

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146332.3 - phase: 0
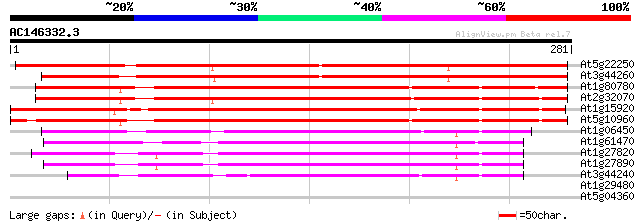
(281 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g22250 CCR4-associated factor-like protein 317 3e-87
At3g44260 CCR4-associated factor 1-like protein 308 2e-84
At1g80780 CCR4-associated factor, putative\0CCR4-associated fact... 239 1e-63
At2g32070 putative CCR4-associated factor protein 232 1e-61
At1g15920 unknown protein 230 6e-61
At5g10960 CCR4-ASSOCIATED FACTOR -like protein 224 3e-59
At1g06450 CCR4-associated like factor 149 1e-36
At1g61470 unknown 127 7e-30
At1g27820 hypothetical protein 127 9e-30
At1g27890 hypothetical protein 126 1e-29
At3g44240 CCR4-associated factor 1-like protein 123 1e-28
At1g29480 hypothetical protein 31 0.85
At5g04360 pullulanase-like protein (starch debranching enzyme) 29 3.2
>At5g22250 CCR4-associated factor-like protein
Length = 278
Score = 317 bits (813), Expect = 3e-87
Identities = 167/282 (59%), Positives = 206/282 (72%), Gaps = 12/282 (4%)
Query: 4 VKEEDASNKPIKIRQVWAENLEREFDLIRDFVHMFPYVSMDTEFPGVVVAPNFDPNIPYH 63
+K E + I IR VWA NLE EFDLIR V +P++SMDTEFPGV+ + D
Sbjct: 2 IKSEADLSDVIVIRDVWAYNLESEFDLIRGIVEDYPFISMDTEFPGVIYKADLDV----- 56
Query: 64 LRHMDPSEQYSFLKANVDNLNLIQLGLTLTDANGNLP---GDVAYSYIWEFNFKDFDVDR 120
LR +P+ Y+ LK+NVD L+LIQ+GLTL+DA+GNLP G YIWEFNF+DFDV+R
Sbjct: 57 LRRGNPNYLYNLLKSNVDALSLIQVGLTLSDADGNLPDLGGQKNRRYIWEFNFRDFDVER 116
Query: 121 DLQNPDSIELLRRQGIDFKRNLIYGVDSLEFAKLFRLKSGLV-NSGVSWVTFHSSYDFGY 179
D PDSIELLRR GIDF+RN GV+S FA+L + SGL+ N VSWVTFHS+YDFGY
Sbjct: 117 DPHAPDSIELLRRHGIDFERNRREGVESERFAELM-MSSGLICNESVSWVTFHSAYDFGY 175
Query: 180 LVKILTQNYLPSRLEEFLSILTQIFGQNVYDMKYMIKFC--NLYGGLERVATKLKVSRAV 237
LVKILT+ LP L EFL +L FG VYD+K++++FC LYGGL+RVA L+V+RAV
Sbjct: 176 LVKILTRRQLPVALREFLGLLRAFFGDRVYDVKHIMRFCEQRLYGGLDRVARSLEVNRAV 235
Query: 238 GNSHQAASDSLLTWQAFKKMKDIYFVNNGITMHAGVLFGLEV 279
G HQA SDSLLTWQAF++M+D+YFV +G HAGVL+GLEV
Sbjct: 236 GKCHQAGSDSLLTWQAFQRMRDLYFVEDGAEKHAGVLYGLEV 277
>At3g44260 CCR4-associated factor 1-like protein
Length = 280
Score = 308 bits (790), Expect = 2e-84
Identities = 159/269 (59%), Positives = 201/269 (74%), Gaps = 15/269 (5%)
Query: 17 RQVWAENLEREFDLIRDFVHMFPYVSMDTEFPGVVVAPNFDPNIPYHLRHMDPSEQYSFL 76
R+VWAENLE EF+LI + + +P++SMDTEFPGV+ + LR +P + Y+ L
Sbjct: 20 REVWAENLESEFELISEIIDDYPFISMDTEFPGVIFKSD--------LRFTNPDDLYTLL 71
Query: 77 KANVDNLNLIQLGLTLTDANGNLPG---DVAYSYIWEFNFKDFDVDRDLQNPDSIELLRR 133
KANVD L+LIQ+GLTL+D NGNLP D+ +IWEFNF+DFDV RD PDSIELLRR
Sbjct: 72 KANVDALSLIQVGLTLSDVNGNLPDLGDDLHRGFIWEFNFRDFDVARDAHAPDSIELLRR 131
Query: 134 QGIDFKRNLIYGVDSLEFAKLFRLKSGLV-NSGVSWVTFHSSYDFGYLVKILTQNYLPSR 192
QGIDF+RN GV+S FA+L + SGLV N VSWVTFHS+YDFGYL+KILT+ LP
Sbjct: 132 QGIDFERNCRDGVESERFAELM-MSSGLVCNEEVSWVTFHSAYDFGYLMKILTRRELPGA 190
Query: 193 LEEFLSILTQIFGQNVYDMKYMIKFC--NLYGGLERVATKLKVSRAVGNSHQAASDSLLT 250
L EF ++ +FG+ VYD+K+M+KFC L+GGL+RVA L+V+RAVG HQA SDSLLT
Sbjct: 191 LGEFKRVMRVLFGERVYDVKHMMKFCERRLFGGLDRVARTLEVNRAVGKCHQAGSDSLLT 250
Query: 251 WQAFKKMKDIYFVNNGITMHAGVLFGLEV 279
W AF++M+D+YFV +G HAGVL+GLEV
Sbjct: 251 WHAFQRMRDLYFVQDGPEKHAGVLYGLEV 279
>At1g80780 CCR4-associated factor, putative\0CCR4-associated factor,
putative
Length = 274
Score = 239 bits (610), Expect = 1e-63
Identities = 132/270 (48%), Positives = 178/270 (65%), Gaps = 16/270 (5%)
Query: 14 IKIRQVWAENLEREFDLIRDFVHMFPYVSMDTEFPGVVVAP--NFDPNIPYHLRHMDPSE 71
I+IR+VW +NL+ E DLIRD V FPYV+MDTEFPG+VV P F N YH
Sbjct: 10 IQIREVWNDNLQEEMDLIRDVVDDFPYVAMDTEFPGIVVRPVGTFKSNADYH-------- 61
Query: 72 QYSFLKANVDNLNLIQLGLTLTDANGNLPGDVAYSY-IWEFNFKDFDVDRDLQNPDSIEL 130
Y LK NV+ L +IQLGLT ++ GNLP Y IW+FNF++FD+D D+ DSIEL
Sbjct: 62 -YETLKTNVNILKMIQLGLTFSNEQGNLPTCGTDKYCIWQFNFREFDLDSDIFALDSIEL 120
Query: 131 LRRQGIDFKRNLIYGVDSLEFAKLFRLKSGLVNSGVSWVTFHSSYDFGYLVKILTQNYLP 190
L++ GID +N + G+DS FA+L ++N V WVTFHS YDFGYL+K+LT LP
Sbjct: 121 LKQSGIDLAKNTLDGIDSKRFAELLMSSGIVLNENVHWVTFHSGYDFGYLLKLLTCQNLP 180
Query: 191 SRLEEFLSILTQIFGQNVYDMKYMIKFCN-LYGGLERVATKLKVSRAVGNSHQAASDSLL 249
+F ++ ++ VYD+K+++KFCN L+GGL ++A L+V R VG HQA SDSLL
Sbjct: 181 DSQTDFFKLI-NVYFPTVYDIKHLMKFCNSLHGGLNKLAELLEVER-VGICHQAGSDSLL 238
Query: 250 TWQAFKKMKDIYFVNNGITMHAGVLFGLEV 279
T F+K+K+ +FV + ++GVL+GL V
Sbjct: 239 TSCTFRKLKENFFV-GPLHKYSGVLYGLGV 267
>At2g32070 putative CCR4-associated factor protein
Length = 275
Score = 232 bits (592), Expect = 1e-61
Identities = 129/271 (47%), Positives = 177/271 (64%), Gaps = 17/271 (6%)
Query: 14 IKIRQVWAENLEREFDLIRDFVHMFPYVSMDTEFPGVVVAP--NFDPNIPYHLRHMDPSE 71
I+IR+VW +NLE E LIR+ V FP+V+MDTEFPG+V P F N YH
Sbjct: 10 IQIREVWNDNLESEMALIREVVDDFPFVAMDTEFPGIVCRPVGTFKTNTEYH-------- 61
Query: 72 QYSFLKANVDNLNLIQLGLTLTDANGNLP--GDVAYSYIWEFNFKDFDVDRDLQNPDSIE 129
Y LK NV+ L +IQLGLT +D GNLP G IW+FNF++FD++ D+ DSIE
Sbjct: 62 -YETLKTNVNILKMIQLGLTFSDEKGNLPTCGTDNKYCIWQFNFREFDLESDIYATDSIE 120
Query: 130 LLRRQGIDFKRNLIYGVDSLEFAKLFRLKSGLVNSGVSWVTFHSSYDFGYLVKILTQNYL 189
LLR+ GIDF +N +G+DS FA+L ++N V WVTFHS YDFGYL+K+LT L
Sbjct: 121 LLRQSGIDFVKNNEFGIDSKRFAELLMSSGIVLNENVHWVTFHSGYDFGYLLKLLTCQNL 180
Query: 190 PSRLEEFLSILTQIFGQNVYDMKYMIKFCN-LYGGLERVATKLKVSRAVGNSHQAASDSL 248
P F +++ ++ VYD+K+++KFCN L+GGL ++A L V R VG HQA SDSL
Sbjct: 181 PETQTGFFEMIS-VYFPRVYDIKHLMKFCNSLHGGLNKLAELLDVER-VGICHQAGSDSL 238
Query: 249 LTWQAFKKMKDIYFVNNGITMHAGVLFGLEV 279
LT F+K+++ +F+ + + ++GVL+GL V
Sbjct: 239 LTSCTFRKLQENFFIGS-MEKYSGVLYGLGV 268
>At1g15920 unknown protein
Length = 286
Score = 230 bits (587), Expect = 6e-61
Identities = 132/283 (46%), Positives = 180/283 (62%), Gaps = 12/283 (4%)
Query: 1 MTKVKEEDASNKPIKIRQVWAENLEREFDLIRDFVHMFPYVSMDTEFPGVV---VAPNFD 57
M++ + + I+IR+VW NLE+E LI + FPYV+MDTEFPG+V V N +
Sbjct: 1 MSQAPNPEEEDDTIEIREVWNHNLEQEMALIEQSIDDFPYVAMDTEFPGIVCKTVTANPN 60
Query: 58 PNIPYHLRHMDPSEQYSFLKANVDNLNLIQLGLTLTDANGNLPG-DVAYSYIWEFNFKDF 116
PN PY + + Y LKANV+ L LIQLGLTL+D GNLP IW+FNF++F
Sbjct: 61 PN-PYSIHY---EYNYDTLKANVNMLKLIQLGLTLSDEKGNLPTCGTNKQCIWQFNFREF 116
Query: 117 DVDRDLQNPDSIELLRRQGIDFKRNLIYGVDSLEFAKLFRLKSGLVNSGVSWVTFHSSYD 176
+V D+ DSIELLR+ ID ++N GVD+ FA+L ++N + WVTFH YD
Sbjct: 117 NVISDMFALDSIELLRKSAIDLEKNNECGVDAKRFAELLMGSGVVLNDKIHWVTFHCGYD 176
Query: 177 FGYLVKILTQNYLPSRLEEFLSILTQIFGQNVYDMKYMIKFC-NLYGGLERVATKLKVSR 235
FGYL+K+L+ LP + +F + + F VYD+KY++ FC NLYGGLE++A L V R
Sbjct: 177 FGYLLKLLSGKELPEEISDFFDQMEKFF-PVVYDIKYLMGFCTNLYGGLEKIAELLGVKR 235
Query: 236 AVGNSHQAASDSLLTWQAFKKMKDIYFVNNGITMHAGVLFGLE 278
VG SHQA SDSLLT + F KMK+ +F + + ++G LFGL+
Sbjct: 236 -VGISHQAGSDSLLTLRTFIKMKEFFFTGS-LLKYSGFLFGLD 276
>At5g10960 CCR4-ASSOCIATED FACTOR -like protein
Length = 277
Score = 224 bits (572), Expect = 3e-59
Identities = 131/287 (45%), Positives = 178/287 (61%), Gaps = 28/287 (9%)
Query: 1 MTKVKEEDASNKPIKIRQVWAENLEREFDLIRDFVHMFPYVSMDTEFPGVVVAP------ 54
M + +ED+ I IR+VW NL EF LIR+ V F Y++MDTEFPGVV+ P
Sbjct: 1 MAETLKEDS----IMIREVWDYNLVEEFALIREIVDKFSYIAMDTEFPGVVLKPVATFKY 56
Query: 55 NFDPNIPYHLRHMDPSEQYSFLKANVDNLNLIQLGLTLTDANGNLPGDVAYSY-IWEFNF 113
N D N Y LK NVD L LIQ+GLT +D NGNLP + IW+FNF
Sbjct: 57 NNDLN-------------YRTLKENVDLLKLIQVGLTFSDENGNLPTCGTDKFCIWQFNF 103
Query: 114 KDFDVDRDLQNPDSIELLRRQGIDFKRNLIYGVDSLEFAKLFRLKSGLVNSGVSWVTFHS 173
++F++ D+ +SIELLR+ GIDFK+N+ G+D + F +L ++N +SWVTFH
Sbjct: 104 REFNIGEDIYASESIELLRQCGIDFKKNIEKGIDVVRFGELMMSSGIVLNDAISWVTFHG 163
Query: 174 SYDFGYLVKILTQNYLPSRLEEFLSILTQIFGQNVYDMKYMIKFCN-LYGGLERVATKLK 232
YDFGYLVK+LT LP + +F +L ++ VYD+K+++ FCN L+GGL R+A +
Sbjct: 164 GYDFGYLVKLLTCKELPLKQADFFKLL-YVYFPTVYDIKHLMTFCNGLFGGLNRLAELMG 222
Query: 233 VSRAVGNSHQAASDSLLTWQAFKKMKDIYFVNNGITMHAGVLFGLEV 279
V R VG HQA SDSLLT +F+K+K+ YF + + GVL+GL V
Sbjct: 223 VER-VGICHQAGSDSLLTLGSFRKLKERYFPGS-TEKYTGVLYGLGV 267
>At1g06450 CCR4-associated like factor
Length = 360
Score = 149 bits (377), Expect = 1e-36
Identities = 91/249 (36%), Positives = 132/249 (52%), Gaps = 21/249 (8%)
Query: 17 RQVWAENLEREFDLIRDFVHMFPYVSMDTEFPGVVVAPNFDPNIPYHLRHMDPSEQYSFL 76
R+VW N++ E + + + FP ++ DTE+PG++ FD + E Y +
Sbjct: 10 RRVWRSNVDEEMARMAECLKRFPLIAFDTEYPGIIFRTYFDSS---------SDECYRAM 60
Query: 77 KANVDNLNLIQLGLTLTDANGNLPGDVAYSYIWEFNFKDFDVDRDLQNPDSIELLRRQGI 136
K NV+N LIQ G TL +A G + G +WE NF +F D +N SIE LRR G+
Sbjct: 61 KGNVENTKLIQCGFTLFNAKGEIGG------VWEINFSNFGDPSDTRNELSIEFLRRHGL 114
Query: 137 DFKRNLIYGVDSLEFAKLFRLKSGL-VNSGVSWVTFHSSYDFGYLVKILTQNYLPSRLEE 195
D ++ GVD + +L + V +VTF +YDF Y + IL LP E
Sbjct: 115 DLQKIRDEGVDMFGYGFFPKLMTVFRSQKHVEFVTFQGAYDFAYFLSILNHGKLPETHGE 174
Query: 196 FLSILTQIFGQNVYDMKYMIKFCNLYG---GLERVATKLKVSRAVGNSHQAASDSLLTWQ 252
F + + ++FGQ VYD K M FC G GL ++A L+++R VG +H A SDSL+T
Sbjct: 175 FATEVVKVFGQ-VYDTKVMAGFCEGLGEHLGLSKLAQLLQITR-VGRAHHAGSDSLMTAL 232
Query: 253 AFKKMKDIY 261
F K+K +Y
Sbjct: 233 VFIKLKHVY 241
>At1g61470 unknown
Length = 278
Score = 127 bits (319), Expect = 7e-30
Identities = 86/243 (35%), Positives = 121/243 (49%), Gaps = 21/243 (8%)
Query: 18 QVWAENLEREFDLIRDFVHMFPYVSMDTEFPGVVVAPNFDPNIPYHLRHMDPSEQYSFLK 77
+VW N + E + IRD + +++DTEFPG + D + R M K
Sbjct: 4 EVWRWNKQAEMNSIRDCLKHCNSIAIDTEFPGCLKETPMDASDEIRYRDM---------K 54
Query: 78 ANVDNLNLIQLGLTLTDANGNLPGDVAYSYIWEFNFKDFDVDRDLQNPDSIELLRRQGID 137
NVDN +LIQLGLTL + WE N DF+ + L+N SI L+ G+D
Sbjct: 55 FNVDNTHLIQLGLTLFGKG--------ITKTWEINLSDFNESKSLKNDKSIAFLKNNGLD 106
Query: 138 FKRNLIYGVDSLEFAKLFRLKSGLVNSGVSWVTFHSSYDFGYLVKILTQNYLPSRLEEFL 197
+ G+ EF F + + WVTF SYD YL+K LT+ LP +EF
Sbjct: 107 LDKIREEGIGIEEFFMEFSQILNEKHGKMRWVTFQGSYDKAYLLKGLTRKPLPETSKEFD 166
Query: 198 SILTQIFGQNVYDMKYMIKFCNLYG---GLERVATKLKVSRAVGNSHQAASDSLLTWQAF 254
+ Q+ G+ VYD+K M C+ GL+R+A L++ R VG +H A SDS LT + F
Sbjct: 167 ETVQQLLGRFVYDVKKMAGLCSGLSSRFGLQRIADVLQM-RRVGKAHHAGSDSELTARVF 225
Query: 255 KKM 257
K+
Sbjct: 226 TKL 228
>At1g27820 hypothetical protein
Length = 310
Score = 127 bits (318), Expect = 9e-30
Identities = 90/252 (35%), Positives = 128/252 (50%), Gaps = 25/252 (9%)
Query: 12 KPIKIRQVWAENLEREFDLIRDFVHMFPYVSMDTEFPGVVVAPNFDPNIPYHLRHMDPSE 71
K I +VW N E E + IRD + +++DTEFPG + MD SE
Sbjct: 2 KQISSGEVWRWNKEVEMNSIRDCLKHCSSIAIDTEFPGCLKE-----------TPMDASE 50
Query: 72 Q--YSFLKANVDNLNLIQLGLTLTDANGNLPGDVAYSYIWEFNFKDFDVDRDLQNPDSIE 129
+ Y +K NVDN +LIQLG TL D G ++ WE N DFD + +N SI
Sbjct: 51 EIRYRDMKFNVDNTHLIQLGFTLFDRRG-------FAKTWEINLSDFDEHKCFKNDKSIA 103
Query: 130 LLRRQGIDFKRNLIYGVDSLEFAKLFRLKSGLVNSGVSWVTFHSSYDFGYLVKILTQNY- 188
L+ G++ + G+ EF + F + ++WV F SYD YLVK LT
Sbjct: 104 FLKSNGLNLDKIREEGIGIDEFFRDFSQILTEKDGKITWVNFQGSYDNAYLVKGLTGGKP 163
Query: 189 LPSRLEEFLSILTQIFGQNVYDMKYMIKFCNLYG---GLERVATKLKVSRAVGNSHQAAS 245
LP EEF + Q+ G+ V+D+K + + C+ GL+R+A L++ R VG +H A S
Sbjct: 164 LPETKEEFHETVQQLLGKFVFDVKKIAESCSGLSSQFGLQRIADVLQMKR-VGKAHHAGS 222
Query: 246 DSLLTWQAFKKM 257
DS LT + F K+
Sbjct: 223 DSELTARVFTKL 234
>At1g27890 hypothetical protein
Length = 302
Score = 126 bits (317), Expect = 1e-29
Identities = 89/246 (36%), Positives = 126/246 (51%), Gaps = 25/246 (10%)
Query: 18 QVWAENLEREFDLIRDFVHMFPYVSMDTEFPGVVVAPNFDPNIPYHLRHMDPSEQ--YSF 75
+VW N E E D IRD + F +++DTEFPG + MD SE+ Y
Sbjct: 3 EVWRWNKEVEMDSIRDCLKHFSSIAIDTEFPGCLKE-----------TPMDASEEIRYRD 51
Query: 76 LKANVDNLNLIQLGLTLTDANGNLPGDVAYSYIWEFNFKDFDVDRDLQNPDSIELLRRQG 135
+K NVDN +LIQLG TL D G + WE N DF+ + +N SI L+ G
Sbjct: 52 MKFNVDNTHLIQLGFTLFDRRG-------ITKTWEINLSDFNEHKCFKNDKSIAFLKSNG 104
Query: 136 IDFKRNLIYGVDSLEFAKLFRLKSGLVNSGVSWVTFHSSYDFGYLVKILTQNY-LPSRLE 194
++ + G+ EF + F + ++WV F SYD YLVK LT LP E
Sbjct: 105 LNLDKIGEEGIGIEEFFRDFSQILKEKDGKITWVNFQGSYDNAYLVKGLTGGKPLPETKE 164
Query: 195 EFLSILTQIFGQNVYDMKYMIKFCNLYG---GLERVATKLKVSRAVGNSHQAASDSLLTW 251
EF + Q+ G+ V+D+K + + C+ GL+R+A L++ R VG +H A SDS LT
Sbjct: 165 EFHETVEQLLGKFVFDVKKIAESCSGLSSRFGLQRIADVLQMKR-VGKAHHAGSDSELTA 223
Query: 252 QAFKKM 257
+ F K+
Sbjct: 224 RVFTKL 229
>At3g44240 CCR4-associated factor 1-like protein
Length = 239
Score = 123 bits (309), Expect = 1e-28
Identities = 82/231 (35%), Positives = 115/231 (49%), Gaps = 21/231 (9%)
Query: 30 LIRDFVHMFPYVSMDTEFPGVVVAPNFDPNIPYHLRHMDPSEQYSFLKANVDNLNLIQLG 89
LI D + + ++++DTEFP + +H E+Y + +VD LIQLG
Sbjct: 3 LIEDCLRSYRFIAIDTEFPSTLRETT---------QHATDEERYMDMSFSVDRAKLIQLG 53
Query: 90 LTLTDANGNLPGDVAYSYIWEFNFKDFDVDRDLQNPDSIELLRRQGIDFKRNLIYGVDSL 149
LTL D NG + G WE NF DF VD D +N SIE LRR G+D ++ G+
Sbjct: 54 LTLFDINGRIGGT------WEINFSDFGVD-DARNEKSIEFLRRNGLDLRKIREEGIRIE 106
Query: 150 EFAKLFRLKSGLVNSGVSWVTFHSSYDFGYLVKILTQNYLPSRLEEFLSILTQIFGQNVY 209
F ++WVTFH SYD YL+K T LP E F + ++ G +VY
Sbjct: 107 GFFSEMFWMLKKTRRNITWVTFHGSYDIAYLLKGFTGEALPVTSERFSKAVARVLG-SVY 165
Query: 210 DMKYMIKFCNLYG---GLERVATKLKVSRAVGNSHQAASDSLLTWQAFKKM 257
D+K M C GLE +A + ++R VG +H A S++ LT F K+
Sbjct: 166 DLKVMAGRCEGLSSRLGLETLAHEFGLNR-VGTAHHAGSNNELTAMVFAKV 215
>At1g29480 hypothetical protein
Length = 225
Score = 30.8 bits (68), Expect = 0.85
Identities = 27/97 (27%), Positives = 47/97 (47%), Gaps = 9/97 (9%)
Query: 6 EEDASNKPIKIRQVWAENLEREFDLIRDFVHMFPYVSMDT-----EFPGVVVAPNFDPNI 60
+E +NK +R + A+ E+ R+ H Y++ +V +FD ++
Sbjct: 92 QETKTNKKPDLRDLIAKAQEK-IQNKREINHRREYITRQRIAARLALNQMVATVSFDDHL 150
Query: 61 PYHLRHMDPSEQYSFLKANVDNLNLIQLGLTLTDANG 97
YH R M+ Y+F++ VDNLN+ L + +D NG
Sbjct: 151 NYH-REME-QRGYTFIQDQVDNLNMFSL-WSRSDYNG 184
>At5g04360 pullulanase-like protein (starch debranching enzyme)
Length = 965
Score = 28.9 bits (63), Expect = 3.2
Identities = 16/54 (29%), Positives = 27/54 (49%)
Query: 76 LKANVDNLNLIQLGLTLTDANGNLPGDVAYSYIWEFNFKDFDVDRDLQNPDSIE 129
L + D ++ L + L +G+LP DV + N+K F V +DL D ++
Sbjct: 119 LSFSEDGIDGYDLRIKLEAESGSLPADVIEKFPHIRNYKSFKVPKDLDIRDLVK 172
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.322 0.140 0.417
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,412,933
Number of Sequences: 26719
Number of extensions: 275869
Number of successful extensions: 641
Number of sequences better than 10.0: 13
Number of HSP's better than 10.0 without gapping: 12
Number of HSP's successfully gapped in prelim test: 1
Number of HSP's that attempted gapping in prelim test: 589
Number of HSP's gapped (non-prelim): 13
length of query: 281
length of database: 11,318,596
effective HSP length: 98
effective length of query: 183
effective length of database: 8,700,134
effective search space: 1592124522
effective search space used: 1592124522
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 59 (27.3 bits)
Medicago: description of AC146332.3


