

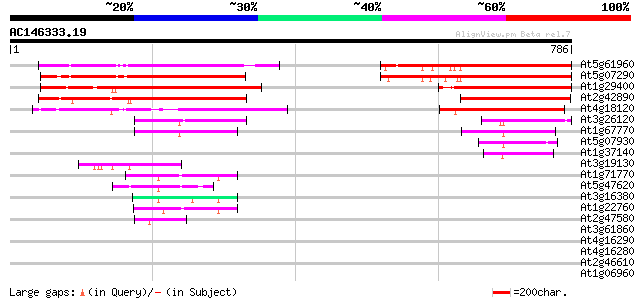
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146333.19 - phase: 0 /pseudo
(786 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g61960 Mei2-like protein 288 8e-78
At5g07290 Mei2-like protein 285 9e-77
At1g29400 putative RNA-binding protein MEI2 (At1g29400) 249 4e-66
At2g42890 MEI2-like protein (MEI2) 216 4e-56
At4g18120 predicted protein 215 7e-56
At3g26120 RNA-binding protein, putative 127 3e-29
At1g67770 terminal ear1-like protein 114 2e-25
At5g07930 putative protein 91 2e-18
At1g37140 hypothetical protein 82 9e-16
At3g19130 nuclear acid binding protein, putative 47 4e-05
At1g71770 polyadenylate-binding protein 5 47 5e-05
At5g47620 RNA-binding protein-like 45 1e-04
At3g16380 putative poly(A) binding protein 44 4e-04
At1g22760 putative polyA-binding protein, PAB3 43 6e-04
At2g47580 small nuclear ribonucleoprotein U1A 43 8e-04
At3g61860 ARGININE/SERINE-RICH SPLICING FACTOR RSP31 42 0.002
At4g16290 FCA delta protein 41 0.003
At4g16280 FCA gamma protein 41 0.003
At2g46610 arginine/serine-rich splicing factor like protein 41 0.003
At1g06960 putative spliceosomal protein U2B 40 0.005
>At5g61960 Mei2-like protein
Length = 915
Score = 288 bits (737), Expect = 8e-78
Identities = 163/314 (51%), Positives = 195/314 (61%), Gaps = 49/314 (15%)
Query: 520 WSNSY-----PPPRTIWPNSPSYFDGIYAASTLQRLNQLPMSPSHMITTVLPTNNHHIQS 574
WSNS P +WPNSPS+ + I + + +P M+ +HHI S
Sbjct: 554 WSNSNTQQQNPSSGMMWPNSPSHINSI--PTQRPPVTVFSRAPPIMVNMASSPVHHHIGS 611
Query: 575 PP-----FWDRRYTYAAEPITP-------------------HCVDFVPHNMFPHFG--LN 608
P FWDRR Y AE + H +D H F G ++
Sbjct: 612 APVLNSPFWDRRQAYVAESLESSGFHIGSHGSMGIPGSSPSHPMDIGSHKTFSVGGNRMD 671
Query: 609 VHNQRGMV---------FPGRN---HMINSFDT----YKRVRSRRNVGASNLADMKRYEL 652
V++Q ++ FPGR+ M SFD+ Y+ + RR+ +S+ AD K YEL
Sbjct: 672 VNSQNAVLRSPQQLSHLFPGRSPMGSMPGSFDSPNERYRNLSHRRSESSSSNADKKLYEL 731
Query: 653 DIDCIKRGEDNRTTLMIKNIPNKYTSKMLLAAIDEHHKGAYDFVYLPIDFRNKCNVGYAF 712
D+D I RGED RTTLMIKNIPNKYTSKMLL+AIDEH KG YDF+YLPIDF+NKCNVGYAF
Sbjct: 732 DVDRILRGEDRRTTLMIKNIPNKYTSKMLLSAIDEHCKGTYDFLYLPIDFKNKCNVGYAF 791
Query: 713 INMTSPSLIVPFYQGFNGKKWEKFNSEKVASLAYARIQGKAALVAHFQNSSLMNEDKRCR 772
IN+ P IVPF++ FNGKKWEKFNSEKVA+L YARIQGK AL+AHFQNSSLMNEDKRCR
Sbjct: 792 INLIEPEKIVPFFKAFNGKKWEKFNSEKVATLTYARIQGKTALIAHFQNSSLMNEDKRCR 851
Query: 773 PILIDTDGPNAGDQ 786
PIL TDGPNAGDQ
Sbjct: 852 PILFHTDGPNAGDQ 865
Score = 244 bits (624), Expect = 1e-64
Identities = 155/339 (45%), Positives = 198/339 (57%), Gaps = 20/339 (5%)
Query: 41 SRHESSLFSSSLSDLFSKKLRLSANNAFYGHSVDTIASNYEEEKLSDSLEELEAQIIGNL 100
S+ ESSLFSSS+SDLFS+KLRL ++ S +T+ ++ EEE S+SLEE+EAQ IGNL
Sbjct: 86 SQWESSLFSSSMSDLFSRKLRLQGSDMLSTMSANTVVTHREEEP-SESLEEIEAQTIGNL 144
Query: 101 LPDEDDLLSGVTDGNNYIICDSNGDDIDELDLFSSNGGFDL-GDVENPSSIERNSEIISG 159
LPDEDDL + VT + GD++DE DLFSS GG +L GD+ + S RN E G
Sbjct: 145 LPDEDDLFAEVTGEVGRKSRANTGDELDEFDLFSSVGGMELDGDIFSSVS-HRNGE--RG 201
Query: 160 VRNSSIAGENSYGEHPSRTLFVRNIDSDVKDSVLKALFEQFGDIHTFDRTCKHQGSAMIS 219
NS GE + GE PSRTL V NI S+V+D LK LFEQFGDI CK++G M+S
Sbjct: 202 GNNS--VGELNRGEIPSRTLLVGNISSNVEDYELKVLFEQFGDIQALHTACKNRGFIMVS 259
Query: 220 YYDIRAAQNAMRALNNRLFGRKKFDIHYPIPKDSPSRNGVNQGTLEVFLYDSSISNTELQ 279
Y DIRAAQNA RAL N+L K DI Y I K++PS+ ++G L V DSSISN EL
Sbjct: 260 YCDIRAAQNAARALQNKLLRGTKLDIRYSISKENPSQKDTSKGALLVNNLDSSISNQELN 319
Query: 280 HILNVYGGIKEIHENPRSQRHKLIEFYDFRAADAALHGINRNDTTMKRLKVDQMQSTNSE 339
++ YG +KEI IEF+D RAA AAL G+N + K+L+
Sbjct: 320 RLVKSYGEVKEIRRTMHDNSQIYIEFFDVRAAAAALGGLNGLEVAGKKLQ---------- 369
Query: 340 SNIIQPMHPEFKQECDLCLHQKSPLLKPTTSFQGREHVH 378
+ P +PE + C + P TS+ H
Sbjct: 370 ---LVPTYPEGTRYTSQCAANDTEGCLPKTSYSNTSSGH 405
>At5g07290 Mei2-like protein
Length = 907
Score = 285 bits (728), Expect = 9e-77
Identities = 163/315 (51%), Positives = 194/315 (60%), Gaps = 49/315 (15%)
Query: 520 WSNSYPPPRT-----IWPNSPSYFDGIYAASTLQRLNQLPMSPSHMITTVLPTNNHHIQS 574
WSNS +WPNSPS +G+ + + + + M+ +HHI S
Sbjct: 550 WSNSNSQQHNQSSGMMWPNSPSRVNGV-PSQRIPPVTAFSRASPLMVNMASSPVHHHIGS 608
Query: 575 PP-----FWDRRYTYAAEP-------------------ITPHCVDFVPHNMFPHFGLN-- 608
P FWDRR Y AE H +DF H +F H G N
Sbjct: 609 APVLNSPFWDRRQAYVAESPESSGFHLGSPGSMGFPGSSPSHPMDFGSHKVFSHVGGNRM 668
Query: 609 --------VHNQRGM--VFPGRNHMIN---SFDT----YKRVRSRRNVGASNLADMKRYE 651
+ + R M +F GR+ M++ SFD Y+ + RR+ S+ A+ K YE
Sbjct: 669 EANSKNAVLRSSRQMPHLFTGRSPMLSVSGSFDLPNERYRNLSHRRSESNSSNAEKKLYE 728
Query: 652 LDIDCIKRGEDNRTTLMIKNIPNKYTSKMLLAAIDEHHKGAYDFVYLPIDFRNKCNVGYA 711
LD+D I RGED+RTTLMIKNIPNKYTSKMLLAAIDE+ KG YDF+YLPIDF+NKCNVGYA
Sbjct: 729 LDVDRILRGEDSRTTLMIKNIPNKYTSKMLLAAIDEYCKGTYDFLYLPIDFKNKCNVGYA 788
Query: 712 FINMTSPSLIVPFYQGFNGKKWEKFNSEKVASLAYARIQGKAALVAHFQNSSLMNEDKRC 771
FIN+ P IVPFY+ FNGKKWEKFNSEKVASLAY RIQGK+AL+AHFQNSSLMNEDKRC
Sbjct: 789 FINLIEPENIVPFYKAFNGKKWEKFNSEKVASLAYGRIQGKSALIAHFQNSSLMNEDKRC 848
Query: 772 RPILIDTDGPNAGDQ 786
RPIL T GPNAGDQ
Sbjct: 849 RPILFHTAGPNAGDQ 863
Score = 221 bits (563), Expect = 1e-57
Identities = 137/288 (47%), Positives = 180/288 (61%), Gaps = 12/288 (4%)
Query: 44 ESSLFSSSLSDLFSKKLRLSANNAFYGHSVDTIASNYEEEKLSDSLEELEAQIIGNLLPD 103
ESSLFSSSLSDLFS+KLRL ++ + +++N EEE S+SLEE+EAQ IGNLLPD
Sbjct: 87 ESSLFSSSLSDLFSRKLRLPRSD-----KLAFMSANREEEP-SESLEEMEAQTIGNLLPD 140
Query: 104 EDDLLSGVTDGNNYIICDSNGDDIDELDLFSSNGGFDL-GDVENPSSIERNSEIISGVRN 162
EDDL + V + + GDD+D+ DLFSS GG +L GDV + S +R+ + S V
Sbjct: 141 EDDLFAEVVGEGVHKSRANGGDDLDDCDLFSSVGGMELDGDVFSSVS-QRDGKRGSNV-- 197
Query: 163 SSIAGENSYGEHPSRTLFVRNIDSDVKDSVLKALFEQFGDIHTFDRTCKHQGSAMISYYD 222
S E+ GE SR LFVRN+DS ++D L LF+QFGD+ K++G M+SYYD
Sbjct: 198 -STVAEHPQGEILSRILFVRNVDSSIEDCELGVLFKQFGDVRALHTAGKNRGFIMVSYYD 256
Query: 223 IRAAQNAMRALNNRLFGRKKFDIHYPIPKDSPSRNGVNQGTLEVFLYDSSISNTELQHIL 282
IRAAQ A RAL+ RL +K DI Y IPK++P N ++G L V DSSISN EL I
Sbjct: 257 IRAAQKAARALHGRLLRGRKLDIRYSIPKENPKENS-SEGALWVNNLDSSISNEELHGIF 315
Query: 283 NVYGGIKEIHENPRSQRHKLIEFYDFRAADAALHGINRNDTTMKRLKV 330
+ YG I+E+ IEF+D R A AL G+N + ++LK+
Sbjct: 316 SSYGEIREVRRTMHENSQVYIEFFDVRKAKVALQGLNGLEVAGRQLKL 363
>At1g29400 putative RNA-binding protein MEI2 (At1g29400)
Length = 800
Score = 249 bits (636), Expect = 4e-66
Identities = 143/324 (44%), Positives = 198/324 (60%), Gaps = 20/324 (6%)
Query: 44 ESSLFSSSLSDLFSKKLRLSANNAFYGHSVDTIASNYEEEKLSDSLEELEAQIIGNLLPD 103
+++LFSSSL KL+LS N + DT S K ++S ++ E+ IGNLLPD
Sbjct: 27 DATLFSSSLPVFPRGKLQLSDNRDGFSLIDDTAVSR--TNKFNESADDFESHSIGNLLPD 84
Query: 104 EDDLLSGVTDGNNYIICDSNGDDIDELDLFSSNGGFDLG-------DVENP-----SSIE 151
E+DLL+G+ D + D D+ DLF S GG +L + P SS+
Sbjct: 85 EEDLLTGMMDDLDL----GELPDADDYDLFGSGGGMELDADFRDNLSMSGPPRLSLSSLG 140
Query: 152 RNSEIISGVRNSS--IAGENSYGEHPSRTLFVRNIDSDVKDSVLKALFEQFGDIHTFDRT 209
N+ + N + +AGE+ YGEHPSRTLFVRNI+S+V+DS L ALFEQ+GDI T T
Sbjct: 141 GNAIPQFNIPNGAGTVAGEHPYGEHPSRTLFVRNINSNVEDSELTALFEQYGDIRTLYTT 200
Query: 210 CKHQGSAMISYYDIRAAQNAMRALNNRLFGRKKFDIHYPIPKDSPSRNGVNQGTLEVFLY 269
CKH+G MISYYDIR+A+ AMR+L N+ R+K DIH+ IPKD+PS +NQGTL VF
Sbjct: 201 CKHRGFVMISYYDIRSARMAMRSLQNKPLRRRKLDIHFSIPKDNPSEKDMNQGTLVVFNL 260
Query: 270 DSSISNTELQHILNVYGGIKEIHENPRSQRHKLIEFYDFRAADAALHGINRNDTTMKRLK 329
D SISN +L I +G IKEI E P + HK +EFYD R A+AAL +NR + KR+K
Sbjct: 261 DPSISNDDLHGIFGAHGEIKEIRETPHKRHHKFVEFYDVRGAEAALKALNRCEIAGKRIK 320
Query: 330 VDQMQSTNSESNIIQPMHPEFKQE 353
V+ + + +++ ++ + + +
Sbjct: 321 VEPSRPGGARRSLMLQLNQDLEND 344
Score = 232 bits (591), Expect = 7e-61
Identities = 122/189 (64%), Positives = 146/189 (76%), Gaps = 14/189 (7%)
Query: 601 MFPHFGLNVHNQRGMVFPGRNHMINSFDT-YKRVRSRRNVGASNLADM-KRYELDIDCIK 658
MF GLN PGR + FD+ Y+ R RR SN + K+++LD++ I
Sbjct: 598 MFLSSGLN---------PGR--FASGFDSLYENGRPRRVENNSNQVESRKQFQLDLEKIL 646
Query: 659 RGEDNRTTLMIKNIPNKYTSKMLLAAIDEHHKGAYDFVYLPIDFRNKCNVGYAFINMTSP 718
GED+RTTLMIKNIPNKYTSKMLLAAIDE ++G Y+F+YLPIDF+NKCNVGYAFINM +P
Sbjct: 647 NGEDSRTTLMIKNIPNKYTSKMLLAAIDEKNQGTYNFLYLPIDFKNKCNVGYAFINMLNP 706
Query: 719 SLIVPFYQGFNGKKWEKFNSEKVASLAYARIQGKAALVAHFQNSSLMNEDKRCRPILIDT 778
LI+PFY+ FNGKKWEKFNSEKVASLAYARIQGK+AL+AHFQNSSLMNED RCRPI+ DT
Sbjct: 707 ELIIPFYEAFNGKKWEKFNSEKVASLAYARIQGKSALIAHFQNSSLMNEDMRCRPIIFDT 766
Query: 779 -DGPNAGDQ 786
+ P + +Q
Sbjct: 767 PNNPESVEQ 775
>At2g42890 MEI2-like protein (MEI2)
Length = 843
Score = 216 bits (550), Expect = 4e-56
Identities = 131/311 (42%), Positives = 190/311 (60%), Gaps = 23/311 (7%)
Query: 41 SRHESSLFSSSLSDLFSKKLRLSANNAFYGHSVDTIASNYEEEKLS----DSLEELEAQI 96
S + S+FSSSL LF +KL ++ ++++ S D + N + + DSLE++E
Sbjct: 45 SSSDLSMFSSSLPTLFHEKLNMTDSDSWL--SFDESSPNLNKLVIGNSEKDSLEDVEPDA 102
Query: 97 IGNLLP-DEDDLLSGVTDGNNYIICDSNGDDIDELDLFSSNGGFDLGDVENPSS--IERN 153
+ LLP DE++LL G+ D N+ DD++E D+F + GG +L DVE+ + ++ +
Sbjct: 103 LEILLPEDENELLPGLIDELNFTGLPDELDDLEECDVFCTGGGMEL-DVESQDNHAVDAS 161
Query: 154 SEIIS--GVRNSSI-------AG----ENSYGEHPSRTLFVRNIDSDVKDSVLKALFEQF 200
IS G N+ + AG E+ GEHPSRTLFVRNI+S V+DS L ALFE F
Sbjct: 162 GMQISDRGAANAFVPRKRPNTAGRVSVEHPNGEHPSRTLFVRNINSSVEDSELSALFEPF 221
Query: 201 GDIHTFDRTCKHQGSAMISYYDIRAAQNAMRALNNRLFGRKKFDIHYPIPKDSPSRNGVN 260
G+I + CK +G MISYYDIRAA AMRAL N L ++ DIH+ IPK++PS +N
Sbjct: 222 GEIRSLYTACKSRGFVMISYYDIRAAHAAMRALQNTLLRKRTLDIHFSIPKENPSEKDMN 281
Query: 261 QGTLEVFLYDSSISNTELQHILNVYGGIKEIHENPRSQRHKLIEFYDFRAADAALHGINR 320
QGTL +F D+++SN EL + YG I+EI E P + H+ IE+YD R A+ AL +NR
Sbjct: 282 QGTLVIFNVDTTVSNDELLQLFGAYGEIREIRETPNRRFHRFIEYYDVRDAETALKALNR 341
Query: 321 NDTTMKRLKVD 331
++ K +K++
Sbjct: 342 SEIGGKCIKLE 352
Score = 175 bits (444), Expect = 8e-44
Identities = 89/155 (57%), Positives = 109/155 (69%), Gaps = 1/155 (0%)
Query: 632 RVRSRRNVGASNLADMKRYELDIDCIKRGEDNRTTLMIKNIPNKYTSKMLLAAIDEHHKG 691
R+ + + + D RY +D+D I G++ RTTL+IKNIPNKYT KML+A IDE HKG
Sbjct: 647 RIHNHESHNQNQFIDGGRYHIDLDRIASGDEIRTTLIIKNIPNKYTYKMLVAEIDEKHKG 706
Query: 692 AYDFVYLPIDFRNKCNVGYAFINMTSPSLIVPFYQGFNGKKWEKFNSEKVASLAYARIQG 751
YDF+ LP DF+NKCN+G+AFINM SP IVPF Q FNGK WEKFNS KVASLAYA IQG
Sbjct: 707 DYDFLCLPTDFKNKCNMGHAFINMVSPLHIVPFQQTFNGKIWEKFNSGKVASLAYAEIQG 766
Query: 752 KAALVAHFQNSSLMNEDKRCRP-ILIDTDGPNAGD 785
K+AL ++ Q S M E K+ P + DG +A D
Sbjct: 767 KSALASYMQTPSSMKEQKQLFPEVSYHDDGQDAND 801
>At4g18120 predicted protein
Length = 729
Score = 215 bits (548), Expect = 7e-56
Identities = 109/183 (59%), Positives = 134/183 (72%), Gaps = 8/183 (4%)
Query: 603 PHFGLNVHNQRGMVFPGRNH-------MINSFDTYKRVRSRRNVGASNLADMK-RYELDI 654
P+F + +R +F G M++ D + +++ N AD+K +++LD+
Sbjct: 513 PNFKMLSAPRRSQLFTGNGSYLWPAATMVSIDDPLEDGSNQQFDSNGNQADIKIQFQLDL 572
Query: 655 DCIKRGEDNRTTLMIKNIPNKYTSKMLLAAIDEHHKGAYDFVYLPIDFRNKCNVGYAFIN 714
I RGED RTTLMIKNIPNKYT MLLAAIDE + G YDF+YLPIDF+NKCNVGYAFIN
Sbjct: 573 SKIMRGEDPRTTLMIKNIPNKYTRNMLLAAIDEKNSGTYDFLYLPIDFKNKCNVGYAFIN 632
Query: 715 MTSPSLIVPFYQGFNGKKWEKFNSEKVASLAYARIQGKAALVAHFQNSSLMNEDKRCRPI 774
M SP + Y+ FNGKKW+KFNSEKVASLAYARIQGKAAL+AHFQNSSLMNED+RC+PI
Sbjct: 633 MVSPKFTIALYEAFNGKKWDKFNSEKVASLAYARIQGKAALIAHFQNSSLMNEDRRCQPI 692
Query: 775 LID 777
+ D
Sbjct: 693 VFD 695
Score = 197 bits (501), Expect = 2e-50
Identities = 140/384 (36%), Positives = 205/384 (52%), Gaps = 68/384 (17%)
Query: 33 SKEDDIVASRHESSLFSSSLSDLFSKKLRLSANNAFYGHSVDTIASNYEEEK--LSDSLE 90
S+ D AS ++SLFSSSL + + + + Y SVD +AS + + + L+
Sbjct: 9 SRSDHFHASS-DASLFSSSLPLIQHQNINPRDS---YHQSVDEMASGLDHFSGGIGNMLD 64
Query: 91 ELEAQIIGNLLPD-EDDLLSGVTDGNNYIICDSNGDDIDELDLFSSNGGFDL-------- 141
+ ++ IGN+LPD E++L SG+ D N + DD+++ DLF S GG +L
Sbjct: 65 DGDSHPIGNMLPDDEEELFSGLMDDLNLSSLPATLDDLEDYDLFGSGGGLELETDPYDSL 124
Query: 142 -----------GDVEN--PSSIERNSEIISGVRNSSIAGENSYGEHPSRTLFVRNIDSDV 188
+V+N P +I +N GV SIAGE+ YGEHPSRTLFVRNI+S+V
Sbjct: 125 NKGFSRMGFADSNVDNVMPQNIFQN-----GV--GSIAGEHPYGEHPSRTLFVRNINSNV 177
Query: 189 KDSVLKALFEQFGDIHTFDRTCKHQGSAMISYYDIRAAQNAMRALNNRLFGRKKFDIHYP 248
+DS L+ALFE C+H + +L ++K DIH+
Sbjct: 178 EDSELQALFE----------LCEHYKA--------------------KLLKKRKLDIHFS 207
Query: 249 IPKDSPSRNGVNQGTLEVFLYDSSISNTELQHILNVYGGIKEIHENPRSQRHKLIEFYDF 308
IPKD+PS VNQGTL VF S+SN +L++I VYG IKEI E P + HK +EF+D
Sbjct: 208 IPKDNPSEKDVNQGTLVVFNLAPSVSNRDLENIFGVYGEIKEIRETPNKRHHKFVEFFDV 267
Query: 309 RAADAALHGINRNDTTMKRLKVDQMQSTNSESNIIQPMHPEFKQECDLCL--HQKSPLL- 365
R+ADAAL +NR + KR+K++ + + N++ M+PE +Q+ H +SPL
Sbjct: 268 RSADAALKALNRTEIAGKRIKLEHSRPGGARRNMMLQMNPELEQDDSYSYLNHVESPLAS 327
Query: 366 KPTTSFQGREHVHALSI*KQSYIF 389
P +++ H L +S IF
Sbjct: 328 SPIGNWRNSPIDHPLQSFSKSPIF 351
>At3g26120 RNA-binding protein, putative
Length = 615
Score = 127 bits (318), Expect = 3e-29
Identities = 67/145 (46%), Positives = 86/145 (59%), Gaps = 22/145 (15%)
Query: 662 DNRTTLMIKNIPNKYTSKMLLAAID------------EHHK--------GAYDFVYLPID 701
D RTTLMIKNIPNKY+ K+LL +D EH+K +YDFVYLP+D
Sbjct: 405 DPRTTLMIKNIPNKYSQKLLLDMLDKHCIHINEAITEEHNKHESHHQPYSSYDFVYLPMD 464
Query: 702 FRNKCNVGYAFINMTSPSLIVPFYQGFNGKKWEKFNSEKVASLAYARIQGKAALVAHFQN 761
F NKCNVGY F+NMTSP FY+ F+G++WE FNS K+ + YAR+QG L HF++
Sbjct: 465 FNNKCNVGYGFVNMTSPEAAWRFYKAFHGQRWEVFNSHKICQITYARVQGLEDLKEHFKS 524
Query: 762 SSLMNEDKRCRPILIDTDGPNAGDQ 786
S E + P++ P G Q
Sbjct: 525 SKFPCEAELYLPVVFSP--PRDGKQ 547
Score = 79.3 bits (194), Expect = 8e-15
Identities = 50/180 (27%), Positives = 84/180 (45%), Gaps = 24/180 (13%)
Query: 175 PSRTLFVRNIDSDVKDSVLKALFEQFGDIHTFDRTCKHQGSAMISYYDIRAAQNAMRALN 234
P+R+L + ++ DV +S ++ E +GD+ +G + +YDIR A+ A+R +
Sbjct: 104 PTRSLSLISVPRDVTESTVRRDLEVYGDVRGVQMERISEGIVTVHFYDIRDAKRAVREVC 163
Query: 235 NR-----------------------LFGRKKFDIHYPIPKDSPSRNGVNQGTLEVFLYDS 271
R + GR + + +P S G NQGTL +F D
Sbjct: 164 GRHMQQQARGGSVWSSPSTSSARGFVSGRPVW-AQFVVPATSAVPGGCNQGTLVIFNLDP 222
Query: 272 SISNTELQHILNVYGGIKEIHENPRSQRHKLIEFYDFRAADAALHGINRNDTTMKRLKVD 331
+S+ L+ I VYG IKE+ E P + + +EFYD R A A +N + K++ ++
Sbjct: 223 EVSSITLRQIFQVYGPIKELRETPYKKHQRFVEFYDVRDAARAFDRMNGKEIGGKQVVIE 282
Score = 41.2 bits (95), Expect = 0.002
Identities = 20/73 (27%), Positives = 35/73 (47%)
Query: 178 TLFVRNIDSDVKDSVLKALFEQFGDIHTFDRTCKHQGSAMISYYDIRAAQNAMRALNNRL 237
TL + N+D +V L+ +F+ +G I T + + +YD+R A A +N +
Sbjct: 214 TLVIFNLDPEVSSITLRQIFQVYGPIKELRETPYKKHQRFVEFYDVRDAARAFDRMNGKE 273
Query: 238 FGRKKFDIHYPIP 250
G K+ I + P
Sbjct: 274 IGGKQVVIEFSRP 286
>At1g67770 terminal ear1-like protein
Length = 527
Score = 114 bits (286), Expect = 2e-25
Identities = 64/149 (42%), Positives = 85/149 (56%), Gaps = 19/149 (12%)
Query: 634 RSRRNVGASNLADMKRYELDIDCIKRGE--DNRTTLMIKNIPNKYTSKMLLAAIDEHHK- 690
+ ++ NL D + ++ + I GE D RTT+MIKNIPNKYT K+LL +D H K
Sbjct: 305 KKKKKYVKKNLGD-PYFMINENAITGGEFRDGRTTVMIKNIPNKYTQKLLLKMLDTHCKD 363
Query: 691 --------------GAYDFVYLPIDFRNKCNVGYAFINMTSPSLIVPFYQGFNGKKWEKF 736
+YDFVYLPIDF NK NVGY F+NMTSP + Y+ F+ + W F
Sbjct: 364 CNQSVIKEGNKTPMSSYDFVYLPIDFSNKSNVGYGFVNMTSPEAVWRLYKSFHNQHWRDF 423
Query: 737 -NSEKVASLAYARIQGKAALVAHFQNSSL 764
+ K+ + YARIQG +L HF+N L
Sbjct: 424 TTTRKICEVTYARIQGLESLREHFKNVRL 452
Score = 69.7 bits (169), Expect = 6e-12
Identities = 39/163 (23%), Positives = 75/163 (45%), Gaps = 18/163 (11%)
Query: 175 PSRTLFVRNIDSDVKDSVLKALFEQFGDIHTFDRTCKHQGSAMISYYDIRAAQNAMRALN 234
P+R + + + + V ++ L+ E FG++ H+G + +Y++ +Q A +
Sbjct: 78 PTRAVMLLQVPATVTETSLRRDMELFGEVRGVQMERAHEGIVIFHFYNLINSQRAFNEIR 137
Query: 235 NR------------------LFGRKKFDIHYPIPKDSPSRNGVNQGTLEVFLYDSSISNT 276
R L H+ P+ + G NQG+L + + ++S++
Sbjct: 138 YRHMQQQEQQQHFHFTTARGLVSGHSLWAHFVFPQLNAVPEGNNQGSLVIMNLEPTVSSS 197
Query: 277 ELQHILNVYGGIKEIHENPRSQRHKLIEFYDFRAADAALHGIN 319
L+HI VYG +K++ E P + + +EF+D R A AL +N
Sbjct: 198 TLRHIFQVYGEVKQVRETPCKREQRFVEFFDVRDAAKALRVMN 240
Score = 40.8 bits (94), Expect = 0.003
Identities = 17/73 (23%), Positives = 37/73 (50%)
Query: 178 TLFVRNIDSDVKDSVLKALFEQFGDIHTFDRTCKHQGSAMISYYDIRAAQNAMRALNNRL 237
+L + N++ V S L+ +F+ +G++ T + + ++D+R A A+R +N ++
Sbjct: 184 SLVIMNLEPTVSSSTLRHIFQVYGEVKQVRETPCKREQRFVEFFDVRDAAKALRVMNGKV 243
Query: 238 FGRKKFDIHYPIP 250
K I + P
Sbjct: 244 ISGKPMVIQFSRP 256
>At5g07930 putative protein
Length = 282
Score = 90.9 bits (224), Expect = 2e-18
Identities = 48/124 (38%), Positives = 69/124 (54%), Gaps = 14/124 (11%)
Query: 657 IKRGEDNRTTLMIKNIPNKYTSKMLLAAIDEHHK-------------GAYDFVYLPIDFR 703
++ D+ TT+M++NIPN+YT +M++ +D+H + AYDF+YLPIDFR
Sbjct: 127 VESNGDHITTVMLRNIPNRYTREMMIQFMDKHCEEANKSGKNEEFTISAYDFIYLPIDFR 186
Query: 704 NKCNVGYAFINMTSPSLIVPFYQGFNGKKWEKFNSEKVASLAYARIQGKAALVAHFQNSS 763
N GYAF+N T+ + F N K W F S+K + YARIQ LV FQ+ +
Sbjct: 187 TTMNKGYAFVNFTNAKAVSKFKAACNNKPWCHFYSKKELEITYARIQAN-ELVKRFQHMT 245
Query: 764 LMNE 767
E
Sbjct: 246 YPEE 249
>At1g37140 hypothetical protein
Length = 233
Score = 82.4 bits (202), Expect = 9e-16
Identities = 42/105 (40%), Positives = 59/105 (56%), Gaps = 6/105 (5%)
Query: 664 RTTLMIKNIPNKYTSKMLLAAIDEH-----HKGAYDFVYLPIDFRNKCNVGYAFINMTSP 718
RT++M+KNIPN LL +D H K +YDF+YLP+DF + N+GYAF+N TS
Sbjct: 83 RTSVMVKNIPNCLGRMDLLRILDNHCRKHNEKSSYDFLYLPMDFGKRANLGYAFVNFTSS 142
Query: 719 SLIVPFYQGFNGKKWEKFN-SEKVASLAYARIQGKAALVAHFQNS 762
F + F W+ +K+ + A+ QGK L HF+NS
Sbjct: 143 LAAERFRREFENFSWDNIGFRKKICEITVAKYQGKEELTRHFRNS 187
>At3g19130 nuclear acid binding protein, putative
Length = 435
Score = 47.0 bits (110), Expect = 4e-05
Identities = 45/179 (25%), Positives = 81/179 (45%), Gaps = 36/179 (20%)
Query: 97 IGNLLPDEDDLLSGVTDGNNY-------IICDSN------------GDD------IDELD 131
+G+L PD D+L T + Y ++ DSN GD+ + E++
Sbjct: 206 VGDLSPDVTDVLLHETFSDRYPSVKSAKVVIDSNTGRSKGYGFVRFGDENERSRALTEMN 265
Query: 132 -LFSSNGGFDLG------DVENPSSIERNSEIISGVRNSSIA---GENSYGEHPSRTLFV 181
+ SN +G + N + I++G S+ + G S GE + T+FV
Sbjct: 266 GAYCSNRQMRVGIATPKRAIANQQQHSSQAVILAGGHGSNGSMGYGSQSDGESTNATIFV 325
Query: 182 RNIDSDVKDSVLKALFEQFGDIHTFDRTCKHQGSAMISYYDIRAAQNAMRALNNRLFGR 240
ID DV D L+ F QFG++ + + +G + + D ++A++A+ +LN + G+
Sbjct: 326 GGIDPDVIDEDLRQPFSQFGEVVSV-KIPVGKGCGFVQFADRKSAEDAIESLNGTVIGK 383
>At1g71770 polyadenylate-binding protein 5
Length = 668
Score = 46.6 bits (109), Expect = 5e-05
Identities = 40/165 (24%), Positives = 71/165 (42%), Gaps = 10/165 (6%)
Query: 163 SSIAGENSYGEHPSRTLFVRNIDSDVKDSVLKALFEQFGDIHTF----DRTCKHQGSAMI 218
++ A + HP+ +L+V ++D V +S L LF Q +H D T + G A +
Sbjct: 31 AAAAAAEALQTHPNSSLYVGDLDPSVNESHLLDLFNQVAPVHNLRVCRDLTHRSLGYAYV 90
Query: 219 SYYDIRAAQNAMRALNNRLFGRKKFDIHYPIPKDSPSRNGVNQGTLEVFLYDSSISNTEL 278
++ + A AM +LN + I + PS +G + + D+SI N L
Sbjct: 91 NFANPEDASRAMESLNYAPI--RDRPIRIMLSNRDPSTRLSGKGNVFIKNLDASIDNKAL 148
Query: 279 QHILNVYGGIKE----IHENPRSQRHKLIEFYDFRAADAALHGIN 319
+ +G I + RS+ + ++F A AA+ +N
Sbjct: 149 YETFSSFGTILSCKVAMDVVGRSKGYGFVQFEKEETAQAAIDKLN 193
Score = 39.7 bits (91), Expect = 0.007
Identities = 19/67 (28%), Positives = 37/67 (54%), Gaps = 4/67 (5%)
Query: 179 LFVRNIDSDVKDSVLKALFEQFGDIHTFDRTCKHQGSA----MISYYDIRAAQNAMRALN 234
L+++N+D V D LK +F ++G++ + QG + ++Y + A AM+ +N
Sbjct: 330 LYLKNLDDSVNDEKLKEMFSEYGNVTSCKVMMNSQGLSRGFGFVAYSNPEEALLAMKEMN 389
Query: 235 NRLFGRK 241
++ GRK
Sbjct: 390 GKMIGRK 396
Score = 32.7 bits (73), Expect = 0.81
Identities = 32/136 (23%), Positives = 63/136 (45%), Gaps = 19/136 (13%)
Query: 172 GEHPSRT-LFVRNIDSDVKDSVLKALFEQFGDIHTF----DRTCKHQGSAMISYYDIRAA 226
G PS T ++V+N+ ++ D LK F ++GDI + D++ + +++ AA
Sbjct: 219 GAVPSFTNVYVKNLPKEITDDELKKTFGKYGDISSAVVMKDQSGNSRSFGFVNFVSPEAA 278
Query: 227 QNAMRALNNRLFG---------RKKFD----IHYPIPKDSPSRNGVNQGT-LEVFLYDSS 272
A+ +N G +KK D + ++ SR QG+ L + D S
Sbjct: 279 AVAVEKMNGISLGEDVLYVGRAQKKSDREEELRRKFEQERISRFEKLQGSNLYLKNLDDS 338
Query: 273 ISNTELQHILNVYGGI 288
+++ +L+ + + YG +
Sbjct: 339 VNDEKLKEMFSEYGNV 354
>At5g47620 RNA-binding protein-like
Length = 431
Score = 45.4 bits (106), Expect = 1e-04
Identities = 39/147 (26%), Positives = 63/147 (42%), Gaps = 14/147 (9%)
Query: 144 VENPSSIERNSEIISGVRNSSIAGENSYGEHPSRTLFVRNIDSDVKDSVLKALFEQFGDI 203
VE ++ R+ ++ NSS+ G S G S+ +FV + S V ++ K F QFG I
Sbjct: 75 VEAKKAVPRDDHVVFNKSNSSLQG--SPGPSNSKKIFVGGLASSVTEAEFKKYFAQFGMI 132
Query: 204 HTF-----DRTCKHQGSAMISYYDIRAAQNAMRALNNRLFGRKKFDIHYPIPKDSPSRNG 258
RT + +G ISY A ++ + L G K ++ +PKD
Sbjct: 133 TDVVVMYDHRTQRPRGFGFISYDSEEAVDKVLQKTFHELNG-KMVEVKLAVPKDMALNTM 191
Query: 259 VNQGTLEVFLYDSSISNTELQHILNVY 285
NQ + +S + + +LN Y
Sbjct: 192 RNQMNV------NSFGTSRISSLLNEY 212
>At3g16380 putative poly(A) binding protein
Length = 537
Score = 43.9 bits (102), Expect = 4e-04
Identities = 39/169 (23%), Positives = 67/169 (39%), Gaps = 21/169 (12%)
Query: 172 GEHPSRTLFVRNIDSDVKDSVLKALFEQFGDIHTF----DRTCKHQGSAMISYYDIRAAQ 227
G S ++V+N+ V D L LF Q+G + + D + +G +++ + A+
Sbjct: 197 GNQDSTNVYVKNLIETVTDDCLHTLFSQYGTVSSVVVMRDGMGRSRGFGFVNFCNPENAK 256
Query: 228 NAMRALNNRLFGRKKFDIHYPIPKDSP-------------SRNGVNQGTLEVFLYDSSIS 274
AM +L G KK + + KD ++ + L V S++
Sbjct: 257 KAMESLCGLQLGSKKLFVGKALKKDERREMLKQKFSDNFIAKPNMRWSNLYVKNLSESMN 316
Query: 275 NTELQHILNVYGGIKE----IHENPRSQRHKLIEFYDFRAADAALHGIN 319
T L+ I YG I HEN RS+ + F + + A +N
Sbjct: 317 ETRLREIFGCYGQIVSAKVMCHENGRSKGFGFVCFSNCEESKQAKRYLN 365
Score = 39.3 bits (90), Expect = 0.009
Identities = 27/118 (22%), Positives = 57/118 (47%), Gaps = 5/118 (4%)
Query: 179 LFVRNIDSDVKDSVLKALFEQFGDI---HTFDRTCKHQGSAMISYYDIRAAQNAMRALNN 235
L+V+N+DS + S L+ +F FG I + + +G + + ++A +A AL+
Sbjct: 114 LYVKNLDSSITSSCLERMFCPFGSILSCKVVEENGQSKGFGFVQFDTEQSAVSARSALHG 173
Query: 236 RLFGRKKFDIHYPIPKDSPSRNGVNQGTLEVFLYD--SSISNTELQHILNVYGGIKEI 291
+ KK + I KD + NQ + V++ + ++++ L + + YG + +
Sbjct: 174 SMVYGKKLFVAKFINKDERAAMAGNQDSTNVYVKNLIETVTDDCLHTLFSQYGTVSSV 231
>At1g22760 putative polyA-binding protein, PAB3
Length = 660
Score = 43.1 bits (100), Expect = 6e-04
Identities = 41/156 (26%), Positives = 71/156 (45%), Gaps = 14/156 (8%)
Query: 174 HPSRTLFVRNIDSDVKDSVLKALFEQFGDIHTFDRTCKHQ-----GSAMISYYDIRAAQN 228
HP+ +L+ ++D V ++ L LF+ ++ + R C+ Q G A I++ + A
Sbjct: 46 HPNSSLYAGDLDPKVTEAHLFDLFKHVANVVSV-RVCRDQNRRSLGYAYINFSNPNDAYR 104
Query: 229 AMRALN-NRLFGRKKFDIHYPIPKDSPSRNGVNQGTLEVFLYDSSISNTELQHILNVYGG 287
AM ALN LF R I + PS +G + + D+SI N L + +G
Sbjct: 105 AMEALNYTPLFDR---PIRIMLSNRDPSTRLSGKGNIFIKNLDASIDNKALFETFSSFGT 161
Query: 288 IKE----IHENPRSQRHKLIEFYDFRAADAALHGIN 319
I + RS+ + ++F +A AA+ +N
Sbjct: 162 ILSCKVAMDVTGRSKGYGFVQFEKEESAQAAIDKLN 197
Score = 39.7 bits (91), Expect = 0.007
Identities = 19/77 (24%), Positives = 39/77 (49%), Gaps = 4/77 (5%)
Query: 169 NSYGEHPSRTLFVRNIDSDVKDSVLKALFEQFGDIHTFDRTCKHQGSA----MISYYDIR 224
N + + L+++N+D V D LK +F ++G++ + QG + ++Y +
Sbjct: 324 NRFEKSQGANLYLKNLDDSVDDEKLKEMFSEYGNVTSSKVMLNPQGMSRGFGFVAYSNPE 383
Query: 225 AAQNAMRALNNRLFGRK 241
A A+ +N ++ GRK
Sbjct: 384 EALRALSEMNGKMIGRK 400
Score = 36.6 bits (83), Expect = 0.056
Identities = 37/184 (20%), Positives = 75/184 (40%), Gaps = 9/184 (4%)
Query: 179 LFVRNIDSDVKDSVLKALFEQFGDIHT----FDRTCKHQGSAMISYYDIRAAQNAMRALN 234
+F++N+D+ + + L F FG I + D T + +G + + +AQ A+ LN
Sbjct: 138 IFIKNLDASIDNKALFETFSSFGTILSCKVAMDVTGRSKGYGFVQFEKEESAQAAIDKLN 197
Query: 235 NRLFGRKKFDIHYPIPKDSPSR--NGVNQGTLEVFLYD--SSISNTELQHILNVYGGIKE 290
L K+ + + I + +R N V++ + I EL+ +G I
Sbjct: 198 GMLMNDKQVFVGHFIRRQERARDENTPTPRFTNVYVKNLPKEIGEDELRKTFGKFGVISS 257
Query: 291 -IHENPRSQRHKLIEFYDFRAADAALHGINRNDTTMKRLKVDQMQSTNSESNIIQPMHPE 349
+ +S + F +F +AA + + + V + +S + + +
Sbjct: 258 AVVMRDQSGNSRCFGFVNFECTEAAASAVEKMNGISLGDDVLYVGRAQKKSEREEELRRK 317
Query: 350 FKQE 353
F+QE
Sbjct: 318 FEQE 321
>At2g47580 small nuclear ribonucleoprotein U1A
Length = 250
Score = 42.7 bits (99), Expect = 8e-04
Identities = 26/79 (32%), Positives = 43/79 (53%), Gaps = 6/79 (7%)
Query: 175 PSRTLFVRNIDSDVKDSVLK----ALFEQFGDIHTF--DRTCKHQGSAMISYYDIRAAQN 228
P++T+++ N++ VK LK A+F QFG I +T KH+G A + + + +A
Sbjct: 16 PNQTIYINNLNEKVKLDELKKSLNAVFSQFGKILEILAFKTFKHKGQAWVVFDNTESAST 75
Query: 229 AMRALNNRLFGRKKFDIHY 247
A+ +NN F K+ I Y
Sbjct: 76 AIAKMNNFPFYDKEMRIQY 94
>At3g61860 ARGININE/SERINE-RICH SPLICING FACTOR RSP31
Length = 264
Score = 41.6 bits (96), Expect = 0.002
Identities = 23/66 (34%), Positives = 39/66 (58%), Gaps = 3/66 (4%)
Query: 177 RTLFVRNIDSDVKDSVLKALFEQFGDIHTFDRTCKHQGSAMISYYDIRAAQNAMRALNNR 236
R +FV N + + + S L+ LF+++G + DR G A + + D R A++A+R L+N
Sbjct: 2 RPVFVGNFEYETRQSDLERLFDKYGRV---DRVDMKSGYAFVYFEDERDAEDAIRKLDNF 58
Query: 237 LFGRKK 242
FG +K
Sbjct: 59 PFGYEK 64
>At4g16290 FCA delta protein
Length = 533
Score = 40.8 bits (94), Expect = 0.003
Identities = 44/192 (22%), Positives = 84/192 (42%), Gaps = 24/192 (12%)
Query: 144 VENPSSIERNSEIISGVRNSSIAGENSYGEHPSRTLFVRNIDSDVKDSVLKALFEQFGDI 203
V+ P S ++ IS + S G + + LFV ++ + ++ FEQ G++
Sbjct: 89 VQQPLSGQKRGYPISD--HGSFTGTDVSDRSSTVKLFVGSVPRTATEEEIRPYFEQHGNV 146
Query: 204 HTF-----DRTCKHQGSAMISYYDIRAAQNAMRALNNRLF---GRKKFDIHYPIPKDSPS 255
RT + QG + Y + A A+RAL+N++ G + Y
Sbjct: 147 LEVALIKDKRTGQQQGCCFVKYATSKDADRAIRALHNQITLPGGTGPVQVRYA----DGE 202
Query: 256 RNGVNQGTLEVFLYDSSI----SNTELQHILNVYGGIKEIH----ENPRSQRHKLIEFYD 307
R + GTLE L+ S+ + E++ I +G +++++ E +S+ +++
Sbjct: 203 RERI--GTLEFKLFVGSLNKQATEKEVEEIFLQFGHVEDVYLMRDEYRQSRGCGFVKYSS 260
Query: 308 FRAADAALHGIN 319
A AA+ G+N
Sbjct: 261 KETAMAAIDGLN 272
>At4g16280 FCA gamma protein
Length = 747
Score = 40.8 bits (94), Expect = 0.003
Identities = 44/192 (22%), Positives = 84/192 (42%), Gaps = 24/192 (12%)
Query: 144 VENPSSIERNSEIISGVRNSSIAGENSYGEHPSRTLFVRNIDSDVKDSVLKALFEQFGDI 203
V+ P S ++ IS + S G + + LFV ++ + ++ FEQ G++
Sbjct: 89 VQQPLSGQKRGYPISD--HGSFTGTDVSDRSSTVKLFVGSVPRTATEEEIRPYFEQHGNV 146
Query: 204 HTF-----DRTCKHQGSAMISYYDIRAAQNAMRALNNRLF---GRKKFDIHYPIPKDSPS 255
RT + QG + Y + A A+RAL+N++ G + Y
Sbjct: 147 LEVALIKDKRTGQQQGCCFVKYATSKDADRAIRALHNQITLPGGTGPVQVRYA----DGE 202
Query: 256 RNGVNQGTLEVFLYDSSI----SNTELQHILNVYGGIKEIH----ENPRSQRHKLIEFYD 307
R + GTLE L+ S+ + E++ I +G +++++ E +S+ +++
Sbjct: 203 RERI--GTLEFKLFVGSLNKQATEKEVEEIFLQFGHVEDVYLMRDEYRQSRGCGFVKYSS 260
Query: 308 FRAADAALHGIN 319
A AA+ G+N
Sbjct: 261 KETAMAAIDGLN 272
>At2g46610 arginine/serine-rich splicing factor like protein
Length = 250
Score = 40.8 bits (94), Expect = 0.003
Identities = 24/73 (32%), Positives = 39/73 (52%), Gaps = 5/73 (6%)
Query: 177 RTLFVRNIDSDVKDSVLKALFEQFGDIHTFDRTCKHQGSAMISYYDIRAAQNAMRALNNR 236
R ++V N D D + S L+ LF +FG + D G A + + D R A++A+R +N
Sbjct: 2 RHVYVGNFDYDTRHSDLERLFSKFGRVKRVD---MKSGYAFVYFEDERDAEDAIRRTDNT 58
Query: 237 LF--GRKKFDIHY 247
F GR+K + +
Sbjct: 59 TFGYGRRKLSVEW 71
>At1g06960 putative spliceosomal protein U2B
Length = 228
Score = 40.0 bits (92), Expect = 0.005
Identities = 25/80 (31%), Positives = 44/80 (54%), Gaps = 8/80 (10%)
Query: 175 PSRTLFVRNIDSDVKDSVLK----ALFEQFG---DIHTFDRTCKHQGSAMISYYDIRAAQ 227
P++++++++I+ +K LK LF QFG D+ +T K +G A + + ++ AA
Sbjct: 8 PNQSIYIKHINEKIKKEELKRSLYCLFSQFGRLLDVVAL-KTPKLRGQAWVVFTEVTAAS 66
Query: 228 NAMRALNNRLFGRKKFDIHY 247
NA+R + N F K I Y
Sbjct: 67 NAVRQMQNFPFYDKPMRIQY 86
Score = 31.6 bits (70), Expect = 1.8
Identities = 17/59 (28%), Positives = 27/59 (44%)
Query: 175 PSRTLFVRNIDSDVKDSVLKALFEQFGDIHTFDRTCKHQGSAMISYYDIRAAQNAMRAL 233
P+ LF+ N+ + +L+ LFEQ+ G A + Y D + AM+AL
Sbjct: 152 PNNILFIHNLPIETNSMMLQLLFEQYPGFKEIRMIEAKPGIAFVEYEDDVQSSMAMQAL 210
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.324 0.139 0.419
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 18,316,669
Number of Sequences: 26719
Number of extensions: 827760
Number of successful extensions: 2965
Number of sequences better than 10.0: 114
Number of HSP's better than 10.0 without gapping: 28
Number of HSP's successfully gapped in prelim test: 86
Number of HSP's that attempted gapping in prelim test: 2805
Number of HSP's gapped (non-prelim): 188
length of query: 786
length of database: 11,318,596
effective HSP length: 107
effective length of query: 679
effective length of database: 8,459,663
effective search space: 5744111177
effective search space used: 5744111177
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 64 (29.3 bits)
Medicago: description of AC146333.19


