

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146342.2 + phase: 0
(340 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
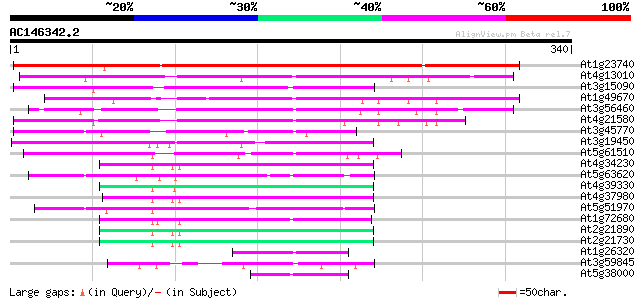
Score E
Sequences producing significant alignments: (bits) Value
At1g23740 auxin-induced protein like 276 1e-74
At4g13010 unknown protein 134 9e-32
At3g15090 zinc-binding dehydrogenase, putative 103 1e-22
At1g49670 ARP protein (At1g49670; F14J22.10) 87 2e-17
At3g56460 quinone reductase-like protein 85 5e-17
At4g21580 putative NADPH quinone oxidoreductase 79 3e-15
At3g45770 nuclear receptor binding factor-like protein 66 2e-11
At3g19450 putative cinnamyl alcohol dehydrogenase 2 65 7e-11
At5g61510 quinone oxidoreductase - like protein 62 3e-10
At4g34230 cinnamyl alcohol dehydrogenase like protein 61 1e-09
At5g63620 alcohol dehydrogenase-like protein 57 1e-08
At4g39330 cinnamyl-alcohol dehydrogenase CAD1 56 2e-08
At4g37980 cinnamyl-alcohol dehydrogenase ELI3-1 55 5e-08
At5g51970 sorbitol dehydrogenase-like protein 55 7e-08
At1g72680 putative cinnamyl-alcohol dehydrogenase 52 5e-07
At2g21890 putative cinnamyl-alcohol dehydrogenase 51 1e-06
At2g21730 putative cinnamyl-alcohol dehydrogenase 50 1e-06
At1g26320 allyl alcohol dehydrogenase like protein 43 2e-04
At3g59845 allyl alcohol dehydrogenase-like protein 42 4e-04
At5g38000 oxidoreductase-like protein 42 6e-04
>At1g23740 auxin-induced protein like
Length = 386
Score = 276 bits (706), Expect = 1e-74
Identities = 144/310 (46%), Positives = 205/310 (65%), Gaps = 5/310 (1%)
Query: 3 KAWFYEEYGPKEVLKL-GDFPIPSPLENQLLVQVYAAALNPIDSKRRMRPIFPSD--FPA 59
KAW Y +YG +VLKL + +P E+Q+L++V AAALNP+D+KRR +D P
Sbjct: 79 KAWVYSDYGGVDVLKLESNIVVPEIKEDQVLIKVVAAALNPVDAKRRQGKFKATDSPLPT 138
Query: 60 VPGCDMAGVVIGKGVNVKKFDIGDEVYGNIQDFNSMEKPKQLGTLAQFIVVEENLVARKP 119
VPG D+AGVV+ G VK GDEVY N+ + ++E PKQ G+LA++ VEE L+A KP
Sbjct: 139 VPGYDVAGVVVKVGSAVKDLKEGDEVYANVSE-KALEGPKQFGSLAEYTAVEEKLLALKP 197
Query: 120 KRLSFEEAASLPLAVQTAIEGFKTGDFKKGETMFVVGGAGGVGTLVLQLAKLMFGASYVV 179
K + F +AA LPLA++TA EG +F G+++ V+ GAGGVG+LV+QLAK ++GAS V
Sbjct: 198 KNIDFAQAAGLPLAIETADEGLVRTEFSAGKSILVLNGAGGVGSLVIQLAKHVYGASKVA 257
Query: 180 SSCSTPKLKFVKQFGADKVVDYTKTKYEDIEEKFDFIYDTVGDCKKSFVVAKKDGAIVDI 239
++ ST KL+ V+ GAD +DYTK ED+ +K+D ++D +G C K+ V K+ G +V +
Sbjct: 258 ATASTEKLELVRSLGADLAIDYTKENIEDLPDKYDVVFDAIGMCDKAVKVIKEGGKVVAL 317
Query: 240 TWPASHERAVYSSLTVCGEILEKLRPYLERGELKAVIDPKGEYDFENVIDAFGYIETGRA 299
T + + +T G++L+KL PY+E G++K V+DPKG + F V DAF Y+ET A
Sbjct: 318 TGAVTPPGFRF-VVTSNGDVLKKLNPYIESGKVKPVVDPKGPFPFSRVADAFSYLETNHA 376
Query: 300 WGKVVVTCFP 309
GKVVV P
Sbjct: 377 TGKVVVYPIP 386
>At4g13010 unknown protein
Length = 329
Score = 134 bits (336), Expect = 9e-32
Identities = 98/328 (29%), Positives = 154/328 (46%), Gaps = 40/328 (12%)
Query: 7 YEEYGPKEV-LKLGDFPIPSPLENQLLVQVYAAALNPID---SKRRMRPIFPSDFPAVPG 62
Y YG L+ P+P+P N++ +++ A +LNP+D K +RP P FP +P
Sbjct: 11 YNSYGGGAAGLEHVQVPVPTPKSNEVCLKLEATSLNPVDWKIQKGMIRPFLPRKFPCIPA 70
Query: 63 CDMAGVVIGKGVNVKKFDIGDEVYGNIQDFNSMEKPKQLGTLAQFIVVEENLVARKPKRL 122
D+AG V+ G VK F GD+V + G LA+F V E L ++P+ +
Sbjct: 71 TDVAGEVVEVGSGVKNFKAGDKVVAVLSHLGG-------GGLAEFAVATEKLTVKRPQEV 123
Query: 123 SFEEAASLPLAVQTAIE------GFKTGDFKKGETMFVVGGAGGVGTLVLQLAKLMFGAS 176
EAA+LP+A TA++ G K K + V +GGVG +QLAKL +
Sbjct: 124 GAAEAAALPVAGLTALQALTNPAGLKLDGTGKKANILVTAASGGVGHYAVQLAKL--ANA 181
Query: 177 YVVSSCSTPKLKFVKQFGADKVVDYTKTKYEDIEEKFDFIYDTVGDCKKSFVVA------ 230
+V ++C ++FVK GAD+V+DY + ++ YD V C +
Sbjct: 182 HVTATCGARNIEFVKSLGADEVLDYKTPEGAALKSPSGKKYDAVVHCANGIPFSVFEPNL 241
Query: 231 KKDGAIVDIT------WPASHERAVYSS-------LTVCGEILEKLRPYLERGELKAVID 277
++G ++DIT W + ++ S L E LE + ++ G++K VID
Sbjct: 242 SENGKVIDITPGPNAMWTYAVKKITMSKKQLVPLLLIPKAENLEFMVNLVKEGKVKTVID 301
Query: 278 PKGEYDFENVIDAFGYIETGRAWGKVVV 305
K + DA+ G A GK++V
Sbjct: 302 SK--HPLSKAEDAWAKSIDGHATGKIIV 327
>At3g15090 zinc-binding dehydrogenase, putative
Length = 366
Score = 103 bits (257), Expect = 1e-22
Identities = 68/225 (30%), Positives = 114/225 (50%), Gaps = 14/225 (6%)
Query: 3 KAWFYEEYGPKEVLKLGD-FPIPSPLENQLLVQVYAAALNPIDSKRRM---RPIFPSDFP 58
+A +G EV +L + P+P+ N++LV+ A ++NP+D + R R +F P
Sbjct: 33 RAVILPRFGGPEVFELRENVPVPNLNPNEVLVKAKAVSVNPLDCRIRAGYGRSVFQPHLP 92
Query: 59 AVPGCDMAGVVIGKGVNVKKFDIGDEVYGNIQDFNSMEKPKQLGTLAQFIVVEENLVARK 118
+ G D++G V G +VK +G EV+G ++ GT + ++ E+ + K
Sbjct: 93 IIVGRDVSGEVAAIGTSVKSLKVGQEVFG------ALHPTALRGTYTDYGILSEDELTEK 146
Query: 119 PKRLSFEEAASLPLAVQTAIEGFKT-GDFKKGETMFVVGGAGGVGTLVLQLAKLMFGASY 177
P +S EA+++P A TA K+ +G+ + V GG G VG +QLA + +
Sbjct: 147 PSSISHVEASAIPFAALTAWRALKSNARITEGQRLLVFGGGGAVGFSAIQLA--VASGCH 204
Query: 178 VVSSCSTPKLKFVKQFGADKVVDYTKTKYE-DIEEKFDFIYDTVG 221
V +SC + GA++ VDYT E ++ KFD + DT+G
Sbjct: 205 VTASCVGQTKDRILAAGAEQAVDYTTEDIELAVKGKFDAVLDTIG 249
>At1g49670 ARP protein (At1g49670; F14J22.10)
Length = 629
Score = 86.7 bits (213), Expect = 2e-17
Identities = 87/326 (26%), Positives = 139/326 (41%), Gaps = 52/326 (15%)
Query: 22 PIPSPL-ENQLLVQVYAAALNPIDSKRRMRPIFPSDFPAVP---GCDMAGVVIGKGVNVK 77
P+ P+ +Q+L+++ A +N D F P +P G + G++ G +VK
Sbjct: 309 PLQLPIGPHQVLLKIIYAGVNASDVNFSSGRYFTGGSPKLPFDAGFEGVGLIAAVGESVK 368
Query: 78 KFDIGDEVYGNIQDFNSMEKPKQLGTLAQFIVVEENLVARKPKRLSFEEAASLPLAVQTA 137
++G + F G +++++V V P R E A L +
Sbjct: 369 NLEVGTPAA--VMTF---------GAYSEYMIVSSKHVLPVP-RPDPEVVAMLTSGLTAL 416
Query: 138 IEGFKTGDFKKGETMFVVGGAGGVGTLVLQLAKLMFGASYVVSSC-STPKLKFVKQFGAD 196
I K G K GET+ V AGG G +QLAKL + V+++C + K K +K+ G D
Sbjct: 417 IALEKAGQMKSGETVLVTAAAGGTGQFAVQLAKL--SGNKVIATCGGSEKAKLLKELGVD 474
Query: 197 KVVDYTKTKYEDIEEK-----FDFIYDTVGD-----CKKSFVVAKKDGAIVDIT------ 240
+V+DY + + +K + IY++VG C + V + I I+
Sbjct: 475 RVIDYKSENIKTVLKKEFPKGVNIIYESVGGQMFDMCLNALAVYGRLIVIGMISQYQGEK 534
Query: 241 ------WPASHERAVYSSLTVCG-----------EILEKLRPYLERGELKAVIDPKGEYD 283
+P E+ + S TV G + L+KL G+LK ID K
Sbjct: 535 GWEPAKYPGLCEKILAKSQTVAGFFLVQYSQLWKQNLDKLFNLYALGKLKVGIDQKKFIG 594
Query: 284 FENVIDAFGYIETGRAWGKVVVTCFP 309
V DA Y+ +G++ GKVVV P
Sbjct: 595 LNAVADAVEYLHSGKSTGKVVVCIDP 620
>At3g56460 quinone reductase-like protein
Length = 348
Score = 85.1 bits (209), Expect = 5e-17
Identities = 94/333 (28%), Positives = 146/333 (43%), Gaps = 58/333 (17%)
Query: 12 PKEVLKLGDFPIPS-PLENQLLVQVYAAALN-----PIDSKRRMRPIFPSDFPAVPGCDM 65
P EV K PIPS + + V+V A +LN I K + +P P +PG D
Sbjct: 23 PVEVSKT--HPIPSLNSDTSVRVRVIATSLNYANYLQILGKYQEKP----PLPFIPGSDY 76
Query: 66 AGVVIGKGVNVKKFDIGDEVYGNIQDFNSMEKPKQLGTLAQFIVVEENLVARKPKRLSFE 125
+G+V G V KF +GD V LG+ AQFIV +++ + P+R
Sbjct: 77 SGIVDAIGPAVTKFRVGDRVCSFAD----------LGSFAQFIVADQSRLFLVPERCDMV 126
Query: 126 EAASLPLAVQTA-IEGFKTGDFKKGETMFVVGGAGGVGTLVLQLAKLMFGASYVVSSCST 184
AA+LP+A T+ + G+ + V+G AGGVG +Q+ K+ GA + + T
Sbjct: 127 AAAALPVAFGTSHVALVHRARLTSGQVLLVLGAAGGVGLAAVQIGKVC-GAIVIAVARGT 185
Query: 185 PKLKFVKQFGADKVVDY-TKTKYEDIEE--------KFDFIYDTVGD--CKKSFVVAKKD 233
K++ +K G D VVD T+ ++E D +YD VG K+S V K
Sbjct: 186 EKIQLLKSMGVDHVVDLGTENVISSVKEFIKTRKLKGVDVLYDPVGGKLTKESMKVLKWG 245
Query: 234 GAIVDITWPASH------ERAVYSSLTVCG---------------EILEKLRPYLERGEL 272
I+ I + + A+ + TV G + +++L +L RG +
Sbjct: 246 AQILVIGFASGEIPVIPANIALVKNWTVHGLYWGSYRIHQPNVLEDSIKELLSWLSRGLI 305
Query: 273 KAVIDPKGEYDFENVIDAFGYIETGRAWGKVVV 305
I Y AFG ++ +A GKV++
Sbjct: 306 --TIHISHTYSLSQANLAFGDLKDRKAIGKVMI 336
>At4g21580 putative NADPH quinone oxidoreductase
Length = 325
Score = 79.3 bits (194), Expect = 3e-15
Identities = 83/308 (26%), Positives = 135/308 (42%), Gaps = 47/308 (15%)
Query: 3 KAWFYEEYGPKEVLKLGDFPIPSPLENQLLVQVYAAALNPIDSKRRM---RPIFPSDFPA 59
KA E G EVL+L D P ++++L++V A ALN D+ +R+ P P
Sbjct: 2 KAIVISEPGKPEVLQLRDVADPEVKDDEVLIRVLATALNRADTLQRLGLYNP--PPGSSP 59
Query: 60 VPGCDMAGVVIGKGVNVKKFDIGDEVYGNIQDFNSMEKPKQLGTLAQFIVVEENLVARKP 119
G + +G + G V ++ +GD+V + G A+ + V + P
Sbjct: 60 YLGLECSGTIESVGKGVSRWKVGDQVCALLSG----------GGYAEKVSVPAGQIFPIP 109
Query: 120 KRLSFEEAASLP-LAVQTAIEGFKTGDFKKGETMFVVGGAGGVGTLVLQLAKLMFGASYV 178
+S ++AA+ P +A F G GE+ + GG+ G+GT +Q+AK + G
Sbjct: 110 AGISLKDAAAFPEVACTVWSTVFMMGRLSVGESFLIHGGSSGIGTFAIQIAKHL-GVRVF 168
Query: 179 VSSCSTPKLKFVKQFGADKVVDY------TKTKYEDIEEKFDFIYDTVGD--CKKSFVVA 230
V++ S KL K+ GAD ++Y K K E + D I D +G +K+
Sbjct: 169 VTAGSDEKLAACKELGADVCINYKTEDFVAKVKAETDGKGVDVILDCIGAPYLQKNLDSL 228
Query: 231 KKDG--AIVDITWPASHERAVYS----SLTVCG----------------EILEKLRPYLE 268
DG I+ + A+ E + S LTV G E+ + + P +E
Sbjct: 229 NFDGRLCIIGLMGGANAEIKLSSLLPKRLTVLGAALRPRSPENKAVVVREVEKNVWPAIE 288
Query: 269 RGELKAVI 276
G++K VI
Sbjct: 289 AGKVKPVI 296
>At3g45770 nuclear receptor binding factor-like protein
Length = 375
Score = 66.2 bits (160), Expect = 2e-11
Identities = 64/218 (29%), Positives = 101/218 (45%), Gaps = 23/218 (10%)
Query: 3 KAWFYEEYG-PKEVLKLGDFPIPSPLENQLLVQVYAAALNPIDSKRRMRPIFP--SDFPA 59
KA YEE+G P V +L + P EN + V++ AA +NP D R+ ++P PA
Sbjct: 46 KAIVYEEHGSPDSVTRLVNLPPVEVKENDVCVKMIAAPINPSDI-NRIEGVYPVRPPVPA 104
Query: 60 VPGCDMAGVVIGKGVNVKKFDIGDEVYGNIQDFNSMEKPKQLGTLAQFIVVEENLVARKP 119
V G + G V G NV F GD V + P GT ++V EE++ +
Sbjct: 105 VGGYEGVGEVYAVGSNVNGFSPGDWV---------IPSPPSSGTWQTYVVKEESVWHKID 155
Query: 120 KRLSFEEAASL---PLAVQTAIEGFKTGDFKKGETMFVVGGAGGVGTLVLQLAKLMFGAS 176
K E AA++ PL +E F + G+++ G VG V+QLA+L G S
Sbjct: 156 KECPMEYAATITVNPLTALRMLEDFV--NLNSGDSVVQNGATSIVGQCVIQLARLR-GIS 212
Query: 177 YV----VSSCSTPKLKFVKQFGADKVVDYTKTKYEDIE 210
+ + S + +K GAD+V ++ ++++
Sbjct: 213 TINLIRDRAGSDEAREQLKALGADEVFSESQLNVKNVK 250
>At3g19450 putative cinnamyl alcohol dehydrogenase 2
Length = 365
Score = 64.7 bits (156), Expect = 7e-11
Identities = 65/251 (25%), Positives = 107/251 (41%), Gaps = 38/251 (15%)
Query: 2 QKAWFYEEYGPKEVLKLGDFPIPSPLENQLLVQVYAAALNPIDSKRRMRPIFPSDFPAVP 61
+KA + P VL + + S + + ++V + D + + S++P VP
Sbjct: 9 KKALGWAARDPSGVLSPYSYTLRSTGADDVYIKVICCGICHTDIHQIKNDLGMSNYPMVP 68
Query: 62 GCDMAGVVIGKGVNVKKFDIGD---------------------EVYGN--IQDFNSM--- 95
G ++ G V+ G +V KF +GD E Y N I +N +
Sbjct: 69 GHEVVGEVLEVGSDVSKFTVGDVVGVGVVVGCCGSCKPCSSELEQYCNKRIWSYNDVYTD 128
Query: 96 EKPKQLGTLAQFIVVEENLVARKPKRLSFEEAASLPLAVQTAIE-----GFKTGDFKKGE 150
KP Q G A ++V + V + P+ ++ E+AA L A T G K G
Sbjct: 129 GKPTQ-GGFADTMIVNQKFVVKIPEGMAVEQAAPLLCAGVTVYSPLSHFGLMASGLKGG- 186
Query: 151 TMFVVGGAGGVGTLVLQLAKLMFGASYVVSSCSTPKLKFVKQFGADK-VVDYTKTKYEDI 209
+ G GGVG + +++AK M V+SS K + ++ GAD VV + + +
Sbjct: 187 ----ILGLGGVGHMGVKIAKAMGHHVTVISSSDKKKEEAIEHLGADDYVVSSDPAEMQRL 242
Query: 210 EEKFDFIYDTV 220
+ D+I DTV
Sbjct: 243 ADSLDYIIDTV 253
>At5g61510 quinone oxidoreductase - like protein
Length = 406
Score = 62.4 bits (150), Expect = 3e-10
Identities = 70/243 (28%), Positives = 104/243 (41%), Gaps = 29/243 (11%)
Query: 9 EYGPKEVLKLGDFPIPSPLENQLLVQVYAAALNPIDSKRRMRPIFPSDFPAVPGCDMAGV 68
E+G EVLK D + P E ++ V+ A LN ID R P+ P PG + G
Sbjct: 91 EHGGPEVLKWEDVEVGEPKEGEIRVKNKAIGLNFIDVYFRKGVYKPASMPFTPGMEAVGE 150
Query: 69 VIGKGVNVKKFDIGDEV--YGNIQDFNSMEKPKQLGTLAQFIVVEENLVARKPKRLSFEE 126
V+ G + IGD V GN +G A+ ++ + V P +
Sbjct: 151 VVAVGSGLTGRMIGDLVAYAGN-----------PMGAYAEEQILPADKVVPVPSSIDPIV 199
Query: 127 AASLPLAVQTA----IEGFKTGDFKKGETMFVVGGAGGVGTLVLQLAKLMFGASYVVSSC 182
AAS+ L TA FK + G T+ V AGGVG+L+ Q A + GA+ + +
Sbjct: 200 AASIMLKGMTAQFLLRRCFKV---EPGHTILVHAAAGGVGSLLCQWANAL-GATVIGTVS 255
Query: 183 STPKLKFVKQFGADKVVDYTK----TKYEDIE--EKFDFIYDTVG--DCKKSFVVAKKDG 234
+ K K+ G V+ Y ++ DI + + +YD+VG K S K G
Sbjct: 256 TNEKAAQAKEDGCHHVIMYKNEDFVSRVNDITSGKGVNVVYDSVGKDTFKGSLACLKSRG 315
Query: 235 AIV 237
+V
Sbjct: 316 YMV 318
>At4g34230 cinnamyl alcohol dehydrogenase like protein
Length = 357
Score = 60.8 bits (146), Expect = 1e-09
Identities = 51/192 (26%), Positives = 88/192 (45%), Gaps = 26/192 (13%)
Query: 55 SDFPAVPGCDMAGVVIGKGVNVKKFDIGDEV-----YGNIQDFNSMEK------PKQL-- 101
S++P VPG ++ G V+ G +V KF +GD V G + E+ PK++
Sbjct: 61 SNYPMVPGHEVVGEVVEVGSDVSKFTVGDIVGVGCLVGCCGGCSPCERDLEQYCPKKIWS 120
Query: 102 ------------GTLAQFIVVEENLVARKPKRLSFEEAASLPLAVQTAIEGFKTGDFKKG 149
G A+ VV + V + P+ ++ E+AA L A T K+
Sbjct: 121 YNDVYINGQPTQGGFAKATVVHQKFVVKIPEGMAVEQAAPLLCAGVTVYSPLSHFGLKQP 180
Query: 150 ETMFVVGGAGGVGTLVLQLAKLMFGASYVVSSCSTPKLKFVKQFGADK-VVDYTKTKYED 208
+ G GGVG + +++AK M V+SS + + + ++ GAD V+ + K +
Sbjct: 181 GLRGGILGLGGVGHMGVKIAKAMGHHVTVISSSNKKREEALQDLGADDYVIGSDQAKMSE 240
Query: 209 IEEKFDFIYDTV 220
+ + D++ DTV
Sbjct: 241 LADSLDYVIDTV 252
>At5g63620 alcohol dehydrogenase-like protein
Length = 427
Score = 57.4 bits (137), Expect = 1e-08
Identities = 65/257 (25%), Positives = 108/257 (41%), Gaps = 53/257 (20%)
Query: 12 PKEVLKLGDFPIPSPLENQLLVQVYAAALNPIDSKRRMRPIFPSDFPAVPGCDMAGVVIG 71
P + L + +F IP P N++L++ A + D M+ P P V G ++ G V+
Sbjct: 64 PNKPLTIEEFHIPRPKSNEILIKTKACGVCHSDL-HVMKGEIPFASPCVIGHEITGEVVE 122
Query: 72 KGVN-----VKKFDIGDEVYGNI------------------QDFNSMEKPK--------- 99
G + +F IG V G +DF + + K
Sbjct: 123 HGPLTDHKIINRFPIGSRVVGAFIMPCGTCSYCAKGHDDLCEDFFAYNRAKGTLYDGETR 182
Query: 100 -------------QLGTLAQFIVVEENLVARKPKRLSFEEAASLPLAVQTAIEGFK-TGD 145
+G +A++ V + +A P+ L + E+A L AV TA +
Sbjct: 183 LFLRHDDSPVYMYSMGGMAEYCVTPAHGLAPLPESLPYSESAILGCAVFTAYGAMAHAAE 242
Query: 146 FKKGETMFVVGGAGGVGTLVLQLAKLMFGASYVVS-SCSTPKLKFVKQFGADKVVDYTKT 204
+ G+++ V+G GGVG+ LQ+A+ FGAS +++ KL+ K GA +V+ K
Sbjct: 243 IRPGDSIAVIG-IGGVGSSCLQIAR-AFGASDIIAVDVQDDKLQKAKTLGATHIVNAAK- 299
Query: 205 KYEDIEEKFDFIYDTVG 221
ED E+ I +G
Sbjct: 300 --EDAVERIREITGGMG 314
>At4g39330 cinnamyl-alcohol dehydrogenase CAD1
Length = 360
Score = 56.2 bits (134), Expect = 2e-08
Identities = 51/192 (26%), Positives = 77/192 (39%), Gaps = 26/192 (13%)
Query: 55 SDFPAVPGCDMAGVVIGKGVNVKKFDIGDEV-----YGNIQDFNSMEKP----------- 98
S +P VPG ++ G+ G NV KF GD V G+ Q S ++
Sbjct: 64 SYYPVVPGHEIVGIATKVGKNVTKFKEGDRVGVGVISGSCQSCESCDQDLENYCPQMSFT 123
Query: 99 ---------KQLGTLAQFIVVEENLVARKPKRLSFEEAASLPLAVQTAIEGFKTGDFKKG 149
K G ++ IVV++ V R P+ L + A L A T K +
Sbjct: 124 YNAIGSDGTKNYGGYSENIVVDQRFVLRFPENLPSDSGAPLLCAGITVYSPMKYYGMTEA 183
Query: 150 ETMFVVGGAGGVGTLVLQLAKLMFGASYVVSSCSTPKLKFVKQFGADK-VVDYTKTKYED 208
V G GG+G + +++ K V+SS ST + + GAD +V K +
Sbjct: 184 GKHLGVAGLGGLGHVAVKIGKAFGLKVTVISSSSTKAEEAINHLGADSFLVTTDPQKMKA 243
Query: 209 IEEKFDFIYDTV 220
D+I DT+
Sbjct: 244 AIGTMDYIIDTI 255
>At4g37980 cinnamyl-alcohol dehydrogenase ELI3-1
Length = 357
Score = 55.1 bits (131), Expect = 5e-08
Identities = 53/190 (27%), Positives = 79/190 (40%), Gaps = 26/190 (13%)
Query: 57 FPAVPGCDMAGVVIGKGVNVKKFDIGDEV-----YGNIQDFNSMEK------PKQL---- 101
+P VPG ++ GVV G VKKF+ GD+V G+ + +S PK +
Sbjct: 62 YPLVPGHEIVGVVTEVGAKVKKFNAGDKVGVGYMAGSCRSCDSCNDGDENYCPKMILTSG 121
Query: 102 ----------GTLAQFIVVEENLVARKPKRLSFEEAASLPLAVQTAIEGFKTGDFKKGET 151
G + +V E+ + R P L + AA L A T K K
Sbjct: 122 AKNFDDTMTHGGYSDHMVCAEDFIIRIPDNLPLDGAAPLLCAGVTVYSPMKYHGLDKPGM 181
Query: 152 MFVVGGAGGVGTLVLQLAKLMFGASYVVSSCSTPKLKFVKQFGADK-VVDYTKTKYEDIE 210
V G GG+G + ++ AK M V+S+ + + V + GAD +V + +D
Sbjct: 182 HIGVVGLGGLGHVAVKFAKAMGTKVTVISTSERKRDEAVTRLGADAFLVSRDPKQMKDAM 241
Query: 211 EKFDFIYDTV 220
D I DTV
Sbjct: 242 GTMDGIIDTV 251
>At5g51970 sorbitol dehydrogenase-like protein
Length = 364
Score = 54.7 bits (130), Expect = 7e-08
Identities = 60/230 (26%), Positives = 97/230 (42%), Gaps = 30/230 (13%)
Query: 16 LKLGDFPIPSPLENQLLVQVYAAALNPIDSKRRMRPIFPSDF----PAVPGCDMAGVVIG 71
LK+ F +PS + + V++ A + D ++ + +DF P V G + AG++
Sbjct: 29 LKIQPFLLPSVGPHDVRVRMKAVGICGSDV-HYLKTMRCADFVVKEPMVIGHECAGIIEE 87
Query: 72 KGVNVKKFDIGDEV------------------YGNIQDFNSMEKPKQLGTLAQFIVVEEN 113
G VK +GD V Y + P G+LA +V +
Sbjct: 88 VGEEVKHLVVGDRVALEPGISCWRCNLCREGRYNLCPEMKFFATPPVHGSLANQVVHPAD 147
Query: 114 LVARKPKRLSFEEAASL-PLAVQT-AIEGFKTGDFKKGETMFVVGGAGGVGTLVLQLAKL 171
L + P+ +S EE A PL+V A + G ET +V GAG +G + + A+
Sbjct: 148 LCFKLPENVSLEEGAMCEPLSVGVHACRRAEVGP----ETNVLVMGAGPIGLVTMLAARA 203
Query: 172 MFGASYVVSSCSTPKLKFVKQFGADKVVDYTKTKYEDIEEKFDFIYDTVG 221
V+ +L KQ GAD++V T T ED+ + + I +G
Sbjct: 204 FSVPRIVIVDVDENRLAVAKQLGADEIVQVT-TNLEDVGSEVEQIQKAMG 252
>At1g72680 putative cinnamyl-alcohol dehydrogenase
Length = 355
Score = 52.0 bits (123), Expect = 5e-07
Identities = 52/192 (27%), Positives = 81/192 (42%), Gaps = 28/192 (14%)
Query: 55 SDFPAVPGCDMAGVVIGKGVNVKKFDIGDEV----YGN-------------------IQD 91
S +P VPG ++AG+V G NV++F +GD V Y N +
Sbjct: 62 SKYPLVPGHEIAGIVTKVGPNVQRFKVGDHVGVGTYVNSCRECEYCNEGQEVNCAKGVFT 121
Query: 92 FNSMEKPKQL--GTLAQFIVVEENLVARKPKRLSFEEAASLPLAVQTAIEGFKTGDFKKG 149
FN ++ + G + IVV E + P E AA L A T + +
Sbjct: 122 FNGIDHDGSVTKGGYSSHIVVHERYCYKIPVDYPLESAAPLLCAGITVYAPMMRHNMNQP 181
Query: 150 ETMFVVGGAGGVGTLVLQLAKLMFGASYVVSSCS-TPKLKFVKQFGADK-VVDYTKTKYE 207
V G GG+G + ++ K FG S V S S + K + + GA+ V+ + +
Sbjct: 182 GKSLGVIGLGGLGHMAVKFGK-AFGLSVTVFSTSISKKEEALNLLGAENFVISSDHDQMK 240
Query: 208 DIEEKFDFIYDT 219
+E+ DF+ DT
Sbjct: 241 ALEKSLDFLVDT 252
>At2g21890 putative cinnamyl-alcohol dehydrogenase
Length = 375
Score = 50.8 bits (120), Expect = 1e-06
Identities = 52/193 (26%), Positives = 77/193 (38%), Gaps = 27/193 (13%)
Query: 55 SDFPAVPGCDMAGVVIGKGVNVKKFDIGDEV-----YGNIQDFNSMEK------PKQL-- 101
S +P +PG ++ G+ G NV KF GD V G+ Q S + PK +
Sbjct: 58 SRYPIIPGHEIVGIATKVGKNVTKFKEGDRVGVGVIIGSCQSCESCNQDLENYCPKVVFT 117
Query: 102 ------------GTLAQFIVVEENLVARKPKRLSFEEAASLPLAVQTAIEGFK-TGDFKK 148
G + IVV+ V P L + A L A T K G K+
Sbjct: 118 YNSRSSDGTRNQGGYSDVIVVDHRFVLSIPDGLPSDSGAPLLCAGITVYSPMKYYGMTKE 177
Query: 149 GETMFVVGGAGGVGTLVLQLAKLMFGASYVVSSCSTPKLKFVKQFGADK-VVDYTKTKYE 207
V G GG+G + +++ K V+S S + + + + GAD +V K +
Sbjct: 178 SGKRLGVNGLGGLGHIAVKIGKAFGLRVTVISRSSEKEREAIDRLGADSFLVTTDSQKMK 237
Query: 208 DIEEKFDFIYDTV 220
+ DFI DTV
Sbjct: 238 EAVGTMDFIIDTV 250
>At2g21730 putative cinnamyl-alcohol dehydrogenase
Length = 376
Score = 50.4 bits (119), Expect = 1e-06
Identities = 52/194 (26%), Positives = 77/194 (38%), Gaps = 28/194 (14%)
Query: 55 SDFPAVPGCDMAGVVIGKGVNVKKFDIGDEV-----YGNIQDFNSMEK------PKQL-- 101
S +P +PG ++ G+ G NV KF GD V G+ Q S + PK +
Sbjct: 58 SRYPIIPGHEIVGIATKVGKNVTKFKEGDRVGVGVIIGSCQSCESCNQDLENYCPKVVFT 117
Query: 102 -------------GTLAQFIVVEENLVARKPKRLSFEEAASLPLAVQTAIEGFK-TGDFK 147
G + IVV+ V P L + A L A T K G K
Sbjct: 118 YNSRSSDGTSRNQGGYSDVIVVDHRFVLSIPDGLPSDSGAPLLCAGITVYSPMKYYGMTK 177
Query: 148 KGETMFVVGGAGGVGTLVLQLAKLMFGASYVVSSCSTPKLKFVKQFGADK-VVDYTKTKY 206
+ V G GG+G + +++ K V+S S + + + + GAD +V K
Sbjct: 178 ESGKRLGVNGLGGLGHIAVKIGKAFGLRVTVISRSSEKEREAIDRLGADSFLVTTDSQKM 237
Query: 207 EDIEEKFDFIYDTV 220
++ DFI DTV
Sbjct: 238 KEAVGTMDFIIDTV 251
>At1g26320 allyl alcohol dehydrogenase like protein
Length = 351
Score = 43.1 bits (100), Expect = 2e-04
Identities = 32/72 (44%), Positives = 41/72 (56%), Gaps = 3/72 (4%)
Query: 136 TAIEGF-KTGDFKKGETMFVVGGAGGVGTLVLQLAKLMFGASYVVSSCSTPKLKFVK-QF 193
TA GF + KKGET+FV +G VG LV Q AKLM G V S+ S K+ +K +F
Sbjct: 148 TAYAGFYEVCSPKKGETVFVSAASGAVGQLVGQFAKLM-GCYVVGSAGSKEKVYLLKTKF 206
Query: 194 GADKVVDYTKTK 205
G D +Y + K
Sbjct: 207 GFDDAFNYKEEK 218
>At3g59845 allyl alcohol dehydrogenase-like protein
Length = 346
Score = 42.4 bits (98), Expect = 4e-04
Identities = 46/184 (25%), Positives = 80/184 (43%), Gaps = 45/184 (24%)
Query: 60 VPGCDMAGVVIGKGVNVK--KFDIGDEVYG--NIQDFNSMEKPKQLGTLAQFIVVEENLV 115
+PG +AG+ + + ++ F GD ++G N ++F+++ A ++ N+
Sbjct: 77 IPGKPIAGIGVSQVIDSDDPSFTKGDMIWGIVNWEEFSTINP-------AAIFKIDVNI- 128
Query: 116 ARKPKRLSFEEAASLPLAVQTAIEG----------FKTGDFKKGETMFVVGGAGGVGTLV 165
++PL+ T I G F+ KKG+T+FV +G VG LV
Sbjct: 129 -------------NVPLSYYTGILGMIGLTAYAGFFEICSPKKGDTVFVSAASGAVGQLV 175
Query: 166 LQLAKLMFGASYVVSSCSTPKL--KFVKQFGADKVVDYTKTKYED------IEEKFDFIY 217
Q AKLM YVV S + + + +FG D +Y + D + + D +
Sbjct: 176 GQFAKLM--GCYVVGSAGSKQKVDLLLNKFGYDDAFNYKEEPDLDSALKRCVPKGIDIYF 233
Query: 218 DTVG 221
+ VG
Sbjct: 234 ENVG 237
>At5g38000 oxidoreductase-like protein
Length = 353
Score = 41.6 bits (96), Expect = 6e-04
Identities = 27/60 (45%), Positives = 35/60 (58%), Gaps = 2/60 (3%)
Query: 147 KKGETMFVVGGAGGVGTLVLQLAKLMFGASYVVSSCSTPKLKFVK-QFGADKVVDYTKTK 205
KKGET+FV +G VG LV Q AK M G V S+ S K+ +K +FG D +Y + K
Sbjct: 162 KKGETVFVSAASGAVGQLVGQFAK-MAGCYVVGSASSEEKVDLLKTKFGYDDAFNYKEEK 220
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.139 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,962,535
Number of Sequences: 26719
Number of extensions: 352974
Number of successful extensions: 927
Number of sequences better than 10.0: 49
Number of HSP's better than 10.0 without gapping: 34
Number of HSP's successfully gapped in prelim test: 15
Number of HSP's that attempted gapping in prelim test: 847
Number of HSP's gapped (non-prelim): 68
length of query: 340
length of database: 11,318,596
effective HSP length: 100
effective length of query: 240
effective length of database: 8,646,696
effective search space: 2075207040
effective search space used: 2075207040
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC146342.2


