (non-prelim): 20
length of query: 379
length of database: 11,318,596
effective HSP length: 101
effective length of query: 278
effective length of database: 8,619,977
effective search space: 2396353606
effective search space used: 2396353606
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 61 (28.1 bits)
Medicago: description of AC146343.8


