

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146332.5 + phase: 0
(220 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
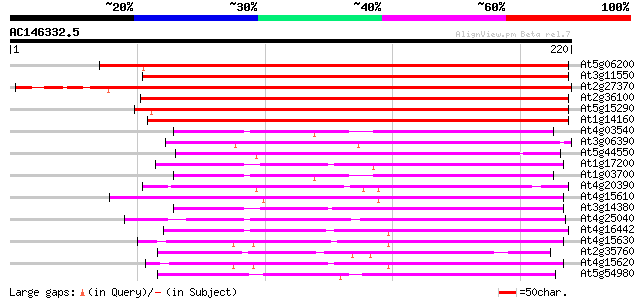
Score E
Sequences producing significant alignments: (bits) Value
At5g06200 putative protein 210 4e-55
At3g11550 unknown protein 205 2e-53
At2g27370 unknown protein 202 1e-52
At2g36100 unknown protein 195 2e-50
At5g15290 unknown protein 171 2e-43
At1g14160 unknown protein 140 7e-34
At4g03540 hypothetical protein 80 1e-15
At3g06390 unknown protein 79 2e-15
At5g44550 unknown protein 77 7e-15
At1g17200 unknown protein 74 8e-14
At1g03700 putative protein 73 1e-13
At4g20390 putative protein 71 5e-13
At4g15610 unknown protein 70 7e-13
At3g14380 unknown protein 66 2e-11
At4g25040 unknown protein 59 2e-09
At4g16442 unknown protein 59 3e-09
At4g15630 unknown protein 59 3e-09
At2g35760 unknown protein 56 2e-08
At4g15620 unknown protein (At4g15620) 53 1e-07
At5g54980 unknown protein (At5g54980) 49 3e-06
>At5g06200 putative protein
Length = 202
Score = 210 bits (535), Expect = 4e-55
Identities = 109/191 (57%), Positives = 139/191 (72%), Gaps = 7/191 (3%)
Query: 36 PPHAATVVTTKATPLQ-------KGGMKKGIAILDFILRLGAIGAALGAAVIMGTNEQIL 88
P +++V+ KA L GG K+G++I DF+LRL AI AAL AA MGT+++ L
Sbjct: 11 PAESSSVIKGKAPLLGLARDHTGSGGYKRGLSIFDFLLRLAAIVAALAAAATMGTSDETL 70
Query: 89 PFFTQFLQFHAQWDDFPMFKFFVVANGAAAGFLILSLPFSIVCIVRPLAAGPRFLLVIVD 148
PFFTQFLQF A +DD P F+FFVVA AG+L+LSLPFS+V IVRPLA PR LL+++D
Sbjct: 71 PFFTQFLQFEASYDDLPTFQFFVVAIAIVAGYLVLSLPFSVVTIVRPLAVAPRLLLLVLD 130
Query: 149 LVLMALVVAAASSAAAVVYLAHNGSQDANWNAICQQFTDFCQGSSLAVVASFVASVFLAC 208
+AL AAAS+AAA+VYLAHNG+ + NW ICQQF DFCQ +S AVV++F + FLA
Sbjct: 131 TAALALDTAAASAAAAIVYLAHNGNTNTNWLPICQQFGDFCQKTSGAVVSAFASVTFLAI 190
Query: 209 LVVVSSVALKR 219
LVV+S V+LKR
Sbjct: 191 LVVISGVSLKR 201
>At3g11550 unknown protein
Length = 204
Score = 205 bits (521), Expect = 2e-53
Identities = 102/167 (61%), Positives = 128/167 (76%)
Query: 53 GGMKKGIAILDFILRLGAIGAALGAAVIMGTNEQILPFFTQFLQFHAQWDDFPMFKFFVV 112
GG +G+AI DF+LRL AI AAL AA MGT+++ LPFFTQFLQF A +DD P F+FFV+
Sbjct: 37 GGYNRGLAIFDFLLRLAAIVAALAAAATMGTSDETLPFFTQFLQFEASYDDLPTFQFFVI 96
Query: 113 ANGAAAGFLILSLPFSIVCIVRPLAAGPRFLLVIVDLVLMALVVAAASSAAAVVYLAHNG 172
A G+L+LSLP S+V I+RPLA PR LL+++D ++AL AAASSAAA+ YLAH+G
Sbjct: 97 AMALVGGYLVLSLPISVVTILRPLATAPRLLLLVLDTGVLALNTAAASSAAAISYLAHSG 156
Query: 173 SQDANWNAICQQFTDFCQGSSLAVVASFVASVFLACLVVVSSVALKR 219
+Q+ NW ICQQF DFCQ SS AVV++FV+ VF LVV+S VALKR
Sbjct: 157 NQNTNWLPICQQFGDFCQKSSGAVVSAFVSVVFFTILVVISGVALKR 203
>At2g27370 unknown protein
Length = 221
Score = 202 bits (514), Expect = 1e-52
Identities = 111/220 (50%), Positives = 144/220 (65%), Gaps = 9/220 (4%)
Query: 3 SRREVEESSTAPILESKRTRSNGKGKSIDGDHSPP--HAATVVTTKATPLQKGGMKKGIA 60
SRRE EE PI++ + GKGK+ +PP + + K + G K+G+A
Sbjct: 8 SRREEEE----PIVQRPKL-DKGKGKA--HVFAPPMNYNRIMDKHKQEKMSPAGWKRGVA 60
Query: 61 ILDFILRLGAIGAALGAAVIMGTNEQILPFFTQFLQFHAQWDDFPMFKFFVVANGAAAGF 120
I DF+LRL A A+ AA M T E+ LPFFTQFLQF A + D P FV+ N G+
Sbjct: 61 IFDFVLRLIAAITAMAAAAKMATTEETLPFFTQFLQFQADYTDLPTMSSFVIVNSIVGGY 120
Query: 121 LILSLPFSIVCIVRPLAAGPRFLLVIVDLVLMALVVAAASSAAAVVYLAHNGSQDANWNA 180
L LSLPFSIVCI+RPLA PR L++ D V+M L + AAS++AA+VYLAHNG+ +NW
Sbjct: 121 LTLSLPFSIVCILRPLAVPPRLFLILCDTVMMGLTLMAASASAAIVYLAHNGNSSSNWLP 180
Query: 181 ICQQFTDFCQGSSLAVVASFVASVFLACLVVVSSVALKRT 220
+CQQF DFCQG+S AVVASF+A+ L LV++S+ ALKRT
Sbjct: 181 VCQQFGDFCQGTSGAVVASFIAATLLMFLVILSAFALKRT 220
>At2g36100 unknown protein
Length = 206
Score = 195 bits (495), Expect = 2e-50
Identities = 93/168 (55%), Positives = 124/168 (73%)
Query: 52 KGGMKKGIAILDFILRLGAIGAALGAAVIMGTNEQILPFFTQFLQFHAQWDDFPMFKFFV 111
+GG K+G+AI DF+LRL AI +GAA +M T E+ LPFFTQFLQF A +DD P F++FV
Sbjct: 38 RGGAKRGLAIFDFLLRLAAIAVTIGAASVMYTAEETLPFFTQFLQFQAGYDDLPAFQYFV 97
Query: 112 VANGAAAGFLILSLPFSIVCIVRPLAAGPRFLLVIVDLVLMALVVAAASSAAAVVYLAHN 171
+A A +L+LSLPFSIV IVRP A PR +L+I D +++ L +AA++AA++ YLAHN
Sbjct: 98 IAVAVVASYLVLSLPFSIVSIVRPHAVAPRLILLICDTLVVTLNTSAAAAAASITYLAHN 157
Query: 172 GSQDANWNAICQQFTDFCQGSSLAVVASFVASVFLACLVVVSSVALKR 219
G+Q NW ICQQF DFCQ S AVVA +A +F L+++S++ALKR
Sbjct: 158 GNQSTNWLPICQQFGDFCQNVSTAVVADSIAILFFIVLIIISAIALKR 205
>At5g15290 unknown protein
Length = 187
Score = 171 bits (434), Expect = 2e-43
Identities = 84/171 (49%), Positives = 121/171 (70%), Gaps = 1/171 (0%)
Query: 50 LQKGG-MKKGIAILDFILRLGAIGAALGAAVIMGTNEQILPFFTQFLQFHAQWDDFPMFK 108
+QK G M + IAIL+FILR+ A +G+A++MGT + LPFFTQF++F A+++D P
Sbjct: 15 IQKSGLMSRRIAILEFILRIVAFFNTIGSAILMGTTHETLPFFTQFIRFQAEYNDLPALT 74
Query: 109 FFVVANGAAAGFLILSLPFSIVCIVRPLAAGPRFLLVIVDLVLMALVVAAASSAAAVVYL 168
FFVVAN +G+LILSL + V IV+ R LL+I+D+ ++ L+ + ASSAAA+VYL
Sbjct: 75 FFVVANAVVSGYLILSLTLAFVHIVKRKTQNTRILLIILDVAMLGLLTSGASSAAAIVYL 134
Query: 169 AHNGSQDANWNAICQQFTDFCQGSSLAVVASFVASVFLACLVVVSSVALKR 219
AHNG+ NW AICQQF FC+ S +++ SF+A V L L+++S++AL R
Sbjct: 135 AHNGNNKTNWFAICQQFNSFCERISGSLIGSFIAIVLLILLILLSAIALSR 185
>At1g14160 unknown protein
Length = 209
Score = 140 bits (352), Expect = 7e-34
Identities = 70/165 (42%), Positives = 105/165 (63%)
Query: 55 MKKGIAILDFILRLGAIGAALGAAVIMGTNEQILPFFTQFLQFHAQWDDFPMFKFFVVAN 114
+ KGI++L F+LRL A+ +G+A+ MGT + + +Q + ++ D P FFVVAN
Sbjct: 44 INKGISVLGFVLRLFAVFGTIGSALAMGTTHESVVSLSQLVLLKVKYSDLPTLMFFVVAN 103
Query: 115 GAAAGFLILSLPFSIVCIVRPLAAGPRFLLVIVDLVLMALVVAAASSAAAVVYLAHNGSQ 174
+ G+L+LSLP SI I A R +L++VD V++ALV + AS+A A VYLAH G+
Sbjct: 104 AISGGYLVLSLPVSIFHIFSTQAKTSRIILLVVDTVMLALVSSGASAATATVYLAHEGNT 163
Query: 175 DANWNAICQQFTDFCQGSSLAVVASFVASVFLACLVVVSSVALKR 219
ANW ICQQF FC+ S +++ SF A + L +V+ S+++L R
Sbjct: 164 TANWPPICQQFDGFCERISGSLIGSFCAVILLMLIVINSAISLSR 208
>At4g03540 hypothetical protein
Length = 164
Score = 79.7 bits (195), Expect = 1e-15
Identities = 50/151 (33%), Positives = 79/151 (52%), Gaps = 13/151 (8%)
Query: 65 ILRLGAIGAALGAAVIMGTNEQILPFFTQFLQFHAQWDDFPMFKFFVVANGAAA--GFLI 122
+LRL A GAAL A ++M T+ + F + A++ D FK+FV+AN + FL+
Sbjct: 12 VLRLAAFGAALAALIVMITSRERASFLA--ISLEAKYTDMAAFKYFVIANAVVSVYSFLV 69
Query: 123 LSLPFSIVCIVRPLAAGPRFLLVIVDLVLMALVVAAASSAAAVVYLAHNGSQDANWNAIC 182
L LP + +V++DLV+ L+ ++ S+A AV + G+ +A W IC
Sbjct: 70 LFLPKESLLWK---------FVVVLDLVMTMLLTSSLSAALAVAQVGKKGNANAGWLPIC 120
Query: 183 QQFTDFCQGSSLAVVASFVASVFLACLVVVS 213
Q FC + A++A FVA V L++ S
Sbjct: 121 GQVPKFCDQITGALIAGFVALVLYVLLLLYS 151
>At3g06390 unknown protein
Length = 199
Score = 79.0 bits (193), Expect = 2e-15
Identities = 49/164 (29%), Positives = 89/164 (53%), Gaps = 6/164 (3%)
Query: 62 LDFILRLGAIGAALGAAVIMGTNEQI----LPFFTQFLQFHAQWDDFPMFKFFVVANGAA 117
+D I R+ A L A ++M T++Q LP + A+++D P F +FVVA A
Sbjct: 37 IDIITRVLLFSATLTALIVMVTSDQTEMTQLPGVSSPAPVSAEFNDSPAFIYFVVALVVA 96
Query: 118 AGFLILSLPFSIVCIVRP-LAAGPRFLLVIVDLVLMALVVAAASSAAAVVYLAHNGSQDA 176
+ + ++S SI +++P A L +D+V++ ++ +A +A V Y+A G+++
Sbjct: 97 SFYALISTLVSISLLLKPEFTAQFSIYLASLDMVMLGILASATGTAGGVAYIALKGNEEV 156
Query: 177 NWNAICQQFTDFCQGSSLAVVASFVASVFLACLVVVSSVALKRT 220
WN IC + FC+ + ++ S AS+ L L + S+++ KRT
Sbjct: 157 GWNKICNVYDKFCRYIATSLALSLFASLLLLVLSIWSALS-KRT 199
>At5g44550 unknown protein
Length = 197
Score = 77.0 bits (188), Expect = 7e-15
Identities = 48/158 (30%), Positives = 82/158 (51%), Gaps = 8/158 (5%)
Query: 66 LRLGAIGAALGAAVIMGTNEQILPFFTQFL-------QFHAQWDDFPMFKFFVVANGAAA 118
LRL A A L AA++MG N++ F + F A++D P F FFVVAN +
Sbjct: 22 LRLLAFSATLSAAIVMGLNKETKTFIVGKVGNTPIQATFTAKFDHTPAFVFFVVANAMVS 81
Query: 119 GFLILSLPFSIVCIVRPLAAGPRFLLVIVDLVLMALVVAAASSAAAVVYLAHNGSQDANW 178
+L + I + I+D++ + L+ AAA++AA + + NG++ A W
Sbjct: 82 FHNLLMIALQIFGGKMEFTGFRLLSVAILDMLNVTLISAAANAAAFMAEVGKNGNKHARW 141
Query: 179 NAICQQFTDFCQGSSLAVVASFVASVFLACLVVVSSVA 216
+ IC +F +C + A++A+F A V L ++ +S++
Sbjct: 142 DKICDRFATYCDHGAGALIAAF-AGVILMLIISAASIS 178
>At1g17200 unknown protein
Length = 204
Score = 73.6 bits (179), Expect = 8e-14
Identities = 47/161 (29%), Positives = 81/161 (50%), Gaps = 9/161 (5%)
Query: 58 GIAILDFILRLGAIGAALGAAVIMGTNEQILPFFTQFLQFHAQWDDFPMFKFFVVANGAA 117
GI + +LRL +G + A V+M + + F + + + F++ V ANG
Sbjct: 29 GIRTAETMLRLAPVGLCVAALVVMLKDSETNEFGS------ISYSNLTAFRYLVHANGIC 82
Query: 118 AGFLILSLPFSIVCIVRPLAAGPR-FLLVIVDLVLMALVVAAASSAAAVVYLAHNGSQDA 176
AG+ +LS +I + R + PR + +D +L LV+AA + +A V+YLA+NG
Sbjct: 83 AGYSLLSA--AIAAMPRSSSTMPRVWTFFCLDQLLTYLVLAAGAVSAEVLYLAYNGDSAI 140
Query: 177 NWNAICQQFTDFCQGSSLAVVASFVASVFLACLVVVSSVAL 217
W+ C + FC ++ +V+ +F F L ++SS L
Sbjct: 141 TWSDACSSYGGFCHRATASVIITFFVVCFYIVLSLISSYKL 181
>At1g03700 putative protein
Length = 164
Score = 72.8 bits (177), Expect = 1e-13
Identities = 48/151 (31%), Positives = 76/151 (49%), Gaps = 13/151 (8%)
Query: 65 ILRLGAIGAALGAAVIMGTNEQILPFFTQFLQFHAQWDDFPMFKFFVVANGAAA--GFLI 122
+LR A AALGA + M T+ + FF + A++ D FK+FV+AN FL+
Sbjct: 12 VLRFAAFCAALGAVIAMITSRERSSFFV--ISLVAKYSDLAAFKYFVIANAIVTVYSFLV 69
Query: 123 LSLPFSIVCIVRPLAAGPRFLLVIVDLVLMALVVAAASSAAAVVYLAHNGSQDANWNAIC 182
L LP + +V++DL++ L+ ++ S+A AV + G+ +A W IC
Sbjct: 70 LFLPKESLLWK---------FVVVLDLMVTMLLTSSLSAAVAVAQVGKRGNANAGWLPIC 120
Query: 183 QQFTDFCQGSSLAVVASFVASVFLACLVVVS 213
Q FC + A++A VA V L++ S
Sbjct: 121 GQVPRFCDQITGALIAGLVALVLYVFLLIFS 151
>At4g20390 putative protein
Length = 197
Score = 70.9 bits (172), Expect = 5e-13
Identities = 54/176 (30%), Positives = 89/176 (49%), Gaps = 15/176 (8%)
Query: 53 GGMKKGIAILDFILRLGAIGAALGAAVIMGTNEQILPFFTQFL-------QFHAQWDDFP 105
GG K +L LR+ A A L AA++M N++ + A++ P
Sbjct: 10 GGTTKSWKLL-LGLRIFAFMATLAAAIVMSLNKETKTLVVATIGTVPIKATLTAKFQHTP 68
Query: 106 MFKFFVVANGAAAGFLILSLPFSIVCIVRPLA-AGPRFL-LVIVDLVLMALVVAAASSAA 163
F FFV+AN + +L + I R L G R L + I+D++ LV AAA++A
Sbjct: 69 AFVFFVIANVMVSFHNLLMIVVQIFS--RKLEYKGLRLLSIAILDMLNATLVSAAANAAV 126
Query: 164 AVVYLAHNGSQDANWNAICQQFTDFCQGSSLAVVASFVASVFLACLVVVSSVALKR 219
V L NG++ A WN +C +FT +C + A++A+F + +++VS+V++ R
Sbjct: 127 FVAELGKNGNKHAKWNKVCDRFTTYCDHGAGAIIAAFAGVIL---MLLVSAVSISR 179
>At4g15610 unknown protein
Length = 193
Score = 70.5 bits (171), Expect = 7e-13
Identities = 48/182 (26%), Positives = 85/182 (46%), Gaps = 4/182 (2%)
Query: 40 ATVVTTKATPLQKGGMKKGIAILDFILRLGAIGAALGAAVIMGTNEQILPFFTQFLQFH- 98
+T+ T ++T G K ++ +LR A L + V+M T++Q F
Sbjct: 7 STLDTERSTAPGTGTTTKSCSMTQVVLRFVLFAATLTSIVVMVTSKQTKNIFLPGTPIRI 66
Query: 99 --AQWDDFPMFKFFVVANGAAAGFLILSLPFSIVCIVRPLAAGPRFL-LVIVDLVLMALV 155
A++ + P +FVVA A + I+S ++ + + L L I+D V++ +V
Sbjct: 67 PAAEFTNSPALIYFVVALSVACFYSIVSTFVTVSAFKKHSCSAVLLLNLAIMDAVMVGIV 126
Query: 156 VAAASSAAAVVYLAHNGSQDANWNAICQQFTDFCQGSSLAVVASFVASVFLACLVVVSSV 215
+A + V YL G+++ W IC + FC+ A+ S ASV L L ++S +
Sbjct: 127 ASATGAGGGVAYLGLKGNKEVRWGKICHIYDKFCRHVGGAIAVSLFASVVLLLLSIISVL 186
Query: 216 AL 217
+L
Sbjct: 187 SL 188
>At3g14380 unknown protein
Length = 178
Score = 65.9 bits (159), Expect = 2e-11
Identities = 42/153 (27%), Positives = 78/153 (50%), Gaps = 7/153 (4%)
Query: 65 ILRLGAIGAALGAAVIMGTNEQILPFFTQFLQFHAQWDDFPMFKFFVVANGAAAGFLILS 124
+LR+ ++ ++ VIM N F + + + F + V ANG A + +LS
Sbjct: 26 VLRVASMALSITGLVIMIKNSISNEFGS------VSYSNIGAFMYLVSANGVCAAYSLLS 79
Query: 125 LPFSIVCIVRPLAAGPRFLLVIVDLVLMALVVAAASSAAAVVYLAHNGSQDANWNAICQQ 184
+I+ + P++ L ++D V+ +V+AA + +A VYLA+ G+ W++ C
Sbjct: 80 A-LAILALPCPISKVQVRTLFLLDQVVTYVVLAAGAVSAETVYLAYYGNIPITWSSACDS 138
Query: 185 FTDFCQGSSLAVVASFVASVFLACLVVVSSVAL 217
+ FC + ++VV +FV S+ L ++SS L
Sbjct: 139 YGSFCHNALISVVFTFVVSLLYMLLSLISSYRL 171
>At4g25040 unknown protein
Length = 170
Score = 59.3 bits (142), Expect = 2e-09
Identities = 42/173 (24%), Positives = 82/173 (47%), Gaps = 12/173 (6%)
Query: 46 KATPLQKGGMKKGIAILDFILRLGAIGAALGAAVIMGTNEQILPFFTQFLQFHAQWDDFP 105
+ TPL G++ + +L IGAA+ + +M TN ++ + + F A++
Sbjct: 8 RRTPLLNLGVQVSMRVL-------TIGAAMASMWVMITNREVASVYG--IAFEAKYSYSS 58
Query: 106 MFKFFVVANGAAAGFLILSLPFSIVCIVRPLAAGPRFLLVIVDLVLMALVVAAASSAAAV 165
F++ V A A + +L ++ + + R G F L DL+ ++A S+A A
Sbjct: 59 AFRYLVYAQIAVCAATLFTLVWACLAVRR---RGLVFALFFFDLLTTLTAISAFSAAFAE 115
Query: 166 VYLAHNGSQDANWNAICQQFTDFCQGSSLAVVASFVASVFLACLVVVSSVALK 218
Y+ G++ A W IC +C ++++ SF + + L L V+++ A +
Sbjct: 116 GYVGKYGNKQAGWLPICGYVHGYCSRVTISLAMSFASFILLFILTVLTASAAR 168
>At4g16442 unknown protein
Length = 182
Score = 58.5 bits (140), Expect = 3e-09
Identities = 42/165 (25%), Positives = 79/165 (47%), Gaps = 11/165 (6%)
Query: 61 ILDFILRLGAIGAALGAAVIMGTNEQILPFFTQFLQFHAQWDDFPMFKFFVVANGAAAGF 120
+ + +LR AL A +++ T+ ++ FT ++ A++ D F VVANG AA +
Sbjct: 10 LTELLLRCSISVFALLALILVVTDTEVKLIFT--IKKTAKYTDMKAVVFLVVANGIAAVY 67
Query: 121 LILSLPFSIVCIVRPLAAGPRFLLVIV------DLVLMALVVAAASSAAAVVYLAHNGSQ 174
+L S+ C+V + F + D + L VAA ++ A +A G +
Sbjct: 68 SLLQ---SVRCVVGTMKGKVLFSKPLAWAFFSGDQAMAYLNVAAIAATAESGVIAREGEE 124
Query: 175 DANWNAICQQFTDFCQGSSLAVVASFVASVFLACLVVVSSVALKR 219
D W +C + FC ++ V ++ +AS+ + + +S+ +L R
Sbjct: 125 DLQWMRVCTMYGKFCNQMAIGVSSALLASIAMVFVSCISAFSLFR 169
>At4g15630 unknown protein
Length = 190
Score = 58.5 bits (140), Expect = 3e-09
Identities = 42/176 (23%), Positives = 89/176 (49%), Gaps = 14/176 (7%)
Query: 51 QKGGMKKGIAILDFILRLGAIGAALGAAVIMGTNEQ-------ILPFFTQF-LQFHAQWD 102
++ G +KG L+ +R+ A+ + AA ++G +Q ++P + A+
Sbjct: 19 KESGSRKG---LELTMRVLALVLTMVAATVLGVAKQTKVVPIKLIPTLPPLNVSTTAKAS 75
Query: 103 DFPMFKFFVVANGAAAGFLILSLPFSIVCIVRPLAAGPRFLLVIV-DLVLMALVVAAASS 161
F + + AN A G+ +S+ IV I + + + V++ DL+++AL+ ++ +
Sbjct: 76 YLSAFVYNISANAIACGYTAISIV--IVMISKGKRSKSLLMAVLIGDLMMVALLFSSTGA 133
Query: 162 AAAVVYLAHNGSQDANWNAICQQFTDFCQGSSLAVVASFVASVFLACLVVVSSVAL 217
A A+ + +G++ W +C F FC ++++V + +ASV LVV+ ++ L
Sbjct: 134 AGAIGLMGRHGNKHVMWKKVCGVFGKFCNQAAVSVAITLIASVVFMLLVVLDALKL 189
>At2g35760 unknown protein
Length = 201
Score = 55.8 bits (133), Expect = 2e-08
Identities = 46/160 (28%), Positives = 72/160 (44%), Gaps = 16/160 (10%)
Query: 59 IAILDFILRLGAIGAALGAAVIMGTNEQILPFFTQFLQFHAQWDDFPMFKFFVVANGAAA 118
+ + + ILR AL AA+++ T+ Q+ F +Q A++ D VV NG AA
Sbjct: 27 VRVTELILRCLVCVLALVAAILIATDVQVREIF--MIQKKAKFTDMKALVLLVVVNGIAA 84
Query: 119 GFLILSLPFSIVCIV-----RPLAAGP-RFLLVIVDLVLMALVVAAASSAAAVVYLAHNG 172
G+ SL ++ C+V R L + P + + D + L VA ++AA A G
Sbjct: 85 GY---SLVQAVRCVVGLMKGRVLFSKPLAWAIFFGDQAVAYLCVAGVAAAAQSAAFAKLG 141
Query: 173 SQDANWNAICQQFTDFCQGSSLAVVASFVASVFLACLVVV 212
+ W IC + FC V +AS AC+ +V
Sbjct: 142 EPELQWMKICNMYGKFCN-----QVGEGIASALFACIGMV 176
>At4g15620 unknown protein (At4g15620)
Length = 190
Score = 52.8 bits (125), Expect = 1e-07
Identities = 39/173 (22%), Positives = 87/173 (49%), Gaps = 14/173 (8%)
Query: 54 GMKKGIAILDFILRLGAIGAALGAAVIMGTNEQ-------ILPFFTQF-LQFHAQWDDFP 105
G +KG+ + +R+ A+ + AA ++G +Q ++P + A+
Sbjct: 22 GSRKGV---ELTMRVLALILTMAAATVLGVAKQTKVVSIKLIPTLPPLDITTTAKASYLS 78
Query: 106 MFKFFVVANGAAAGFLILSLPFSIVCIVRPLAAGPRFLLVIV-DLVLMALVVAAASSAAA 164
F + + N A G+ +S+ +I+ I R + ++V++ DLV++AL+ + +A+A
Sbjct: 79 AFVYNISVNAIACGYTAISI--AILMISRGRRSKKLLMVVLLGDLVMVALLFSGTGAASA 136
Query: 165 VVYLAHNGSQDANWNAICQQFTDFCQGSSLAVVASFVASVFLACLVVVSSVAL 217
+ + +G++ W +C F FC ++ ++ + +A+V LVV+ ++ L
Sbjct: 137 IGLMGLHGNKHVMWKKVCGVFGKFCHRAAPSLPLTLLAAVVFMFLVVLDAIKL 189
>At5g54980 unknown protein (At5g54980)
Length = 194
Score = 48.5 bits (114), Expect = 3e-06
Identities = 34/157 (21%), Positives = 69/157 (43%), Gaps = 11/157 (7%)
Query: 59 IAILDFILRLGAIGAALGAAVIMGTNEQILPFFTQFLQFHAQWDDFPMFKFFVVANGAAA 118
+ I+D LRL + ++ + TN + P + +++ K+ V + +A
Sbjct: 29 LKIIDSCLRLSVVPLSVATIWLTVTNHESNPDYGNL-----EYNSIMGLKYMVGVSAISA 83
Query: 119 GFLILSLPFS-IVCIVRPLAAGPRFLLVIVDLVLMALVVAAASSAAAVVYLAHNGSQDAN 177
+ +LS S + C+V +L I D VL ++ + + A +VYL + G +
Sbjct: 84 IYALLSTVSSWVTCLV-----SKAWLFFIPDQVLAYVMTTSVAGATEIVYLLNKGDKIVT 138
Query: 178 WNAICQQFTDFCQGSSLAVVASFVASVFLACLVVVSS 214
W+ +C + +C ++A+ F L V+S+
Sbjct: 139 WSEMCSSYPHYCSKLTIALGLHVFVLFFFLFLSVISA 175
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.325 0.136 0.396
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,430,747
Number of Sequences: 26719
Number of extensions: 164084
Number of successful extensions: 614
Number of sequences better than 10.0: 29
Number of HSP's better than 10.0 without gapping: 24
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 571
Number of HSP's gapped (non-prelim): 29
length of query: 220
length of database: 11,318,596
effective HSP length: 95
effective length of query: 125
effective length of database: 8,780,291
effective search space: 1097536375
effective search space used: 1097536375
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 58 (26.9 bits)
Medicago: description of AC146332.5


