

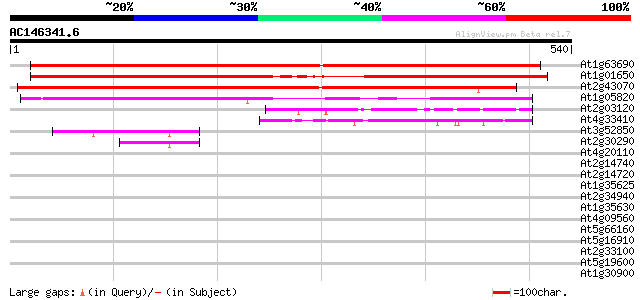
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146341.6 + phase: 0
(540 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g63690 unknown protein 743 0.0
At1g01650 unknown protein 585 e-167
At2g43070 unknown protein 419 e-117
At1g05820 unknown protein 268 6e-72
At2g03120 unknown protein 100 2e-21
At4g33410 unknown protein 100 3e-21
At3g52850 Spot 3 protein and vacuolar sorting receptor homolog/A... 48 1e-05
At2g30290 putative vacuolar sorting receptor 43 5e-04
At4g20110 vacuolar sorting receptor-like protein 41 0.001
At2g14740 putative vacuolar sorting receptor 37 0.021
At2g14720 putative vacuolar sorting receptor 37 0.021
At1g35625 integral membrane protein, putative 37 0.028
At2g34940 putative vacuolar sorting receptor 33 0.40
At1g35630 integral membrane protein like protein 33 0.40
At4g09560 putative protein 32 0.68
At5g66160 ReMembR-H2 protein JR700 (gb|AAF32325.1) 31 1.5
At5g16910 cellulose synthase catalytic subunit -like protein 31 2.0
At2g33100 putative cellulose synthase 30 2.6
At5g19600 sulfate transporter (Sultr3;5) 29 7.6
At1g30900 F17F8.23 29 7.6
>At1g63690 unknown protein
Length = 540
Score = 743 bits (1918), Expect = 0.0
Identities = 365/491 (74%), Positives = 418/491 (84%), Gaps = 2/491 (0%)
Query: 21 SLVLAGDIVHHDDVAPTRPGCENNFVLVKVPTWIDGVENAEYVGVGARFGPTLESKEKHA 80
S V AGDIVH D++AP +PGCEN+FVLVKV TWIDGVEN E+VGVGARFG + SKEK+A
Sbjct: 23 STVTAGDIVHQDNLAPKKPGCENDFVLVKVQTWIDGVENEEFVGVGARFGKRIVSKEKNA 82
Query: 81 NHTRVVMADPPDCCSKPKNKLTNEIILVHRGKCSFTTKANIADEAGASAILIINYRTELF 140
N T +V A+P D C+ KNKL+ ++++V RG C FT KAN A+ AGASA+LIIN + EL+
Sbjct: 83 NQTHLVFANPRDSCTPLKNKLSGDVVIVERGNCRFTAKANNAEAAGASALLIINNQKELY 142
Query: 141 KMVCEENETDVDIGIPAVMLPQDAGLNLERHIQNNSIVSIQLYSPLRPLVDVAEVFLWLM 200
KMVCE +ETD+DI IPAVMLPQDAG +L++ + N+S VS QLYSP RP VDVAEVFLWLM
Sbjct: 143 KMVCEPDETDLDIQIPAVMLPQDAGASLQKMLANSSKVSAQLYSPRRPAVDVAEVFLWLM 202
Query: 201 AVGTILCASYWSAWTAREAAIEQEKLLKDASDEYVAESVGSRGYVEISTTAAILFVVLAS 260
A+GTILCASYWSAW+AREAAIE +KLLKDA DE + G G VEI++ +AI FVVLAS
Sbjct: 203 AIGTILCASYWSAWSAREAAIEHDKLLKDAIDEIPNTNDGGSGVVEINSISAIFFVVLAS 262
Query: 261 CFLVMLYKLMSFWFLEVLVVLFCIGGIEGLQTCLTALLSCFRWFQYPAQTYVKIPFFGAV 320
FLV+LYKLMS+WF+E+LVV+FCIGG+EGLQTCL ALLS RWFQ A TYVK+PF G +
Sbjct: 263 GFLVILYKLMSYWFVELLVVVFCIGGVEGLQTCLVALLS--RWFQRAADTYVKVPFLGPI 320
Query: 321 PYLTLAVTPFCIVFAVVWAVKRQASYAWIGQDILGIALIITVLQIVRIPNLKVGTVLLSC 380
YLTLAV+PFCIVFAV+WAV R S+AWIGQD+LGIALIITVLQIV +PNLKVGTVLLSC
Sbjct: 321 SYLTLAVSPFCIVFAVLWAVYRVHSFAWIGQDVLGIALIITVLQIVHVPNLKVGTVLLSC 380
Query: 381 AFLYDILWVFVSKWWFHESVMIVVARGDKSGEDGIPMLLKLPRLFDPWGGYSIIGFGDII 440
AFLYDI WVFVSK FHESVMIVVARGDKSGEDGIPMLLK+PR+FDPWGGYSIIGFGDI+
Sbjct: 381 AFLYDIFWVFVSKKLFHESVMIVVARGDKSGEDGIPMLLKIPRMFDPWGGYSIIGFGDIL 440
Query: 441 LPGLVVAFSLRYDWLAKKNLRAGYFVWAMTAYGLGLLITYVALNLMDGHGQPALLYIVPF 500
LPGL++AF+LRYDWLA K LR GYF+WAM AYGLGLLITYVALNLMDGHGQPALLYIVPF
Sbjct: 441 LPGLLIAFALRYDWLANKTLRTGYFIWAMVAYGLGLLITYVALNLMDGHGQPALLYIVPF 500
Query: 501 TLGKYNHLTHK 511
TLG L K
Sbjct: 501 TLGTMLTLARK 511
>At1g01650 unknown protein
Length = 491
Score = 585 bits (1508), Expect = e-167
Identities = 310/498 (62%), Positives = 359/498 (71%), Gaps = 58/498 (11%)
Query: 21 SLVLAGDIVHHDDVAPTRPGCENNFVLVKVPTWIDGVENAEYVGVGARFGPTLESKEKHA 80
S V AGDIVHHDD P RPGC NNFVLVKVPT ++G E EYVGVGARFGPTLESKEKHA
Sbjct: 30 SFVCAGDIVHHDDSLPQRPGCNNNFVLVKVPTRVNGSEYTEYVGVGARFGPTLESKEKHA 89
Query: 81 NHTRVVMADPPDCCSKPKNKLTNEIILVHRGKCSFTTKANIADEAGASAILIINYRTELF 140
++ +ADPPDCCS PKNKLT E+ILVHRGKCSFTTK +A+ AGASAILIIN T+LF
Sbjct: 90 TLIKLAIADPPDCCSTPKNKLTGEVILVHRGKCSFTTKTKVAEAAGASAILIINNSTDLF 149
Query: 141 KMVCEENETDVDIGIPAVMLPQDAGLNLERHIQNNSIVSIQLYSPLRPLVDVAEVFLWLM 200
KMVCE+ E +DI IP VMLP DAG +LE +++N+IV++QLYSP RP VDVAEVFLWLM
Sbjct: 150 KMVCEKGENVLDITIPVVMLPVDAGRSLENIVKSNAIVTLQLYSPKRPAVDVAEVFLWLM 209
Query: 201 AVGTILCASYWSAWTAREAAIEQEKLLKDASDEYV-AESVGSRGYVEISTTAAILFVVLA 259
AVGTILCASYWSAWT RE AIEQ+KLLKD SDE + + SRG VE++ +AI
Sbjct: 210 AVGTILCASYWSAWTVREEAIEQDKLLKDGSDELLQLSTTSSRGVVEVTVISAIF----- 264
Query: 260 SCFLVMLYKLMSFWFLEVLVVLFCIGGIEGLQTCLTALLSCFRWFQYPAQTYVKIPFFGA 319
FL + L ++ V++V +GG T L RW
Sbjct: 265 GGFLFPDHALQAY----VILVYRGVGG-----TLL------HRW---------------- 293
Query: 320 VPYLTLAVTPFCIVFAVVWAVKRQASYAWIGQDILGIALIITVLQIVRIPNLKVGTVLLS 379
+R A+ + ++GI+LIITVLQIVR+PNLKVG VLLS
Sbjct: 294 ---------------------RRGAANLFGLSTLMGISLIITVLQIVRVPNLKVGFVLLS 332
Query: 380 CAFLYDILWVFVSKWWFHESVMIVVARGDKSGEDGIPMLLKLPRLFDPWGGYSIIGFGDI 439
CAF+YDI WVFVSKWWF ESVMIVVARGD+SGEDGIPMLLK+PR+FDPWGGYSIIGFGDI
Sbjct: 333 CAFMYDIFWVFVSKWWFRESVMIVVARGDRSGEDGIPMLLKIPRMFDPWGGYSIIGFGDI 392
Query: 440 ILPGLVVAFSLRYDWLAKKNLRAGYFVWAMTAYGLGLLITYVALNLMDGHGQPALLYIVP 499
ILPGL+V F+LRYDWLA K L++GYF+ M+AYGLGLLITY+ALNLMDGHGQPALLYIVP
Sbjct: 393 ILPGLLVTFALRYDWLANKRLKSGYFLGTMSAYGLGLLITYIALNLMDGHGQPALLYIVP 452
Query: 500 FTLGKYNHLTHKFESVNT 517
F LG L HK + T
Sbjct: 453 FILGTLFVLGHKRGDLKT 470
>At2g43070 unknown protein
Length = 543
Score = 419 bits (1076), Expect = e-117
Identities = 219/506 (43%), Positives = 308/506 (60%), Gaps = 27/506 (5%)
Query: 8 YSNIVAYVFLFSVSLVLAGDIVHHDDVAPTRPGCENNFVLVKVPTWIDGVENAEYVGVGA 67
YS +V + L S+ A D+ +D + PGC N F +VKV W+DGVE G+ A
Sbjct: 11 YSALVLILLLLGFSVAAADDVSWTEDSSLESPGCTNKFQMVKVLNWVDGVEGDFLTGLTA 70
Query: 68 RFGPTLESKEKHANHTRVVMADPPDCCSKPKNKLTNEIILVHRGKCSFTTKANIADEAGA 127
+FG L S A DP D CS ++L I L RG C+FT KA A+ AGA
Sbjct: 71 QFGAALPSVPDQALRFPAAFVDPLDSCSHLSSRLDGHIALSIRGNCAFTEKAKHAEAAGA 130
Query: 128 SAILIINYRTELFKMVCEENETDVDIGIPAVMLPQDAGLNLERHIQNNSIVSIQLYSPLR 187
SA+L+IN + +L +M C E +T +++ IP +M+ + +G L + + +N V + LY+P R
Sbjct: 131 SALLVINDKEDLDEMGCMEKDTSLNVSIPVLMISKSSGDALNKSMVDNKNVELLLYAPKR 190
Query: 188 PLVDVAEVFLWLMAVGTILCASYWSAWTAREAAIEQEKLL-KDASDEYVAESVGSRGYVE 246
P VD+ L LMAVGT++ AS WS T + A E +L KD S + + ++
Sbjct: 191 PAVDLTAGLLLLMAVGTVVVASLWSELTDPDQANESYSILAKDVSSAGTRKDDPEKEILD 250
Query: 247 ISTTAAILFVVLASCFLVMLYKLMSFWFLEVLVVLFCIGGIEGLQTCLTALLSCFRWFQY 306
IS T A+ F+V AS FL++L+ MS WF+ VL + FCIGG++G+ + A++ R ++
Sbjct: 251 ISVTGAVFFIVTASIFLLLLFYFMSSWFVWVLTIFFCIGGMQGMHNIIMAVI--LRKCRH 308
Query: 307 PAQTYVKIPFFGAVPYLTLAVTPFCIVFAVVWAVKRQASYAWIGQDILGIALIITVLQIV 366
A+ VK+P G + L+L V C+ FAV W +KR SY+W+GQDILGI L+IT LQ+V
Sbjct: 309 LARKSVKLPLLGTMSVLSLLVNIVCLAFAVFWFIKRHTSYSWVGQDILGICLMITALQVV 368
Query: 367 RIPNLKVGTVLLSCAFLYDILWVFVSKWWFHESVMIVVARGDKSGEDGIPMLLKLPRLFD 426
R+PN+KV TVLL CAF+YDI WVF+S FHESVMIVVA+GD S + IPMLL++PR FD
Sbjct: 369 RLPNIKVATVLLCCAFVYDIFWVFISPLIFHESVMIVVAQGDSSTGESIPMLLRIPRFFD 428
Query: 427 PWGGYSIIGFGDIILPGLVVAFS------------------------LRYDWLAKKNLRA 462
PWGGY +IGFGDI+ PGL+++F+ LRYD + K+ +
Sbjct: 429 PWGGYDMIGFGDILFPGLLISFASRVSFSTILSSNPLPRLILAPLFDLRYDKIKKRVISN 488
Query: 463 GYFVWAMTAYGLGLLITYVALNLMDG 488
GYF+W YG+GLL+TY+ L ++ G
Sbjct: 489 GYFLWLTIGYGIGLLLTYLGLAVILG 514
>At1g05820 unknown protein
Length = 441
Score = 268 bits (685), Expect = 6e-72
Identities = 166/495 (33%), Positives = 241/495 (48%), Gaps = 100/495 (20%)
Query: 11 IVAYVFLFSVSLVLAGDIVHHDDVAPTRPGCENNFVLVKVPTWIDGVENAEYVGVGARFG 70
+ A L+ + L+ G D AP PGC N F +VKV W++G + + A+FG
Sbjct: 11 LAAAAALYLIGLLCVGADTK-DVTAPKIPGCSNEFQMVKVENWVNGENGETFTAMTAQFG 69
Query: 71 PTLESKEKHANHTRVVMADPPDCCSKPKNKLTNEIILVHRGKCSFTTKANIADEAGASAI 130
L S + A V + P D CS +KL+ I L RG+C+FT KA +A GA+A+
Sbjct: 70 TMLPSDKDKAVKLPVALTTPLDSCSNLTSKLSWSIALSVRGECAFTVKAQVAQAGGAAAL 129
Query: 131 LIINYRTELFKMVCEENETDVDIGIPAVMLPQDAGLNLERHIQNNSIVSIQLYSPLRPLV 190
++IN + EL +MVC E +T +++ IP +M+ +G L++ I N V + LY+P P+V
Sbjct: 130 VLINDKEELDEMVCGEKDTSLNVSIPILMITTSSGDALKKSIMQNKKVELLLYAPKSPIV 189
Query: 191 DVAEVFLWLMAVGTILCASYWSAWTAREAAIEQEKLL--KDASDEYVAESVGSRGYVEIS 248
D A VFLWLM+VGT+ AS WS T+ + EQ L K +S+ + ++IS
Sbjct: 190 DYAVVFLWLMSVGTVFVASVWSHVTSPKKNDEQYDELSPKKSSNVDATKGGAEEETLDIS 249
Query: 249 TTAAILFVVLASCFLVMLYKLMSFWFLEVLVVLFCIGGIEGLQTCLTALLSCFRWFQYPA 308
A++ F A
Sbjct: 250 AMGAVI-------------------------------------------------FVISA 260
Query: 309 QTYVKIPFFGAVPYLTLAVTPFCIVFAVVWAVKRQASYAWIGQDILGIALIITVLQIVRI 368
T++ + FF + L +T F ++ + GI ++I VLQ+ R+
Sbjct: 261 STFLVLLFFFMSSWFILILTIFFVIGGMQ-----------------GICMMINVLQVARL 303
Query: 369 PNLKVGTVLLSCAFLYDILWVFVSKWWFHESVMIVVARGDKSGEDGIPMLLKLPRLFDPW 428
PN++V ARG K + IPMLL++PRL DPW
Sbjct: 304 PNIRV-------------------------------ARGSKDTGESIPMLLRIPRLSDPW 332
Query: 429 GGYSIIGFGDIILPGLVVAFSLRYDWLAKKNLRAGYFVWAMTAYGLGLLITYVALNLMDG 488
GGY++IGFGDI+ PGL++ F R+D K + GYF W M YGLGL +TY+ L +M+G
Sbjct: 333 GGYNMIGFGDILFPGLLICFIFRFDKENNKGVSNGYFPWLMFGYGLGLFLTYLGLYVMNG 392
Query: 489 HGQPALLYIVPFTLG 503
HGQPALLY+VP TLG
Sbjct: 393 HGQPALLYLVPCTLG 407
>At2g03120 unknown protein
Length = 344
Score = 100 bits (250), Expect = 2e-21
Identities = 80/270 (29%), Positives = 129/270 (47%), Gaps = 36/270 (13%)
Query: 247 ISTTAAILFVVLASCFLVMLYKLMSFWFLE----VLVVLFCIGGIEGLQTCLTALLSCFR 302
+S A+ F ++ S L+ L+ L F + VL F + GI L L + F
Sbjct: 55 MSKEHAMRFPLVGSAMLLSLFLLFKFLSKDLVNAVLTAYFFVLGIVALSATLLPAIRRFL 114
Query: 303 ---W------FQYPAQTYVKIPFFGAVPYLTLAVTPFCIVFAVVWAVKRQASYAWIGQDI 353
W +++P +++ F + + T FC +A W W+ +I
Sbjct: 115 PNPWNDNLIVWRFPYFKSLEVEFTKSQVVAGIPGTFFCAWYA--WKKH------WLANNI 166
Query: 354 LGIALIITVLQIVRIPNLKVGTVLLSCAFLYDILWVFVSKWWFHESVMIVVARGDKSGED 413
LG++ I ++++ + + K G +LL+ F YDI WVF + VM+ VA KS +
Sbjct: 167 LGLSFCIQGIEMLSLGSFKTGAILLAGLFFYDIFWVFFT------PVMVSVA---KSFDA 217
Query: 414 GIPMLLKLPRLFDPWGGYSIIGFGDIILPGLVVAFSLRYDWLAKKNLRAGYFVWAMTAYG 473
I +L P YS++G GDI++PG+ VA +LR+D + + YF A Y
Sbjct: 218 PIKLLFPTGDALRP---YSMLGLGDIVIPGIFVALALRFD--VSRRRQPQYFTSAFIGYA 272
Query: 474 LGLLITYVALNLMDGHGQPALLYIVPFTLG 503
+G+++T V +N QPALLYIVP +G
Sbjct: 273 VGVILTIVVMNWFQA-AQPALLYIVPAVIG 301
>At4g33410 unknown protein
Length = 372
Score = 99.8 bits (247), Expect = 3e-21
Identities = 83/310 (26%), Positives = 145/310 (46%), Gaps = 64/310 (20%)
Query: 241 SRGYVEISTTAAILFVVLASCFLVMLYKLMSFWFLEVLVVLFCIGGIEGLQTCLTALLSC 300
S + + ++ A++ V++SC L++++ L S + L+ F T + ++ S
Sbjct: 47 SEASITLDSSQALMIPVMSSCSLLLMFYLFSS--VSQLLTAF---------TAIASVSSL 95
Query: 301 FRWFQYPAQTYVKIPFFGAVPYLTLAVTPF-----------CIVFAVVWAVKRQASYAWI 349
F W P Y+K + P+L+ + C + V W + S W+
Sbjct: 96 FYWLS-PYAVYMKTQLGLSDPFLSRCCSKSFTRIQGLLLVACAMTVVAWLI----SGHWV 150
Query: 350 GQDILGIALIITVLQIVRIPNLKVGTVLLSCAFLYDILWVFVSKWWFHESVMIVVARGDK 409
++LGI++ I + VR+PN+K+ +LL C F+YDI WVF S+ +F +VM+ VA
Sbjct: 151 LNNLLGISICIAFVSHVRLPNIKICAMLLVCLFVYDIFWVFFSERFFGANVMVAVATQQA 210
Query: 410 S-------------GEDGIPMLLKLP-RLFDP---WGG---------YSIIGFGDIILPG 443
S G I L+LP ++ P GG + ++G GD+ +P
Sbjct: 211 SNPVHTVANSLNLPGLQLITKKLELPVKIVFPRNLLGGVVPGVSASDFMMLGLGDMAIPA 270
Query: 444 LVVAFSLRYDW---------LAKKNLRAGYFVW-AMTAYGLGLLITYVALNLMDGHGQPA 493
+++A L +D K+ + ++W A+ Y +G L+ +A ++ QPA
Sbjct: 271 MLLALVLCFDHRKTRDVVNIFDLKSSKGHKYIWYALPGYAIG-LVAALAAGVLTHSPQPA 329
Query: 494 LLYIVPFTLG 503
LLY+VP TLG
Sbjct: 330 LLYLVPSTLG 339
>At3g52850 Spot 3 protein and vacuolar sorting receptor
homolog/AtELP1
Length = 623
Score = 48.1 bits (113), Expect = 1e-05
Identities = 43/150 (28%), Positives = 71/150 (46%), Gaps = 9/150 (6%)
Query: 42 ENNFVLVKVPTWIDGVENAEYVGVGA-RFGPTLESKEKH--ANHTRVVMADPPDCCSKPK 98
E N + V P I G+ G ++G TL + +N D K K
Sbjct: 24 EKNNLKVTSPDSIKGIYECAIGNFGVPQYGGTLVGTVVYPKSNQKACKSYSDFDISFKSK 83
Query: 99 NKLTNEIILVHRGKCSFTTKANIADEAGASAILIINYRTE-LFKM-VCEENETDVD---- 152
+L+ RG C FT KA IA +AGA+AIL+ + + E L M EE+++D D
Sbjct: 84 PGRLPTFVLIDRGDCYFTLKAWIAQQAGAAAILVADSKAEPLITMDTPEEDKSDADYLQN 143
Query: 153 IGIPAVMLPQDAGLNLERHIQNNSIVSIQL 182
I IP+ ++ + G +++ + +V+++L
Sbjct: 144 ITIPSALITKTLGDSIKSALSGGDMVNMKL 173
>At2g30290 putative vacuolar sorting receptor
Length = 625
Score = 42.7 bits (99), Expect = 5e-04
Identities = 32/83 (38%), Positives = 44/83 (52%), Gaps = 6/83 (7%)
Query: 106 ILVHRGKCSFTTKANIADEAGASAILIINYRTE-LFKMVCEENET-DVD----IGIPAVM 159
+LV RG C FT KA A AGA+ IL+ + R E L M E+ET D D I IP+ +
Sbjct: 95 VLVDRGDCYFTLKAWNAQRAGAATILVADNRPEQLITMDAPEDETSDADYLQNITIPSAL 154
Query: 160 LPQDAGLNLERHIQNNSIVSIQL 182
+ + G ++ I + V I L
Sbjct: 155 VSRSLGSAIKTAIAHGDPVHISL 177
>At4g20110 vacuolar sorting receptor-like protein
Length = 625
Score = 41.2 bits (95), Expect = 0.001
Identities = 28/84 (33%), Positives = 43/84 (50%), Gaps = 6/84 (7%)
Query: 105 IILVHRGKCSFTTKANIADEAGASAILII-NYRTELFKMVCEENETDVD-----IGIPAV 158
I+L+ RG C F KA A +AGA+A+L+ N L M E D D + IP+V
Sbjct: 93 ILLLDRGGCYFALKAWHAQQAGAAAVLVADNVDEPLLTMDSPEESKDADGFIEKLTIPSV 152
Query: 159 MLPQDAGLNLERHIQNNSIVSIQL 182
++ + G +L + Q + I+L
Sbjct: 153 LIDKSFGDDLRQGFQKGKNIVIKL 176
>At2g14740 putative vacuolar sorting receptor
Length = 628
Score = 37.4 bits (85), Expect = 0.021
Identities = 26/83 (31%), Positives = 41/83 (49%), Gaps = 6/83 (7%)
Query: 106 ILVHRGKCSFTTKANIADEAGASAILII-NYRTELFKMVCEENETD-----VDIGIPAVM 159
+LV RG C F K A +AGASA+L+ N L M E + +I IP+ +
Sbjct: 96 LLVDRGDCFFALKVWNAQKAGASAVLVADNVDEPLITMDTPEEDVSSAKYIENITIPSAL 155
Query: 160 LPQDAGLNLERHIQNNSIVSIQL 182
+ + G L++ I +V++ L
Sbjct: 156 VTKGFGEKLKKAISGGDMVNLNL 178
>At2g14720 putative vacuolar sorting receptor
Length = 628
Score = 37.4 bits (85), Expect = 0.021
Identities = 26/83 (31%), Positives = 41/83 (49%), Gaps = 6/83 (7%)
Query: 106 ILVHRGKCSFTTKANIADEAGASAILII-NYRTELFKMVCEENETD-----VDIGIPAVM 159
+LV RG C F K A +AGASA+L+ N L M E + +I IP+ +
Sbjct: 96 LLVDRGDCFFALKVWNAQKAGASAVLVADNVDEPLITMDTPEEDVSSAKYIENITIPSAL 155
Query: 160 LPQDAGLNLERHIQNNSIVSIQL 182
+ + G L++ I +V++ L
Sbjct: 156 VTKGFGEKLKKAISGGDMVNLNL 178
>At1g35625 integral membrane protein, putative
Length = 279
Score = 37.0 bits (84), Expect = 0.028
Identities = 36/132 (27%), Positives = 61/132 (45%), Gaps = 12/132 (9%)
Query: 88 ADPPDCCSKPKN------KLTNEIILVHRGKCSFTTKANIADEAGASAILIINYRTELFK 141
A+P D CS N K +L+ RG CSF K A EAG A ++ N R E +
Sbjct: 19 AEPLDACSYLTNMAEKGSKFRPSYVLIVRGGCSFEEKIRNAQEAGYKAAIVYNDRYE--E 76
Query: 142 MVCEENETDVDIGIPAVMLPQDAGLNLERHIQNNSIVSIQLYS-PLRPLVDVAEVFLWLM 200
++ N + V I V++ + +G L+ + + + + + +A F+ L+
Sbjct: 77 LLVRRNSSGV--YIHGVLVTRTSGEVLKEYTSRAEMELLLIPGFGISSWSIMAITFVSLL 134
Query: 201 AVGTILCASYWS 212
+ +L ASY+S
Sbjct: 135 VISAVL-ASYFS 145
>At2g34940 putative vacuolar sorting receptor
Length = 618
Score = 33.1 bits (74), Expect = 0.40
Identities = 19/83 (22%), Positives = 44/83 (52%), Gaps = 4/83 (4%)
Query: 104 EIILVHRGKCSFTTKANIADEAGASAILIINYRTELFKMVCEENETDVD----IGIPAVM 159
+I+L+ RG C+F K ++GA+A+L+ + E + + D D + IP+ +
Sbjct: 92 KILLIDRGVCNFALKIWNGQQSGAAAVLLADNIVEPLITMDTPQDEDPDFIDKVKIPSAL 151
Query: 160 LPQDAGLNLERHIQNNSIVSIQL 182
+ + G +L++ ++ V +++
Sbjct: 152 ILRSFGDSLKKALKRGEEVILKM 174
>At1g35630 integral membrane protein like protein
Length = 318
Score = 33.1 bits (74), Expect = 0.40
Identities = 20/73 (27%), Positives = 35/73 (47%), Gaps = 6/73 (8%)
Query: 87 MADPPDCCS------KPKNKLTNEIILVHRGKCSFTTKANIADEAGASAILIINYRTELF 140
+A+P + CS + ++K + +L+ G CSF K A +AG A ++ N +
Sbjct: 55 VAEPLEACSDITNMAEKRSKYRSSYVLIVLGGCSFEEKVRKAQKAGYKAAIVYNDGYDEL 114
Query: 141 KMVCEENETDVDI 153
+ N + VDI
Sbjct: 115 LVPMAGNSSGVDI 127
>At4g09560 putative protein
Length = 431
Score = 32.3 bits (72), Expect = 0.68
Identities = 22/79 (27%), Positives = 37/79 (45%), Gaps = 11/79 (13%)
Query: 65 VGARFGPTLESKEKHANHTRVVMADPPDCCS--------KPKNKLTNEIILVHRGKCSFT 116
V A F P ++ ++ + +A+P D CS K ++ +L+ RG CSF
Sbjct: 36 VEATFTPMIKRSDQGGV---LYVAEPLDACSDLVNTVNVKNGTTVSPPYVLIIRGGCSFE 92
Query: 117 TKANIADEAGASAILIINY 135
K A +AG A ++ +Y
Sbjct: 93 DKIRNAQKAGYKAAIVYDY 111
>At5g66160 ReMembR-H2 protein JR700 (gb|AAF32325.1)
Length = 310
Score = 31.2 bits (69), Expect = 1.5
Identities = 26/93 (27%), Positives = 42/93 (44%), Gaps = 10/93 (10%)
Query: 87 MADPPDCCSKPKNKL--------TNEIILVHRGKCSFTTKANIADEAGASAILIINYRTE 138
+ADP D CS + T + L+ RG+CSF K A +G A+++ +
Sbjct: 58 VADPLDGCSPLLHAAASNWTQHRTTKFALIIRGECSFEDKLLNAQNSGFQAVIVYDNIDN 117
Query: 139 LFKMVCEENETDVDIGIPAVMLPQDAGLNLERH 171
+V + N DI + AV + AG L ++
Sbjct: 118 EDLIVMKVNPQ--DITVDAVFVSNVAGEILRKY 148
>At5g16910 cellulose synthase catalytic subunit -like protein
Length = 1145
Score = 30.8 bits (68), Expect = 2.0
Identities = 20/93 (21%), Positives = 41/93 (43%), Gaps = 9/93 (9%)
Query: 313 KIPFFGAVPYLTLAVTPFCIVFAVVWAVKRQASYAWIGQDILGIALIITVLQIVRIPNLK 372
K+ + YL + + PF +F +V+ S + GQ I+ L + + L
Sbjct: 910 KMKILQRIAYLNVGIYPFTSIFLIVYCFLPALS-LFSGQ------FIVQTLNVTFLVYLL 962
Query: 373 VGTVLLSCAFLYDILW--VFVSKWWFHESVMIV 403
+ ++ L L +I W + + +WW +E ++
Sbjct: 963 IISITLCLLALLEIKWSGISLEEWWRNEQFWLI 995
>At2g33100 putative cellulose synthase
Length = 1036
Score = 30.4 bits (67), Expect = 2.6
Identities = 23/93 (24%), Positives = 42/93 (44%), Gaps = 9/93 (9%)
Query: 313 KIPFFGAVPYLTLAVTPFCIVFAVVWAVKRQASYAWIGQDILGIALIITVLQIVRIPNLK 372
++ F V YL + + PF +F VV+ A + G+ I+ L I + L
Sbjct: 803 RLKFLQRVAYLNVGIYPFTSIFLVVYCF-LPALCLFSGK------FIVQSLDIHFLSYLL 855
Query: 373 VGTVLLSCAFLYDILW--VFVSKWWFHESVMIV 403
TV L+ L ++ W + + +WW +E ++
Sbjct: 856 CITVTLTLISLLEVKWSGIGLEEWWRNEQFWLI 888
>At5g19600 sulfate transporter (Sultr3;5)
Length = 634
Score = 28.9 bits (63), Expect = 7.6
Identities = 31/128 (24%), Positives = 53/128 (41%), Gaps = 20/128 (15%)
Query: 415 IPMLLKLPRLFDPWGGYSIIGFGDIILPGLVVAFS-----------------LRYDWLAK 457
IP ++ L F P Y++ G + + G V A S L +
Sbjct: 105 IPPIIGLYSSFVPPFVYAVFGSSNNLAVGTVAACSLLIAETFGEEMIKNEPELYLHLIFT 164
Query: 458 KNLRAGYFVWAMTAYGLGLLITYVALNLMDGH-GQPALLYIVPFTLGKYN--HLTHKFES 514
L G F +AM LG+L+ +++ + + G G A++ ++ G + H THK +
Sbjct: 165 ATLITGLFQFAMGFLRLGILVDFLSHSTITGFMGGTAIIILLQQLKGIFGLVHFTHKTDV 224
Query: 515 VNTYGGIL 522
V+ IL
Sbjct: 225 VSVLHSIL 232
>At1g30900 F17F8.23
Length = 649
Score = 28.9 bits (63), Expect = 7.6
Identities = 16/58 (27%), Positives = 28/58 (47%), Gaps = 1/58 (1%)
Query: 105 IILVHRGKCSFTTKANIADEAGASAILII-NYRTELFKMVCEENETDVDIGIPAVMLP 161
I+++ RG+C F K ++G +A+L+ N L M E + D I + +P
Sbjct: 92 ILIIDRGECYFALKVWNGQQSGVAAVLVADNVDEPLITMDSPEESKEADDFIEKLNIP 149
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.327 0.142 0.447
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,834,892
Number of Sequences: 26719
Number of extensions: 495248
Number of successful extensions: 1404
Number of sequences better than 10.0: 23
Number of HSP's better than 10.0 without gapping: 14
Number of HSP's successfully gapped in prelim test: 9
Number of HSP's that attempted gapping in prelim test: 1376
Number of HSP's gapped (non-prelim): 29
length of query: 540
length of database: 11,318,596
effective HSP length: 104
effective length of query: 436
effective length of database: 8,539,820
effective search space: 3723361520
effective search space used: 3723361520
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 62 (28.5 bits)
Medicago: description of AC146341.6


