

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146343.4 - phase: 0
(141 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
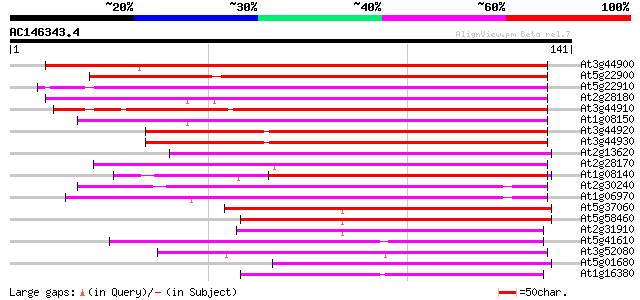
Score E
Sequences producing significant alignments: (bits) Value
At3g44900 putative protein 93 4e-20
At5g22900 Na+/H+ antiporter-like protein 91 2e-19
At5g22910 Na+/H+ antiporter-like protein 88 1e-18
At2g28180 hypothetical protein 83 4e-17
At3g44910 putative protein 82 7e-17
At1g08150 hypothetical protein 80 4e-16
At3g44920 putative protein 79 6e-16
At3g44930 putative protein 76 7e-15
At2g13620 putative Na/H antiporter 75 1e-14
At2g28170 hypothetical protein 73 6e-14
At1g08140 hypothetical protein 70 5e-13
At2g30240 putative Na/H antiporter 65 9e-12
At1g06970 hypothetical protein 61 2e-10
At5g37060 putative transporter protein 59 1e-09
At5g58460 putative protein 57 3e-09
At2g31910 putative Na+/H+ antiporter 56 6e-09
At5g41610 Na+/H+ antiporter-like protein 54 4e-08
At3g52080 putative protein 53 5e-08
At5g01680 putative transporter protein 53 6e-08
At1g16380 putative Na/H antiporter 53 6e-08
>At3g44900 putative protein
Length = 817
Score = 93.2 bits (230), Expect = 4e-20
Identities = 50/127 (39%), Positives = 78/127 (61%), Gaps = 1/127 (0%)
Query: 10 KGQSKKILITWVVYHLVSTIKN-DEFTSWDVMLDDELLKGVKGVYGSVDNVTYEKVEVEN 68
K ++ IT V L+S+ + ++ T WD MLD ELL+ VK + ++ + + V +
Sbjct: 687 KRMARDSRITITVVSLISSEQRANQATDWDRMLDLELLRDVKSNVLAGADIVFSEEVVND 746
Query: 69 TSDTTEFISDIAIQHDFIIVGRRNGIKSPQTQALASWTEYPELGVLGDLLASPDTNTKAS 128
+ T++ + IA ++D IVGR G KS T+ L W+E+ ELG++GDLL S D N +AS
Sbjct: 747 ANQTSQLLKSIANEYDLFIVGREKGRKSVFTEGLEEWSEFEELGIIGDLLTSQDLNCQAS 806
Query: 129 ILVVQQQ 135
+LV+QQQ
Sbjct: 807 VLVIQQQ 813
>At5g22900 Na+/H+ antiporter-like protein
Length = 822
Score = 91.3 bits (225), Expect = 2e-19
Identities = 47/115 (40%), Positives = 71/115 (60%), Gaps = 2/115 (1%)
Query: 21 VVYHLVSTIKNDEFTSWDVMLDDELLKGVKGVYGSVDNVTYEKVEVENTSDTTEFISDIA 80
+V + + K E T WD MLDDELL+ VK ++ ++ Y + +E+ ++T+ + +
Sbjct: 706 IVRLITTDEKARENTVWDKMLDDELLRDVKS--NTLVDIFYSEKAIEDAAETSSLLRSMV 763
Query: 81 IQHDFIIVGRRNGIKSPQTQALASWTEYPELGVLGDLLASPDTNTKASILVVQQQ 135
D IVGR NG S T+ L W+E+ ELG++GDLL S D N +AS+LV+QQQ
Sbjct: 764 SDFDMFIVGRGNGRTSVFTEGLEEWSEFKELGIIGDLLTSQDFNCQASVLVIQQQ 818
>At5g22910 Na+/H+ antiporter-like protein
Length = 800
Score = 88.2 bits (217), Expect = 1e-18
Identities = 49/128 (38%), Positives = 71/128 (55%), Gaps = 3/128 (2%)
Query: 8 WLKGQSKKILITWVVYHLVSTIKNDEFTSWDVMLDDELLKGVKGVYGSVDNVTYEKVEVE 67
W+ GQ+ ++ +T V +S + D+ +WD ++DDE+L +K Y +N Y + V
Sbjct: 672 WM-GQNSRVCLT--VIRFLSGQELDKSKNWDYLVDDEVLNDLKATYSLANNFNYMEKVVN 728
Query: 68 NTSDTTEFISDIAIQHDFIIVGRRNGIKSPQTQALASWTEYPELGVLGDLLASPDTNTKA 127
+ +A HD +IVGR + S LA W E PELGV+GDLLAS D +
Sbjct: 729 GGPAVATTVRLVAEDHDLMIVGRDHEDYSLDLTGLAQWMELPELGVIGDLLASKDLRARV 788
Query: 128 SILVVQQQ 135
S+LVVQQQ
Sbjct: 789 SVLVVQQQ 796
>At2g28180 hypothetical protein
Length = 822
Score = 83.2 bits (204), Expect = 4e-17
Identities = 47/131 (35%), Positives = 75/131 (56%), Gaps = 5/131 (3%)
Query: 10 KGQSKKILITWVVYHLVSTIKNDEFTSWDVMLDD----ELLKGVK-GVYGSVDNVTYEKV 64
K +++ +T V L+++ K+ + T WD MLD EL+K G+ + Y +
Sbjct: 685 KRMARQENVTLTVLRLLASGKSKDATGWDQMLDTVELRELIKSNNAGMVKEETSTIYLEQ 744
Query: 65 EVENTSDTTEFISDIAIQHDFIIVGRRNGIKSPQTQALASWTEYPELGVLGDLLASPDTN 124
E+ + +DT+ + +A +D +VGR G T+ + +W E+ ELGV+GD LASPD
Sbjct: 745 EILDGADTSMLLRSMAFDYDLFVVGRTCGENHEATKGIENWCEFEELGVIGDFLASPDFP 804
Query: 125 TKASILVVQQQ 135
+K S+LVVQQQ
Sbjct: 805 SKTSVLVVQQQ 815
>At3g44910 putative protein
Length = 705
Score = 82.4 bits (202), Expect = 7e-17
Identities = 45/124 (36%), Positives = 75/124 (60%), Gaps = 4/124 (3%)
Query: 12 QSKKILITWVVYHLVSTIKNDEFTSWDVMLDDELLKGVKGVYGSVDNVTYEKVEVENTSD 71
Q+ ++ IT V L+S + E T+WD +LD E+L+ +K + +++ Y + V +
Sbjct: 583 QNPRVKIT--VIRLISD-RETESTNWDYILDHEVLEDLKDTEAT-NSIAYTERIVTGGPE 638
Query: 72 TTEFISDIAIQHDFIIVGRRNGIKSPQTQALASWTEYPELGVLGDLLASPDTNTKASILV 131
+ ++ +D ++VGR +G+ SP L W E PELGV+GDLLAS + +++ S+LV
Sbjct: 639 VATTVRSLSEDYDLMVVGRDHGMASPDFDGLMEWVELPELGVIGDLLASRELDSRVSVLV 698
Query: 132 VQQQ 135
VQQQ
Sbjct: 699 VQQQ 702
>At1g08150 hypothetical protein
Length = 815
Score = 80.1 bits (196), Expect = 4e-16
Identities = 46/122 (37%), Positives = 63/122 (50%), Gaps = 4/122 (3%)
Query: 18 ITWVVYHLVSTIKNDEFTSWDVMLDD----ELLKGVKGVYGSVDNVTYEKVEVENTSDTT 73
+ V L+ + T WD MLD E+L+ G V Y + V + SDT+
Sbjct: 688 VNLTVLKLIPAKMDGMTTGWDQMLDSAEVKEVLRNNNNTVGQHSFVEYVEETVNDGSDTS 747
Query: 74 EFISDIAIQHDFIIVGRRNGIKSPQTQALASWTEYPELGVLGDLLASPDTNTKASILVVQ 133
+ IA D +VGR G+ + AL+ WTE+ ELGV+GDLL S D + S+LVVQ
Sbjct: 748 TLLLSIANSFDLFVVGRSAGVGTDVVSALSEWTEFDELGVIGDLLVSQDFPRRGSVLVVQ 807
Query: 134 QQ 135
QQ
Sbjct: 808 QQ 809
>At3g44920 putative protein
Length = 671
Score = 79.3 bits (194), Expect = 6e-16
Identities = 40/101 (39%), Positives = 63/101 (61%), Gaps = 1/101 (0%)
Query: 35 TSWDVMLDDELLKGVKGVYGSVDNVTYEKVEVENTSDTTEFISDIAIQHDFIIVGRRNGI 94
+ WD +LD+E LK +K + D + E++ V + + + + +A ++D ++VGR + +
Sbjct: 568 SEWDYILDNEGLKDLKSTESNEDILYTERI-VTSVVEVVKAVQLLAEEYDLMVVGRDHDM 626
Query: 95 KSPQTQALASWTEYPELGVLGDLLASPDTNTKASILVVQQQ 135
S L W E PELGV+GDLLA+ D N+K S+LVVQQQ
Sbjct: 627 TSQDLSGLTEWVELPELGVIGDLLAARDLNSKVSVLVVQQQ 667
>At3g44930 putative protein
Length = 731
Score = 75.9 bits (185), Expect = 7e-15
Identities = 39/101 (38%), Positives = 63/101 (61%), Gaps = 1/101 (0%)
Query: 35 TSWDVMLDDELLKGVKGVYGSVDNVTYEKVEVENTSDTTEFISDIAIQHDFIIVGRRNGI 94
+ WD +LD+E LK +K + D E++ V ++ + + + +A ++D ++VGR + +
Sbjct: 628 SDWDYILDNEGLKDLKSTEDNKDIDYIERI-VTSSVEVVKAVQLLAEEYDLMVVGRDHDM 686
Query: 95 KSPQTQALASWTEYPELGVLGDLLASPDTNTKASILVVQQQ 135
S L W E PELGV+GDLLA+ D ++K S+LVVQQQ
Sbjct: 687 TSQDLSGLMEWVELPELGVIGDLLAARDLSSKVSVLVVQQQ 727
>At2g13620 putative Na/H antiporter
Length = 821
Score = 74.7 bits (182), Expect = 1e-14
Identities = 38/96 (39%), Positives = 52/96 (53%)
Query: 41 LDDELLKGVKGVYGSVDNVTYEKVEVENTSDTTEFISDIAIQHDFIIVGRRNGIKSPQTQ 100
LDD+ + + +++ Y + V N +T + + HD IVGR G+ SP T
Sbjct: 695 LDDDYINLFRAENAEYESIVYIEKLVSNGEETVAAVRSMDSSHDLFIVGRGEGMSSPLTA 754
Query: 101 ALASWTEYPELGVLGDLLASPDTNTKASILVVQQQV 136
L W+E PELG +GDLLAS D S+LVVQQ V
Sbjct: 755 GLTDWSECPELGAIGDLLASSDFAATVSVLVVQQYV 790
>At2g28170 hypothetical protein
Length = 617
Score = 72.8 bits (177), Expect = 6e-14
Identities = 40/116 (34%), Positives = 68/116 (58%), Gaps = 2/116 (1%)
Query: 22 VYHLVSTIKNDEFTSWDVMLDDELLKGVKGVYGSVDNVTYEKVE--VENTSDTTEFISDI 79
+ HL+ + +E LD + +K + S +N ++ +E V+ ++T+ + I
Sbjct: 500 ILHLIPQLTTEESEDSVQKLDYDDIKEIMKTEDSNENDSWICIEKSVKEGAETSVILRSI 559
Query: 80 AIQHDFIIVGRRNGIKSPQTQALASWTEYPELGVLGDLLASPDTNTKASILVVQQQ 135
A +D IVGR +G+ S T+ L WTE+ ELG LGD++AS + ++AS+LV+QQQ
Sbjct: 560 AYDYDLFIVGRSSGMNSAVTKGLNEWTEFEELGALGDVIASKEFPSRASVLVLQQQ 615
>At1g08140 hypothetical protein
Length = 1536
Score = 69.7 bits (169), Expect = 5e-13
Identities = 43/115 (37%), Positives = 63/115 (54%), Gaps = 8/115 (6%)
Query: 27 STIKNDEFTSWDVMLDDELLKGVKGVYGSV-----DNVTYEKVEVENTSDTTEFISDIAI 81
S + NDE W+ L + V + GS VTY V + S+T+ + +A
Sbjct: 657 SEMDNDE---WEQQQSINLKESVTSIVGSNIKENDAKVTYIDKAVSDGSETSRILRAMAN 713
Query: 82 QHDFIIVGRRNGIKSPQTQALASWTEYPELGVLGDLLASPDTNTKASILVVQQQV 136
+D IVG +GI + T ++ WTE+ ELG +GDLLAS + + AS+LVVQ+QV
Sbjct: 714 DYDLFIVGSGSGIGTEATSGISEWTEFNELGPIGDLLASHEYPSSASVLVVQKQV 768
Score = 67.0 bits (162), Expect = 3e-12
Identities = 31/70 (44%), Positives = 49/70 (69%)
Query: 66 VENTSDTTEFISDIAIQHDFIIVGRRNGIKSPQTQALASWTEYPELGVLGDLLASPDTNT 125
V + ++T++ + ++ +D IVGRR+G+ + T+ L W E+ ELGV+GDLLAS +
Sbjct: 1466 VGDGTETSKILHSVSYDYDLFIVGRRSGVGTTVTRGLGDWMEFEELGVIGDLLASEYFPS 1525
Query: 126 KASILVVQQQ 135
+AS+LVVQQQ
Sbjct: 1526 RASVLVVQQQ 1535
>At2g30240 putative Na/H antiporter
Length = 831
Score = 65.5 bits (158), Expect = 9e-12
Identities = 36/118 (30%), Positives = 66/118 (55%), Gaps = 5/118 (4%)
Query: 18 ITWVVYHLVSTIKNDEFTSWDVMLDDELLKGVKGVYGSVDNVTYEKVEVENTSDTTEFIS 77
+T + + S +++++++ M + L+ K + + Y + V + +TT+ IS
Sbjct: 674 VTMIHFRHKSALQDEDYSD---MSEYNLISDFKSYAANKGKIHYVEEIVRDGVETTQVIS 730
Query: 78 DIAIQHDFIIVGRRNGIKSPQTQALASWTEYPELGVLGDLLASPDTNTKASILVVQQQ 135
+ +D ++VGR + ++S L W+E PELGV+GD+L SPD + S+LVV QQ
Sbjct: 731 SLGDAYDMVLVGRDHDLESSVLYGLTDWSECPELGVIGDMLTSPDFH--FSVLVVHQQ 786
>At1g06970 hypothetical protein
Length = 829
Score = 60.8 bits (146), Expect = 2e-10
Identities = 35/122 (28%), Positives = 65/122 (52%), Gaps = 3/122 (2%)
Query: 15 KILITWVVYHLVSTIKNDEFTSWDVMLDDE-LLKGVKGVYGSVDNVTYEKVEVENTSDTT 73
++ +T + + S+++ + + L + L+ K S ++Y + V + +TT
Sbjct: 666 EVSVTMIHFRHKSSLQQNHVVDVESELAESYLINDFKNFAMSKPKISYREEIVRDGVETT 725
Query: 74 EFISDIAIQHDFIIVGRRNGIKSPQTQALASWTEYPELGVLGDLLASPDTNTKASILVVQ 133
+ IS + D ++VGR + ++S L W+E PELGV+GD+ AS D + S+LV+
Sbjct: 726 QVISSLGDSFDLVVVGRDHDLESSVLYGLTDWSECPELGVIGDMFASSDFH--FSVLVIH 783
Query: 134 QQ 135
QQ
Sbjct: 784 QQ 785
>At5g37060 putative transporter protein
Length = 859
Score = 58.5 bits (140), Expect = 1e-09
Identities = 32/83 (38%), Positives = 53/83 (63%), Gaps = 1/83 (1%)
Query: 55 SVDNVTYEKVEVENTSDTTEFISDIAIQ-HDFIIVGRRNGIKSPQTQALASWTEYPELGV 113
S + V+Y++V V+N ++T I + + +D I GRR GI + L++W+E +LGV
Sbjct: 749 SNERVSYKEVVVKNGAETLAAIQAMNVNDYDLWITGRREGINPKILEGLSTWSEDHQLGV 808
Query: 114 LGDLLASPDTNTKASILVVQQQV 136
+GD +A+ ++ S+LVVQQQV
Sbjct: 809 IGDTVAASVFASEGSVLVVQQQV 831
>At5g58460 putative protein
Length = 857
Score = 57.0 bits (136), Expect = 3e-09
Identities = 31/79 (39%), Positives = 50/79 (63%), Gaps = 1/79 (1%)
Query: 59 VTYEKVEVENTSDTTEFISDIAIQ-HDFIIVGRRNGIKSPQTQALASWTEYPELGVLGDL 117
V+Y++V V+N ++T I + + +D I GRR GI + L++W+E +LGV+GD
Sbjct: 751 VSYKEVVVKNGAETLAAIQAMNVNDYDLWITGRREGINPKILEGLSTWSEDHQLGVIGDT 810
Query: 118 LASPDTNTKASILVVQQQV 136
+A ++ S+LVVQQQV
Sbjct: 811 VAGSVFASEGSVLVVQQQV 829
>At2g31910 putative Na+/H+ antiporter
Length = 735
Score = 56.2 bits (134), Expect = 6e-09
Identities = 30/79 (37%), Positives = 46/79 (57%), Gaps = 2/79 (2%)
Query: 58 NVTYEKVEVENTSDTTEFISDIAIQ--HDFIIVGRRNGIKSPQTQALASWTEYPELGVLG 115
+VTY + V+N +T I ++ +D IVGR +++P T L W P+LG++G
Sbjct: 611 SVTYVEKVVKNGQETITAILELEDNNSYDLYIVGRGYQVETPVTSGLTDWNSTPDLGIIG 670
Query: 116 DLLASPDTNTKASILVVQQ 134
D L S + +AS+LVVQQ
Sbjct: 671 DTLISSNFTMQASVLVVQQ 689
>At5g41610 Na+/H+ antiporter-like protein
Length = 810
Score = 53.5 bits (127), Expect = 4e-08
Identities = 31/109 (28%), Positives = 60/109 (54%), Gaps = 2/109 (1%)
Query: 26 VSTIKNDEFTSWDVMLDDELLKGVKGVYGSVDNVTYEKVEVENTSDTTEFISDIAIQHDF 85
VS N+ + ++ D+E++ ++ + ++V + + ++EN + + + +
Sbjct: 678 VSNNNNENQSVKNLKSDEEIMSEIRKISSVDESVKFVEKQIENAAVDVRSAIEEVRRSNL 737
Query: 86 IIVGRRNGIKSPQTQALASWTEYPELGVLGDLLASPDTNTKASILVVQQ 134
+VGR G A+ +E PELG +G LL SP+++TKAS+LV+QQ
Sbjct: 738 FLVGRMPG--GEIALAIRENSECPELGPVGSLLISPESSTKASVLVIQQ 784
>At3g52080 putative protein
Length = 732
Score = 53.1 bits (126), Expect = 5e-08
Identities = 32/100 (32%), Positives = 52/100 (52%), Gaps = 2/100 (2%)
Query: 38 DVMLDDELLKGVKGVY-GSVDNVTYEKVEVENTSDTTEFISDIAIQHDFIIVGRRNG-IK 95
++ LDDE Y V+Y + + N+S+T + + ++ +IVGR G
Sbjct: 617 EMKLDDECFAEFYERYIAGGGRVSYMEKHLTNSSETFTALKSLDGEYGLVIVGRGGGRAS 676
Query: 96 SPQTQALASWTEYPELGVLGDLLASPDTNTKASILVVQQQ 135
S T L W + PELG +GD+L+ D + S+L++QQQ
Sbjct: 677 SGLTTGLNDWQQCPELGPIGDVLSGSDFSHNTSMLIIQQQ 716
>At5g01680 putative transporter protein
Length = 780
Score = 52.8 bits (125), Expect = 6e-08
Identities = 28/70 (40%), Positives = 38/70 (54%)
Query: 67 ENTSDTTEFISDIAIQHDFIIVGRRNGIKSPQTQALASWTEYPELGVLGDLLASPDTNTK 126
E SD + + +D I+VG R+ Q L+ W+E ELG +GDLL S D
Sbjct: 704 EEASDLVNLLREEGNNYDLIMVGIRHEKSFEVLQGLSVWSEIEELGEIGDLLVSRDLKLS 763
Query: 127 ASILVVQQQV 136
AS+L VQQQ+
Sbjct: 764 ASVLAVQQQL 773
>At1g16380 putative Na/H antiporter
Length = 785
Score = 52.8 bits (125), Expect = 6e-08
Identities = 27/76 (35%), Positives = 40/76 (52%), Gaps = 1/76 (1%)
Query: 59 VTYEKVEVENTSDTTEFISDIAIQHDFIIVGRRNGIKSPQTQALASWTEYPELGVLGDLL 118
V + + V N T + +I + +VG+ G P T + W E PELG +GD L
Sbjct: 700 VGFIEKRVSNGMQTLTILREIGEMYSLFVVGKNRG-DCPMTSGMNDWEECPELGTVGDFL 758
Query: 119 ASPDTNTKASILVVQQ 134
AS + + AS+LVVQ+
Sbjct: 759 ASSNMDVNASVLVVQR 774
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.316 0.133 0.387
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,222,590
Number of Sequences: 26719
Number of extensions: 125154
Number of successful extensions: 314
Number of sequences better than 10.0: 40
Number of HSP's better than 10.0 without gapping: 31
Number of HSP's successfully gapped in prelim test: 9
Number of HSP's that attempted gapping in prelim test: 270
Number of HSP's gapped (non-prelim): 45
length of query: 141
length of database: 11,318,596
effective HSP length: 89
effective length of query: 52
effective length of database: 8,940,605
effective search space: 464911460
effective search space used: 464911460
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 55 (25.8 bits)
Medicago: description of AC146343.4


