

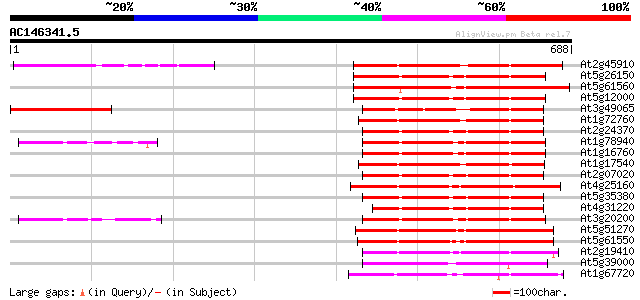
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146341.5 + phase: 0 /pseudo
(688 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g45910 protein kinase- like 293 3e-79
At5g26150 putative protein 218 1e-56
At5g61560 putative protein 212 5e-55
At5g12000 putative receptor - like kinase 212 6e-55
At3g49065 unknown protein 211 8e-55
At1g72760 putative protein kinase 207 2e-53
At2g24370 putative protein kinase 205 8e-53
At1g78940 unknown protein 204 2e-52
At1g16760 hypothetical protein 203 2e-52
At1g17540 protein kinase, putative 201 1e-51
At2g07020 putative protein kinase 201 1e-51
At4g25160 putative Ser/Thr protein kinase 197 1e-50
At5g35380 putative protein 196 4e-50
At4g31220 protein kinase-like protein 195 8e-50
At3g20200 protein kinase like protein 192 5e-49
At5g51270 putative protein 190 2e-48
At5g61550 putative protein 190 3e-48
At2g19410 putative protein kinase 180 3e-45
At5g39000 protein kinase - like protein 155 7e-38
At1g67720 putative receptor protein kinase 151 1e-36
>At2g45910 protein kinase- like
Length = 834
Score = 293 bits (749), Expect = 3e-79
Identities = 148/258 (57%), Positives = 189/258 (72%), Gaps = 15/258 (5%)
Query: 422 DPQNFNRSLSYQ--VEVLSKLKHPNLITLIGVNQESKTLIYEYLPNGSLEDHLSRNGNNN 479
+P + + YQ V+VLSK++HPN+ITLIG E +L+YEYLP GSLED L+ +N
Sbjct: 511 NPNSSQGPVEYQQEVDVLSKMRHPNIITLIGACPEGWSLVYEYLPGGSLEDRLTCK--DN 568
Query: 480 APPLTWQTRIRIATELCSALIFLHSNKPHSIVHGDLKPSNILLDANLVTKLSDFGICRVL 539
+PPL+WQ R+RIATE+C+AL+FLHSNK HS+VHGDLKP+NILLD+NLV+KLSDFG C +L
Sbjct: 569 SPPLSWQNRVRIATEICAALVFLHSNKAHSLVHGDLKPANILLDSNLVSKLSDFGTCSLL 628
Query: 540 SCQNDSSSNNSTTQFWITSFAKGTFAYMDPEFLGTGELTSKSDVYSFGIVLLRLITGKPA 599
S T GT AY+DPE +GELT KSDVYSFGI+LLRL+TG+PA
Sbjct: 629 HPNGSKSVRTDVT---------GTVAYLDPEASSSGELTPKSDVYSFGIILLRLLTGRPA 679
Query: 600 LGIKNEVLYALNNAGGNVKSVLDPLAGDWPIVEAEKLVHFALRCCDMNKKSRPELCSEGW 659
L I NEV YAL+N G + +LDPLAGDWP V+AE+L ALRCC+ ++RP+L +E W
Sbjct: 680 LRISNEVKYALDN--GTLNDLLDPLAGDWPFVQAEQLARLALRCCETVSENRPDLGTEVW 737
Query: 660 RVLEPMKVSCSGTNNFGL 677
RVLEPM+ S G+++F L
Sbjct: 738 RVLEPMRASSGGSSSFHL 755
Score = 151 bits (382), Expect = 1e-36
Identities = 95/247 (38%), Positives = 146/247 (58%), Gaps = 15/247 (6%)
Query: 5 VDVYLAICQRMGVTATKLHIEMDCIEKGIIELISRYNIQNLVMGAASDKYHSRRMTDLRS 64
+D YL ICQ+ GV A K+ IEM+ IE GI++LIS I+ LVMGAA+D+++SRRMTDL+S
Sbjct: 118 LDDYLRICQQRGVRAEKMFIEMESIENGIVQLISELGIRKLVMGAAADRHYSRRMTDLKS 177
Query: 65 KKAIYVCEQSPASCHIQFICKGYLIQTRDCSLENFRVEATSPLVQQMQNSEVGHSPHLIS 124
+KAI+V ++P C I F CKGYLI TR+ ++++ E SP + S L +
Sbjct: 178 RKAIFVRREAPTLCQIWFTCKGYLIHTREATMDDTESEYASP------RPSISASDLLQT 231
Query: 125 RSISHDQELHHHRVRSISSASESGRSMASSVSSSERFSSFETVLTPNLTSDGNESLLDLK 184
S + H RV+S ++S + + S+ SS+E+ V +L +D E D
Sbjct: 232 FSTPESEHQHISRVQS----TDSVQQLVSNGSSTEQSG---RVSDGSLNTDEEERESD-G 283
Query: 185 MSYLSSIKEENLCHSSPPGVLDRGMDESVYDQLEQAIAEAVKARWDAYQETVKRRKAEKD 244
S + HSSP D G+D+S ++ +A +EA ++ +A+ ET++R+KAEK+
Sbjct: 284 SEVTGSATVMSSGHSSPSSFPD-GVDDSFNVKIRKATSEAHSSKQEAFAETLRRQKAEKN 342
Query: 245 VIDTIRK 251
+D IR+
Sbjct: 343 ALDAIRR 349
>At5g26150 putative protein
Length = 703
Score = 218 bits (554), Expect = 1e-56
Identities = 110/236 (46%), Positives = 158/236 (66%), Gaps = 9/236 (3%)
Query: 422 DPQNFNRSLSYQVEVLSKLKHPNLITLIGVNQESKTLIYEYLPNGSLEDHLSRNGNNNAP 481
D + +VEVL ++HP+++ L+G E L+YE++ NGSLED L R GN+ P
Sbjct: 454 DAAQGKKQFQQEVEVLCSIRHPHMVLLLGACPEYGCLVYEFMENGSLEDRLFRTGNS--P 511
Query: 482 PLTWQTRIRIATELCSALIFLHSNKPHSIVHGDLKPSNILLDANLVTKLSDFGICRVLSC 541
PL+W+ R IA E+ +AL FLH KP +VH DLKP+NILLD N V+K+SD G+ R++
Sbjct: 512 PLSWRKRFEIAAEIATALSFLHQAKPEPLVHRDLKPANILLDKNYVSKISDVGLARLV-- 569
Query: 542 QNDSSSNNSTTQFWITSFAKGTFAYMDPEFLGTGELTSKSDVYSFGIVLLRLITGKPALG 601
+S +S TQF +TS A GTF Y+DPE+ TG LT+KSDVYS GI+LL++ITG+P +G
Sbjct: 570 --PASIADSVTQFHMTS-AAGTFCYIDPEYQQTGMLTTKSDVYSLGILLLQIITGRPPMG 626
Query: 602 IKNEVLYALNNAGGNVKSVLDPLAGDWPIVEAEKLVHFALRCCDMNKKSRPELCSE 657
+ ++V A++ G K +LDP+ DWP+ EA+ AL+C ++ K+ RP+L E
Sbjct: 627 LAHQVSRAISK--GTFKEMLDPVVPDWPVQEAQSFATLALKCAELRKRDRPDLGKE 680
Score = 33.1 bits (74), Expect = 0.53
Identities = 31/149 (20%), Positives = 64/149 (42%), Gaps = 13/149 (8%)
Query: 8 YLAICQRMGVTATKLHIEMDCIEKGIIELISRYNIQNLVMGAASDKYHSRR-MTDLRSKK 66
Y C R GV ++ + K I++ ++ + NLV+GA++ +R M +
Sbjct: 75 YRGYCARKGVV-----LDDSDVAKTILDYVNNNLVNNLVLGASTKNTFARSFMFSKPHEV 129
Query: 67 AIYVCEQSPASCHIQFICKGYLIQTR-------DCSLENFRVEATSPLVQQMQNSEVGHS 119
+ + +P C + I KG +Q+ +L RV ++ L+Q + +SE
Sbjct: 130 QSSIMKSTPDFCSVYVISKGGKVQSSRPAQRPITNTLAPPRVPSSGFLIQSLSDSEQDLI 189
Query: 120 PHLISRSISHDQELHHHRVRSISSASESG 148
P + + + E + R+ + ++ G
Sbjct: 190 PRVQRSARNKPNETTYPHNRAAFNTTQKG 218
>At5g61560 putative protein
Length = 815
Score = 212 bits (540), Expect = 5e-55
Identities = 111/269 (41%), Positives = 167/269 (61%), Gaps = 9/269 (3%)
Query: 422 DPQNFNRSLSYQVEVLSKLKHPNLITLIGVNQESKTLIYEYLPNGSLEDHLSRNGNN--- 478
D + + ++E+LSK++HP+L+ L+G E +L+YEY+ NGSLE+ L + N
Sbjct: 480 DKSSLTKQFHQELEILSKIRHPHLLLLLGACPERGSLVYEYMHNGSLEERLMKRRPNVDT 539
Query: 479 -NAPPLTWQTRIRIATELCSALIFLHSNKPHSIVHGDLKPSNILLDANLVTKLSDFGICR 537
PPL W R RIA E+ SAL FLH+N+P IVH DLKP+NILLD N V+K+ D G+ +
Sbjct: 540 PQPPPLRWFERFRIAWEIASALYFLHTNEPRPIVHRDLKPANILLDRNNVSKIGDVGLSK 599
Query: 538 VLSCQNDSSSNNSTTQFWITSFAKGTFAYMDPEFLGTGELTSKSDVYSFGIVLLRLITGK 597
+++ ++++T F T GTF Y+DPE+ TG +T +SD+Y+FGI+LL+L+T +
Sbjct: 600 MVNLD----PSHASTVFNETG-PVGTFFYIDPEYQRTGVVTPESDIYAFGIILLQLVTAR 654
Query: 598 PALGIKNEVLYALNNAGGNVKSVLDPLAGDWPIVEAEKLVHFALRCCDMNKKSRPELCSE 657
A+G+ + + AL + G +LD AGDWP+ EA+++V LRC +M K+ RP+L E
Sbjct: 655 SAMGLAHSIEKALRDQTGKFTEILDKTAGDWPVKEAKEMVMIGLRCAEMRKRDRPDLGKE 714
Query: 658 GWRVLEPMKVSCSGTNNFGLKSCLDKQLN 686
VLE +K S N + +D N
Sbjct: 715 ILPVLERLKEVASIARNMFADNLIDHHHN 743
>At5g12000 putative receptor - like kinase
Length = 703
Score = 212 bits (539), Expect = 6e-55
Identities = 107/236 (45%), Positives = 156/236 (65%), Gaps = 9/236 (3%)
Query: 422 DPQNFNRSLSYQVEVLSKLKHPNLITLIGVNQESKTLIYEYLPNGSLEDHLSRNGNNNAP 481
D + +VEVLS ++HP+++ L+G E L+YE++ NGSLED L R GN+ P
Sbjct: 454 DAAQGKKQFQQEVEVLSSIRHPHMVLLLGACPEYGCLVYEFMDNGSLEDRLFRRGNS--P 511
Query: 482 PLTWQTRIRIATELCSALIFLHSNKPHSIVHGDLKPSNILLDANLVTKLSDFGICRVLSC 541
PL+W+ R +IA E+ +AL FLH KP +VH DLKP+NILLD N V+K+SD G+ R++
Sbjct: 512 PLSWRKRFQIAAEIATALSFLHQAKPEPLVHRDLKPANILLDKNYVSKISDVGLARLV-- 569
Query: 542 QNDSSSNNSTTQFWITSFAKGTFAYMDPEFLGTGELTSKSDVYSFGIVLLRLITGKPALG 601
+S N+ TQ+ +TS A GTF Y+DPE+ TG+LT+KSD++S GI+LL++IT K +G
Sbjct: 570 --PASVANTVTQYHMTS-AAGTFCYIDPEYQQTGKLTTKSDIFSLGIMLLQIITAKSPMG 626
Query: 602 IKNEVLYALNNAGGNVKSVLDPLAGDWPIVEAEKLVHFALRCCDMNKKSRPELCSE 657
+ + V A++ G K +LDP+ DWP+ EA LRC ++ K+ RP+L E
Sbjct: 627 LAHHVSRAIDK--GTFKDMLDPVVPDWPVEEALNFAKLCLRCAELRKRDRPDLGKE 680
Score = 37.7 bits (86), Expect = 0.022
Identities = 20/86 (23%), Positives = 44/86 (50%), Gaps = 1/86 (1%)
Query: 8 YLAICQRMGVTATKLHIEMDCIEKGIIELISRYNIQNLVMGAASDKYHSRRMTDLRSKK- 66
Y C R G++ ++ ++ + K +++ ++ + NLV+G++S +R + +S
Sbjct: 75 YRGYCARKGISMMEVILDDTDVSKAVLDYVNNNLVTNLVLGSSSKSPFARSLKFTKSHDV 134
Query: 67 AIYVCEQSPASCHIQFICKGYLIQTR 92
A V + +P C + I KG + +R
Sbjct: 135 ASSVLKSTPEFCSVYVISKGKVHSSR 160
>At3g49065 unknown protein
Length = 805
Score = 211 bits (538), Expect = 8e-55
Identities = 106/222 (47%), Positives = 154/222 (68%), Gaps = 21/222 (9%)
Query: 433 QVEVLSKLKHPNLITLIGVNQESKTLIYEYLPNGSLEDHLSRNGNNNAPPLTWQTRIRIA 492
+VE+LS+++HPNL+TL+G ES++LIY+Y+PNGSLED S NN P L+W++RIRIA
Sbjct: 503 RVEILSRVRHPNLVTLMGACPESRSLIYQYIPNGSLEDCFS--SENNVPALSWESRIRIA 560
Query: 493 TELCSALIFLHSNKPHSIVHGDLKPSNILLDANLVTKLSDFGICRVLSCQNDSSSNNSTT 552
+E+CSAL+FLHSN P I+HG+LKPS ILLD+NLVTK++D+GI +++ S+
Sbjct: 561 SEICSALLFLHSNIP-CIIHGNLKPSKILLDSNLVTKINDYGISQLIPIDGLDKSD---- 615
Query: 553 QFWITSFAKGTFAYMDPEFLGTGELTSKSDVYSFGIVLLRLITGKPALGIKNEVLYALNN 612
++DP + + E+T +SD+Y+FGI+LL+L+T +P GI +V AL N
Sbjct: 616 ------------PHVDPHYFVSREMTLESDIYAFGIILLQLLTRRPVSGILRDVKCALEN 663
Query: 613 AGGNVKSVLDPLAGDWPIVEAEKLVHFALRCCDMNKKSRPEL 654
N+ +VLD AGDWP+ +KL + A+RCC N +RP+L
Sbjct: 664 --DNISAVLDNSAGDWPVARGKKLANVAIRCCKKNPMNRPDL 703
Score = 103 bits (256), Expect = 4e-22
Identities = 56/124 (45%), Positives = 82/124 (65%)
Query: 1 MQKTVDVYLAICQRMGVTATKLHIEMDCIEKGIIELISRYNIQNLVMGAASDKYHSRRMT 60
+ + ++ YL + + KL I IE+ I+ELI+R+ I+ LVMGAASDK++S +MT
Sbjct: 84 VDELMNSYLQLLSETEIQTDKLCIAGQNIEECIVELIARHKIKWLVMGAASDKHYSWKMT 143
Query: 61 DLRSKKAIYVCEQSPASCHIQFICKGYLIQTRDCSLENFRVEATSPLVQQMQNSEVGHSP 120
DL+SKKAI+VC+++P SCHI F+CKGYLI TR + ++ + PLVQ ++E S
Sbjct: 144 DLKSKKAIFVCKKAPDSCHIWFLCKGYLIFTRASNDDSNNRQTMPPLVQLDSDNETRKSE 203
Query: 121 HLIS 124
L S
Sbjct: 204 KLES 207
>At1g72760 putative protein kinase
Length = 697
Score = 207 bits (527), Expect = 2e-53
Identities = 104/228 (45%), Positives = 150/228 (65%), Gaps = 11/228 (4%)
Query: 428 RSLSYQVEVLSKLKHPNLITLIGVNQESKTLIYEYLPNGSLEDHLSRNGNNNAPPLTWQT 487
+ ++EVLS ++HPN++ L+G E L+YEY+ NG+LED L NN PPL+W+
Sbjct: 420 KQFQQEIEVLSSMRHPNMVILLGACPEYGCLVYEYMENGTLEDRLFCK--NNTPPLSWRA 477
Query: 488 RIRIATELCSALIFLHSNKPHSIVHGDLKPSNILLDANLVTKLSDFGICRVL-SCQNDSS 546
R RIA+E+ + L+FLH KP +VH DLKP+NILLD +L K+SD G+ R++ D+
Sbjct: 478 RFRIASEIATGLLFLHQAKPEPLVHRDLKPANILLDKHLTCKISDVGLARLVPPAVADTY 537
Query: 547 SNNSTTQFWITSFAKGTFAYMDPEFLGTGELTSKSDVYSFGIVLLRLITGKPALGIKNEV 606
SN T A GTF Y+DPE+ TG L KSD+YSFG+VLL++IT +PA+G+ ++V
Sbjct: 538 SNYHMTS------AAGTFCYIDPEYQQTGMLGVKSDLYSFGVVLLQIITAQPAMGLGHKV 591
Query: 607 LYALNNAGGNVKSVLDPLAGDWPIVEAEKLVHFALRCCDMNKKSRPEL 654
A+ N N++ +LDP +WP E +L AL+CC++ KK RP+L
Sbjct: 592 EMAVEN--NNLREILDPTVSEWPEEETLELAKLALQCCELRKKDRPDL 637
>At2g24370 putative protein kinase
Length = 816
Score = 205 bits (521), Expect = 8e-53
Identities = 105/222 (47%), Positives = 149/222 (66%), Gaps = 9/222 (4%)
Query: 433 QVEVLSKLKHPNLITLIGVNQESKTLIYEYLPNGSLEDHLSRNGNNNAPPLTWQTRIRIA 492
+VEVLS ++HPN++ L+G E L+YE++ NGSLED L R GN+ PPL+WQ R RIA
Sbjct: 523 EVEVLSCIRHPNMVLLLGACPECGCLVYEFMANGSLEDRLFRLGNS--PPLSWQMRFRIA 580
Query: 493 TELCSALIFLHSNKPHSIVHGDLKPSNILLDANLVTKLSDFGICRVLSCQNDSSSNNSTT 552
E+ + L+FLH KP +VH DLKP NILLD N V+K+SD G+ R++ + ++ T
Sbjct: 581 AEIGTGLLFLHQAKPEPLVHRDLKPGNILLDRNFVSKISDVGLARLV----PPTVADTVT 636
Query: 553 QFWITSFAKGTFAYMDPEFLGTGELTSKSDVYSFGIVLLRLITGKPALGIKNEVLYALNN 612
Q+ +TS A GTF Y+DPE+ TG L KSD+YS GI+ L+LIT KP +G+ + V AL
Sbjct: 637 QYRMTSTA-GTFCYIDPEYQQTGMLGVKSDIYSLGIMFLQLITAKPPMGLTHYVERALEK 695
Query: 613 AGGNVKSVLDPLAGDWPIVEAEKLVHFALRCCDMNKKSRPEL 654
G + +LDP+ DWP+ + E+ AL+C ++ +K RP+L
Sbjct: 696 --GTLVDLLDPVVSDWPMEDTEEFAKLALKCAELRRKDRPDL 735
Score = 32.3 bits (72), Expect = 0.91
Identities = 39/191 (20%), Positives = 79/191 (40%), Gaps = 15/191 (7%)
Query: 8 YLAICQRMGVTATKLHIEMDCIEKGIIELISRYNIQNLVMGAASDKYHSR--RMTDLRSK 65
+ C R + + +E + K ++E +++ I+ LV+G++S R + TD+
Sbjct: 95 FRCFCTRKDIQCQDVLLEESDVAKALVEYVNQAAIEVLVVGSSSKGGFLRFNKPTDIPGS 154
Query: 66 KAIYVCEQSPASCHIQFICKGYLIQTRDCSLENFRVEATSPLVQQMQNSEV---GHSPHL 122
+ + +P C + I KG + R S T+PL +Q + P
Sbjct: 155 ----ITKNAPDFCTVYIISKGKIQTMRSASRS---APMTAPLRSSVQPPSLKPPQPMPST 207
Query: 123 ISRSISHD-QELHHHRVRSISSASESGRSMASSVSSSERFSSFETVLT--PNLTSDGNES 179
+ S+ D + ++ RS+ S ++ SF + T N S G+ +
Sbjct: 208 SANSLRADRRSFESNQRRSVEDQQRRSMEDQQRRSIDDQSESFRSPFTRRGNGRSYGDLT 267
Query: 180 LLDLKMSYLSS 190
+ + +S++SS
Sbjct: 268 VPESDISFISS 278
>At1g78940 unknown protein
Length = 740
Score = 204 bits (518), Expect = 2e-52
Identities = 106/225 (47%), Positives = 145/225 (64%), Gaps = 10/225 (4%)
Query: 433 QVEVLSKLKHPNLITLIGVNQESKTLIYEYLPNGSLEDHLSRNGNNNAPPLTWQTRIRIA 492
+VEVLS ++HPN++ L+G E L+YEY+ GSLED L GN PP+TWQ R RIA
Sbjct: 480 EVEVLSCIRHPNMVLLLGACPEFGILVYEYMAKGSLEDRLFMRGNT--PPITWQLRFRIA 537
Query: 493 TELCSALIFLHSNKPHSIVHGDLKPSNILLDANLVTKLSDFGICRVLSCQNDSSSNNSTT 552
E+ + L+FLH KP IVH DLKP N+LLD N V+K+SD G+ R++ + + T
Sbjct: 538 AEIATGLLFLHQTKPEPIVHRDLKPGNVLLDYNYVSKISDVGLARLVPAVAE-----NVT 592
Query: 553 QFWITSFAKGTFAYMDPEFLGTGELTSKSDVYSFGIVLLRLITGKPALGIKNEVLYALNN 612
Q+ +TS A GTF Y+DPE+ TG L KSDVYS GI+LL+++T K +G+ V A+
Sbjct: 593 QYRVTS-AAGTFCYIDPEYQQTGMLGVKSDVYSLGIMLLQILTAKQPMGLAYYVEQAIEE 651
Query: 613 AGGNVKSVLDPLAGDWPIVEAEKLVHFALRCCDMNKKSRPELCSE 657
G +K +LDP DWPI EA L +L+C ++ +K RP+L E
Sbjct: 652 --GTLKDMLDPAVPDWPIEEALSLAKLSLQCAELRRKDRPDLGKE 694
Score = 43.1 bits (100), Expect = 5e-04
Identities = 44/175 (25%), Positives = 78/175 (44%), Gaps = 25/175 (14%)
Query: 12 CQRMGVTATKLHIEMDCIEKGIIELISRYNIQNLVMGAASDKYHSRRM-TDLRSKKAIYV 70
C R + + +E + I E +S I+NLV+G+AS RR TDL + V
Sbjct: 87 CSRKEINCRDILLEDADKVRAITEYVSSSAIENLVVGSASRNGFMRRFKTDLPTT----V 142
Query: 71 CEQSPASCHIQFICKGYLIQTRDCSLENFRVEATSPLVQQMQNSEV-GHSPHLISRSISH 129
+ +P C++ I KG + R+ S +P MQ E+ H PH ++
Sbjct: 143 SKSAPDFCNVYVISKGKIASVRNAS-------RPAPYQNSMQQCEIDNHHPHTPDKA--- 192
Query: 130 DQELHHHRVRSISSASESGRSMASSVSSSERFSSFETV---LTPNLTSDGNESLL 181
+ H H + S+ S +S+ ++R+ +++ V L+P+ D + S +
Sbjct: 193 -PKYHDHPNSAGSTPSRPRKSV-----EADRYRNWKLVVMSLSPSSHRDSDISFI 241
>At1g16760 hypothetical protein
Length = 758
Score = 203 bits (517), Expect = 2e-52
Identities = 105/225 (46%), Positives = 146/225 (64%), Gaps = 10/225 (4%)
Query: 433 QVEVLSKLKHPNLITLIGVNQESKTLIYEYLPNGSLEDHLSRNGNNNAPPLTWQTRIRIA 492
+VEVLS ++HPN++ L+G E L+YEY+ GSL+D L R GN PP++WQ R RIA
Sbjct: 493 EVEVLSCIRHPNMVLLLGACPEYGILVYEYMARGSLDDRLFRRGNT--PPISWQLRFRIA 550
Query: 493 TELCSALIFLHSNKPHSIVHGDLKPSNILLDANLVTKLSDFGICRVLSCQNDSSSNNSTT 552
E+ + L+FLH KP IVH DLKP N+LLD N V+K+SD G+ R++ + + T
Sbjct: 551 AEIATGLLFLHQTKPEPIVHRDLKPGNVLLDHNYVSKISDVGLARLVPAVAE-----NVT 605
Query: 553 QFWITSFAKGTFAYMDPEFLGTGELTSKSDVYSFGIVLLRLITGKPALGIKNEVLYALNN 612
Q+ +TS A GTF Y+DPE+ TG L KSDVYS GI+LL+L+T K +G+ V A+
Sbjct: 606 QYRVTS-AAGTFCYIDPEYQQTGMLGVKSDVYSLGIMLLQLLTAKQPMGLAYYVEQAIEE 664
Query: 613 AGGNVKSVLDPLAGDWPIVEAEKLVHFALRCCDMNKKSRPELCSE 657
G +K +LDP DWP+ EA L +L+C ++ +K RP+L E
Sbjct: 665 --GTLKDMLDPAVPDWPLEEALSLAKLSLQCAELRRKDRPDLGKE 707
Score = 36.2 bits (82), Expect = 0.063
Identities = 28/98 (28%), Positives = 49/98 (49%), Gaps = 8/98 (8%)
Query: 2 QKTVDVYLAI---CQRMGVTATKLHIEMDCIEKGIIELISRYNIQNLVMGAASDKYHSRR 58
++T D++++ C R + + +E K I+E ++ I+NLV+GA S RR
Sbjct: 73 KQTKDLFVSFHCYCSRKEIHCLDVVLEDVDKVKAIVEYVTVSAIENLVLGAPSRNSFMRR 132
Query: 59 M-TDLRSKKAIYVCEQSPASCHIQFICKGYLIQTRDCS 95
TDL + V + +P C++ I KG + R+ S
Sbjct: 133 FKTDLPTS----VSKAAPDFCNVYVISKGKISSLRNAS 166
>At1g17540 protein kinase, putative
Length = 740
Score = 201 bits (511), Expect = 1e-51
Identities = 103/230 (44%), Positives = 147/230 (63%), Gaps = 11/230 (4%)
Query: 428 RSLSYQVEVLSKLKHPNLITLIGVNQESKTLIYEYLPNGSLEDHLSRNGNNNAPPLTWQT 487
+ + ++EVLS ++HPN++ L+G E L+YEY+ NG+LED L +N PPL+W+
Sbjct: 459 KQFNQEIEVLSCMRHPNMVILLGACPEYGCLVYEYMENGTLEDRLFCK--DNTPPLSWRA 516
Query: 488 RIRIATELCSALIFLHSNKPHSIVHGDLKPSNILLDANLVTKLSDFGICRVL-SCQNDSS 546
R RIA E+ + L+FLH KP +VH DLKP+NIL+D + +K+SD G+ R++ + DS
Sbjct: 517 RFRIAAEIATGLLFLHQAKPEPLVHRDLKPANILIDRHFTSKISDVGLARLVPAAVADSF 576
Query: 547 SNNSTTQFWITSFAKGTFAYMDPEFLGTGELTSKSDVYSFGIVLLRLITGKPALGIKNEV 606
SN T A GTF Y+DPE+ TG L KSD+YSFG+VLL++IT PA+G+ + V
Sbjct: 577 SNYHMTA------AAGTFCYIDPEYQQTGMLGVKSDLYSFGVVLLQIITAMPAMGLSHRV 630
Query: 607 LYALNNAGGNVKSVLDPLAGDWPIVEAEKLVHFALRCCDMNKKSRPELCS 656
A+ ++ VLDP DWP E L AL+CC++ KK RP+L S
Sbjct: 631 EKAIEKK--KLREVLDPKISDWPEEETMVLAQLALQCCELRKKDRPDLAS 678
Score = 40.8 bits (94), Expect = 0.003
Identities = 38/147 (25%), Positives = 66/147 (44%), Gaps = 16/147 (10%)
Query: 8 YLAICQRMGVTATKLHIEMDCIEKGIIELISRYNIQNLVMGAASDKYHSRRMTDLRSKKA 67
+ C R G+ AT++ + I I++ I+ +I N+V+GA S R + L+ K+
Sbjct: 73 FRGFCARKGIIATEVLLHDIDISSAIVDYITNNSISNIVIGA------SARNSFLKKFKS 126
Query: 68 IYV----CEQSPASCHIQFICKGYLIQTRDCSLENFRVEATSPLVQQMQNSEVGHSPHLI 123
+ V + +P +C + + KG L+ +R S +P Q + H PH
Sbjct: 127 VDVPTTLLKTTPDTCAVFIVSKGKLLTSRSASRPQTPQGPHTPQGPQTPQAP-QHPPH-- 183
Query: 124 SRSISHDQELHHHRVRSISSASESGRS 150
S + + S++SESGRS
Sbjct: 184 ---PSKHPSMMSDPGPTSSTSSESGRS 207
>At2g07020 putative protein kinase
Length = 620
Score = 201 bits (510), Expect = 1e-51
Identities = 104/222 (46%), Positives = 150/222 (66%), Gaps = 9/222 (4%)
Query: 433 QVEVLSKLKHPNLITLIGVNQESKTLIYEYLPNGSLEDHLSRNGNNNAPPLTWQTRIRIA 492
+VEVL+ ++HPN++ L+G E L+YEY+ NGSL+D L R GN+ P L+WQ R RIA
Sbjct: 345 EVEVLTCIRHPNMVLLLGACAEYGCLVYEYMSNGSLDDCLLRRGNS--PVLSWQLRFRIA 402
Query: 493 TELCSALIFLHSNKPHSIVHGDLKPSNILLDANLVTKLSDFGICRVLSCQNDSSSNNSTT 552
E+ ++L FLH KP +VH DLKP+NILLD ++V+K+SD G+ R++ + ++ T
Sbjct: 403 AEIATSLNFLHQLKPEPLVHRDLKPANILLDQHMVSKISDVGLARLV----PPTIDDIAT 458
Query: 553 QFWITSFAKGTFAYMDPEFLGTGELTSKSDVYSFGIVLLRLITGKPALGIKNEVLYALNN 612
+ +TS A GT Y+DPE+ TG L +KSD+YSFGIVLL+++T K +G+ N+V A+
Sbjct: 459 HYRMTSTA-GTLCYIDPEYQQTGMLGTKSDIYSFGIVLLQILTAKTPMGLTNQVEKAIEE 517
Query: 613 AGGNVKSVLDPLAGDWPIVEAEKLVHFALRCCDMNKKSRPEL 654
GN +LDPL DWPI EA L L+C ++ +K RP+L
Sbjct: 518 --GNFAKILDPLVTDWPIEEALILAKIGLQCAELRRKDRPDL 557
>At4g25160 putative Ser/Thr protein kinase
Length = 835
Score = 197 bits (502), Expect = 1e-50
Identities = 110/259 (42%), Positives = 165/259 (63%), Gaps = 9/259 (3%)
Query: 419 VLKDPQN-FNRSLSYQVEVLSKLKHPNLITLIGVNQESKTLIYEYLPNGSLEDHLSRNGN 477
VL+ +N ++ ++E+LSK++HP+L+ L+G E L+YEY+ NGSLED L +
Sbjct: 508 VLQSAENQLSKQFQQELEILSKIRHPHLVLLLGACPEQGALVYEYMENGSLEDRLFQV-- 565
Query: 478 NNAPPLTWQTRIRIATELCSALIFLHSNKPHSIVHGDLKPSNILLDANLVTKLSDFGICR 537
NN+PPL W R RIA E+ +AL+FLH +KP I+H DLKP+NILLD N V+K+ D G+
Sbjct: 566 NNSPPLPWFERFRIAWEVAAALVFLHKSKPKPIIHRDLKPANILLDHNFVSKVGDVGLST 625
Query: 538 VLSCQNDSSSNNSTTQFWITSFAKGTFAYMDPEFLGTGELTSKSDVYSFGIVLLRLITGK 597
++ Q D S T + + GT Y+DPE+ TG ++SKSD+YSFG++LL+L+T K
Sbjct: 626 MV--QVDPLSTKFT--IYKQTSPVGTLCYIDPEYQRTGRISSKSDIYSFGMILLQLLTAK 681
Query: 598 PALGIKNEVLYALNNAGGNVKSVLDPLAGDWPIVEAEKLVHFALRCCDMNKKSRPELCSE 657
PA+ + + V A+++ +K +LD AG+WPI E +L AL C ++ K RP+L +
Sbjct: 682 PAIALTHFVESAMDSNDEFLK-ILDQKAGNWPIEETRELAALALCCTELRGKDRPDLKDQ 740
Query: 658 GWRVLEPM-KVSCSGTNNF 675
LE + KV+ N+F
Sbjct: 741 ILPALENLKKVAEKARNSF 759
Score = 31.2 bits (69), Expect = 2.0
Identities = 23/81 (28%), Positives = 37/81 (45%), Gaps = 5/81 (6%)
Query: 8 YLAICQRMGVTATKLHIEMDCIEKGIIELISRYNIQNLVMGAASDKYHSRRMTDLRSKKA 67
Y + R V L IE D + I E ++R +I +V+G +S + SR+ D+ S
Sbjct: 99 YTKLFVRRKVAVEVLVIESDNVAAAIAEEVTRDSIDRIVIGGSSRSFFSRK-ADICS--- 154
Query: 68 IYVCEQSPASCHIQFICKGYL 88
+ P C + + KG L
Sbjct: 155 -VISALMPNFCTVYVVSKGKL 174
>At5g35380 putative protein
Length = 731
Score = 196 bits (498), Expect = 4e-50
Identities = 103/222 (46%), Positives = 146/222 (65%), Gaps = 9/222 (4%)
Query: 433 QVEVLSKLKHPNLITLIGVNQESKTLIYEYLPNGSLEDHLSRNGNNNAPPLTWQTRIRIA 492
+VEVL+ ++HPN++ L+G E L+YEY+ NGSL+D L R GN+ P L+WQ R RIA
Sbjct: 457 EVEVLTCMRHPNMVLLLGACPEYGCLVYEYMANGSLDDCLFRRGNS--PILSWQLRFRIA 514
Query: 493 TELCSALIFLHSNKPHSIVHGDLKPSNILLDANLVTKLSDFGICRVLSCQNDSSSNNSTT 552
+E+ + L FLH KP +VH DLKP NILLD + V+K+SD G+ R++ S ++ T
Sbjct: 515 SEIATGLHFLHQMKPEPLVHRDLKPGNILLDQHFVSKISDVGLARLV----PPSVADTAT 570
Query: 553 QFWITSFAKGTFAYMDPEFLGTGELTSKSDVYSFGIVLLRLITGKPALGIKNEVLYALNN 612
Q+ +TS A GTF Y+DPE+ TG L +KSD+YSFGI+LL+++T KP +G+ + V A+
Sbjct: 571 QYRMTSTA-GTFFYIDPEYQQTGMLGTKSDIYSFGIMLLQILTAKPPMGLTHHVEKAIEK 629
Query: 613 AGGNVKSVLDPLAGDWPIVEAEKLVHFALRCCDMNKKSRPEL 654
G +LDP DWP EA AL+C + +K RP+L
Sbjct: 630 --GTFAEMLDPAVPDWPFEEALAAAKLALQCAKLRRKDRPDL 669
Score = 32.7 bits (73), Expect = 0.69
Identities = 22/78 (28%), Positives = 38/78 (48%), Gaps = 10/78 (12%)
Query: 12 CQRMGVTATKLHIEMDCIEKGIIELISRYNIQNLVMGAASDKYHSRRMTDLRSKKA---I 68
C R + + +E KGI++ + + I+ L++G S +MT LR K A
Sbjct: 78 CARKDINCQDVVVEDVSAAKGIVDYVQQNAIETLILG-------SSKMTLLRFKAADVSS 130
Query: 69 YVCEQSPASCHIQFICKG 86
V +++P+ C + I KG
Sbjct: 131 TVMKKAPSFCTVYVISKG 148
>At4g31220 protein kinase-like protein
Length = 243
Score = 195 bits (495), Expect = 8e-50
Identities = 100/210 (47%), Positives = 140/210 (66%), Gaps = 9/210 (4%)
Query: 445 LITLIGVNQESKTLIYEYLPNGSLEDHLSRNGNNNAPPLTWQTRIRIATELCSALIFLHS 504
++ L+G E L+YE++ NGSLED L R G++ P L+WQTR RIA E+ + L+FLH
Sbjct: 1 MVLLLGACPECGCLVYEFMANGSLEDRLFRQGDS--PALSWQTRFRIAAEIGTVLLFLHQ 58
Query: 505 NKPHSIVHGDLKPSNILLDANLVTKLSDFGICRVLSCQNDSSSNNSTTQFWITSFAKGTF 564
KP +VH DLKP+NILLD N V+KL+D G+ R++ S N+ TQ+ +TS A GTF
Sbjct: 59 TKPEPLVHRDLKPANILLDRNFVSKLADVGLARLV----PPSVANTVTQYHMTSTA-GTF 113
Query: 565 AYMDPEFLGTGELTSKSDVYSFGIVLLRLITGKPALGIKNEVLYALNNAGGNVKSVLDPL 624
Y+DPE+ TG L KSD+YS GI+ L+LITGKP +G+ + V AL GN+K +LDP
Sbjct: 114 CYIDPEYQQTGMLGVKSDIYSLGIMFLQLITGKPPMGLTHYVERALEK--GNLKDLLDPA 171
Query: 625 AGDWPIVEAEKLVHFALRCCDMNKKSRPEL 654
DWP+ + + AL+C ++ +K RP+L
Sbjct: 172 VSDWPVEDTTEFAKLALKCAEIRRKDRPDL 201
>At3g20200 protein kinase like protein
Length = 780
Score = 192 bits (488), Expect = 5e-49
Identities = 102/225 (45%), Positives = 140/225 (61%), Gaps = 10/225 (4%)
Query: 433 QVEVLSKLKHPNLITLIGVNQESKTLIYEYLPNGSLEDHLSRNGNNNAPPLTWQTRIRIA 492
+VEVLS ++HP+++ LIG E L+YEY+ GSL D L + GN PPL+W+ R RIA
Sbjct: 496 EVEVLSCIRHPHMVLLIGACPEYGVLVYEYMAKGSLADRLYKYGNT--PPLSWELRFRIA 553
Query: 493 TELCSALIFLHSNKPHSIVHGDLKPSNILLDANLVTKLSDFGICRVLSCQNDSSSNNSTT 552
E+ + L+FLH KP IVH DLKP NIL+D N V+K+ D G+ +++ + + T
Sbjct: 554 AEVATGLLFLHQTKPEPIVHRDLKPGNILIDQNYVSKIGDVGLAKLVPAVAE-----NVT 608
Query: 553 QFWITSFAKGTFAYMDPEFLGTGELTSKSDVYSFGIVLLRLITGKPALGIKNEVLYALNN 612
Q ++S A GTF Y+DPE+ TG L KSDVYSFGI+LL L+T K G+ V A+
Sbjct: 609 QCHVSSTA-GTFCYIDPEYQQTGMLGVKSDVYSFGILLLELLTAKRPTGLAYTVEQAMEQ 667
Query: 613 AGGNVKSVLDPLAGDWPIVEAEKLVHFALRCCDMNKKSRPELCSE 657
G K +LDP +WP+ EA L AL+C + +K RP+L E
Sbjct: 668 --GKFKDMLDPAVPNWPVEEAMSLAKIALKCAQLRRKDRPDLGKE 710
Score = 42.4 bits (98), Expect = 9e-04
Identities = 43/177 (24%), Positives = 73/177 (40%), Gaps = 28/177 (15%)
Query: 12 CQRMGVTATKLHIEMDCIEKGIIELISRYNIQNLVMGAASDKYHSR--RMTDLRSKKAIY 69
C R + + +E D I K + E +S I+NL++GA S R +++D S
Sbjct: 81 CSRKEIQCLDVTLEDDNIVKSLAEYVSSGVIENLILGAPSRHGFMRKFKISDTPSN---- 136
Query: 70 VCEQSPASCHIQFICKGYLIQTRDCSLENFRVEATSPLVQQMQNSEVGHSPHLISRSISH 129
V + +P C + I KG + R S SPL+ Q++N H
Sbjct: 137 VAKAAPDFCTVYVISKGKISSVRHASR---AAPYRSPLMGQIEN---------------H 178
Query: 130 DQELHHHRVRSISSASESGRSMASSVSSSERFSSFETVLTPNLTSDGNESLLDLKMS 186
+ +++ + R+ S + +S+ SS E + T N + S DL+ S
Sbjct: 179 SEIINYEKFRNTMSFRDRAPPRSSTASSIEDYGKSPMARTSNYAN----SFFDLEDS 231
>At5g51270 putative protein
Length = 819
Score = 190 bits (483), Expect = 2e-48
Identities = 99/243 (40%), Positives = 156/243 (63%), Gaps = 6/243 (2%)
Query: 425 NFNRSLSYQVEVLSKLKHPNLITLIGVNQESKTLIYEYLPNGSLEDHLSRNGNNNAPPLT 484
+ ++ ++E+LSK++HP+L+ L+G + L+YEY+ NGSLED L + N++ P+
Sbjct: 495 SLSKQFDQELEILSKIRHPHLVLLLGACPDHGALVYEYMENGSLEDRLFQV--NDSQPIP 552
Query: 485 WQTRIRIATELCSALIFLHSNKPHSIVHGDLKPSNILLDANLVTKLSDFGICRVLSCQND 544
W R+RIA E+ SAL+FLH +KP I+H DLKP+NILL+ N V+K+ D G+ ++ +
Sbjct: 553 WFVRLRIAWEVASALVFLHKSKPTPIIHRDLKPANILLNHNFVSKVGDVGLSTMIQAADP 612
Query: 545 SSSNNSTTQFWITSFAKGTFAYMDPEFLGTGELTSKSDVYSFGIVLLRLITGKPALGIKN 604
S+ T + TS GT Y+DPE+ TG ++ KSDVY+FG+++L+L+TG+ A+ +
Sbjct: 613 LST--KFTMYKQTS-PVGTLCYIDPEYQRTGRISPKSDVYAFGMIILQLLTGQQAMALTY 669
Query: 605 EVLYAL-NNAGGNVKSVLDPLAGDWPIVEAEKLVHFALRCCDMNKKSRPELCSEGWRVLE 663
V A+ NN + +LD AG+WPI E +L AL+C ++ K RP+L + VLE
Sbjct: 670 TVETAMENNNDDELIQILDEKAGNWPIEETRQLAALALQCTELRSKDRPDLEDQILPVLE 729
Query: 664 PMK 666
+K
Sbjct: 730 SLK 732
>At5g61550 putative protein
Length = 845
Score = 190 bits (482), Expect = 3e-48
Identities = 104/240 (43%), Positives = 151/240 (62%), Gaps = 8/240 (3%)
Query: 427 NRSLSYQVEVLSKLKHPNLITLIGVNQESKTLIYEYLPNGSLEDHLSRNGNNNAPPLTWQ 486
++ ++E+LSK++HP+L+ L+G E L+YEY+ NGSL+D L N+ PP+ W
Sbjct: 527 SKQFDQELEILSKIRHPHLVLLLGACPERGCLVYEYMDNGSLDDRLMLV--NDTPPIPWF 584
Query: 487 TRIRIATELCSALIFLHSNKPHSIVHGDLKPSNILLDANLVTKLSDFGICRVLSCQNDSS 546
R RIA E+ SAL+FLH +KP I+H DLKP NILLD N V+KL D G+ +++ Q+D S
Sbjct: 585 ERFRIALEVASALVFLHKSKPRPIIHRDLKPGNILLDHNFVSKLGDVGLSTMVN-QDDVS 643
Query: 547 SNNSTTQFWITSFAKGTFAYMDPEFLGTGELTSKSDVYSFGIVLLRLITGKPALGIKNEV 606
S T F TS GT Y+DPE+ TG ++ KSDVYS G+V+L+LIT KPA+ I + V
Sbjct: 644 SR---TIFKQTS-PVGTLCYIDPEYQRTGIISPKSDVYSLGVVILQLITAKPAIAITHMV 699
Query: 607 LYALNNAGGNVKSVLDPLAGDWPIVEAEKLVHFALRCCDMNKKSRPELCSEGWRVLEPMK 666
A+ + ++LD AG WPI + +L L C +M ++ RP+L + LE ++
Sbjct: 700 EEAIGD-DAEFMAILDKKAGSWPISDTRELAALGLCCTEMRRRDRPDLKDQIIPALERLR 758
>At2g19410 putative protein kinase
Length = 801
Score = 180 bits (456), Expect = 3e-45
Identities = 100/247 (40%), Positives = 149/247 (59%), Gaps = 15/247 (6%)
Query: 433 QVEVLSKLKHPNLITLIGVNQESKTLIYEYLPNGSLEDHLSRNGNNNAPPLTWQTRIRIA 492
+VEVLS+L+HP+++ L+G E+ L+YEYL NGSLE+++ N PPL W R R+
Sbjct: 485 EVEVLSQLRHPHVVLLLGACPENGCLVYEYLENGSLEEYIFHR--KNKPPLPWFIRFRVI 542
Query: 493 TELCSALIFLHSNKPHSIVHGDLKPSNILLDANLVTKLSDFGICRVLSCQNDSSSNNSTT 552
E+ L FLHS+KP IVH DLKP NILL+ N V+K++D G+ ++++ D + +N T
Sbjct: 543 FEVACGLAFLHSSKPEPIVHRDLKPGNILLNRNYVSKIADVGLAKLVT---DVAPDNVT- 598
Query: 553 QFWITSFAKGTFAYMDPEFLGTGELTSKSDVYSFGIVLLRLITGKPALGIKNEVLYALNN 612
+ S GT Y+DPE+ TG + KSD+Y+FGI++L+L+T + GI V A+
Sbjct: 599 -MYRNSVLAGTLHYIDPEYHRTGTIRPKSDLYAFGIIILQLLTARNPSGIVPAVENAVKK 657
Query: 613 AGGNVKSVLDPLAGDWPIVEAEKLVHFALRCCDMNKKSRPELCSEGWRVLEPM------K 666
G + +LD DWP+ E E+L L+C + + RP+L SE VL+ + K
Sbjct: 658 --GTLTEMLDKSVTDWPLAETEELARIGLKCAEFRCRDRPDLKSEVIPVLKRLVETANSK 715
Query: 667 VSCSGTN 673
V G+N
Sbjct: 716 VKKEGSN 722
>At5g39000 protein kinase - like protein
Length = 873
Score = 155 bits (392), Expect = 7e-38
Identities = 89/237 (37%), Positives = 131/237 (54%), Gaps = 18/237 (7%)
Query: 433 QVEVLSKLKHPNLITLIGVNQESK--TLIYEYLPNGSLEDHLSRNGNNNAPPLTWQTRIR 490
++E+LSKL+H +L++LIG E L+YEY+P+G+L+DHL R + PPL+W+ R+
Sbjct: 563 ELEMLSKLRHVHLVSLIGYCDEDNEMVLVYEYMPHGTLKDHLFRRDKTSDPPLSWKRRLE 622
Query: 491 IATELCSALIFLHSNKPHSIVHGDLKPSNILLDANLVTKLSDFGICRVLSCQNDSSSNNS 550
I L +LH+ ++I+H D+K +NILLD N VTK+SDFG+ RV S
Sbjct: 623 ICIGAARGLQYLHTGAKYTIIHRDIKTTNILLDENFVTKVSDFGLSRV--------GPTS 674
Query: 551 TTQFWITSFAKGTFAYMDPEFLGTGELTSKSDVYSFGIVLLRLITGKP--ALGIKNEVLY 608
+Q +++ KGTF Y+DPE+ LT KSDVYSFG+VLL ++ +P + E
Sbjct: 675 ASQTHVSTVVKGTFGYLDPEYYRRQVLTEKSDVYSFGVVLLEVLCCRPIRMQSVPPEQAD 734
Query: 609 AL-----NNAGGNVKSVLD-PLAGDWPIVEAEKLVHFALRCCDMNKKSRPELCSEGW 659
+ N G V ++D L+ D EK A+RC RP + W
Sbjct: 735 LIRWVKSNYRRGTVDQIIDSDLSADITSTSLEKFCEIAVRCVQDRGMERPPMNDVVW 791
>At1g67720 putative receptor protein kinase
Length = 914
Score = 151 bits (381), Expect = 1e-36
Identities = 98/277 (35%), Positives = 153/277 (54%), Gaps = 29/277 (10%)
Query: 416 APTVLKDPQN-FNRSLSYQVEVLSKLKHPNLITLIGVNQES--KTLIYEYLPNGSLEDHL 472
A + DP + NR +V +LS++ H NL+ LIG +E+ + L+YEY+ NGSL DHL
Sbjct: 617 AVKITADPSSHLNRQFVTEVALLSRIHHRNLVPLIGYCEEADRRILVYEYMHNGSLGDHL 676
Query: 473 SRNGNNNAPPLTWQTRIRIATELCSALIFLHSNKPHSIVHGDLKPSNILLDANLVTKLSD 532
+G+++ PL W TR++IA + L +LH+ SI+H D+K SNILLD N+ K+SD
Sbjct: 677 --HGSSDYKPLDWLTRLQIAQDAAKGLEYLHTGCNPSIIHRDVKSSNILLDINMRAKVSD 734
Query: 533 FGICRVLSCQNDSSSNNSTTQFWITSFAKGTFAYMDPEFLGTGELTSKSDVYSFGIVLLR 592
FG+ R Q + + ++S AKGT Y+DPE+ + +LT KSDVYSFG+VL
Sbjct: 735 FGLSR----QTEEDLTH------VSSVAKGTVGYLDPEYYASQQLTEKSDVYSFGVVLFE 784
Query: 593 LITGK---------PALGIKNEVLYALNNAGGNVKSVLDP-LAGDWPIVEAEKLVHFALR 642
L++GK P L I + + G+V ++DP +A + I ++ A +
Sbjct: 785 LLSGKKPVSAEDFGPELNIVHWARSLIRK--GDVCGIIDPCIASNVKIESVWRVAEVANQ 842
Query: 643 CCDMNKKSRPELCSEGWRVLEPMKVSCSGTNNFGLKS 679
C + +RP + + + +++ N GLKS
Sbjct: 843 CVEQRGHNRPRMQEVIVAIQDAIRIERGNEN--GLKS 877
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.331 0.141 0.432
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,408,364
Number of Sequences: 26719
Number of extensions: 595598
Number of successful extensions: 5371
Number of sequences better than 10.0: 1013
Number of HSP's better than 10.0 without gapping: 818
Number of HSP's successfully gapped in prelim test: 195
Number of HSP's that attempted gapping in prelim test: 2141
Number of HSP's gapped (non-prelim): 1193
length of query: 688
length of database: 11,318,596
effective HSP length: 106
effective length of query: 582
effective length of database: 8,486,382
effective search space: 4939074324
effective search space used: 4939074324
T: 11
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.9 bits)
S2: 64 (29.3 bits)
Medicago: description of AC146341.5


