

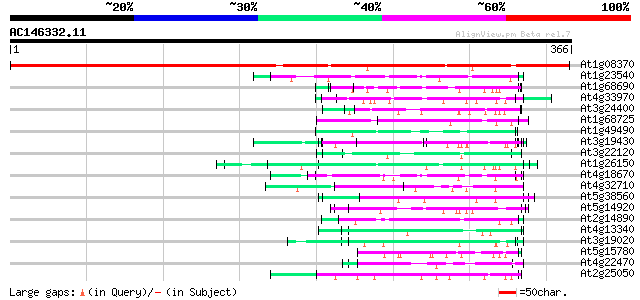
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146332.11 - phase: 0
(366 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g08370 unknown protein 451 e-127
At1g23540 putative serine/threonine protein kinase 55 8e-08
At1g68690 protein kinase, putative 54 2e-07
At4g33970 extensin-like protein 53 2e-07
At3g24400 protein kinase, putative 53 2e-07
At1g68725 putative arabinogalactan protein AGP19 53 2e-07
At1g49490 hypothetical protein 53 3e-07
At3g19430 putative late embryogenesis abundant protein 52 4e-07
At3g22120 unknown protein 52 7e-07
At1g26150 Pto kinase interactor, putative 52 7e-07
At4g18670 extensin-like protein 51 1e-06
At4g32710 putative protein kinase 50 2e-06
At5g38560 putative protein 50 3e-06
At5g14920 unknown protein 49 3e-06
At2g14890 arabinogalactan-protein AGP9 49 3e-06
At4g13340 extensin-like protein 49 4e-06
At3g19020 hypothetical protein 49 4e-06
At5g15780 proline-rich protein 49 6e-06
At4g22470 extensin - like protein 49 6e-06
At2g25050 hypothetical protein 49 6e-06
>At1g08370 unknown protein
Length = 367
Score = 451 bits (1159), Expect = e-127
Identities = 244/375 (65%), Positives = 287/375 (76%), Gaps = 19/375 (5%)
Query: 1 MSQNGKLMPNLDQQSTKLLNLTVLQRIDPFVEEILITAAHVTFYEFNIDLSQWSRKDVEG 60
MSQNGK++PNLDQ ST+LLNLTVLQRIDP++EEILITAAHVTFYEFNI+LSQWSRKDVEG
Sbjct: 1 MSQNGKIIPNLDQNSTRLLNLTVLQRIDPYIEEILITAAHVTFYEFNIELSQWSRKDVEG 60
Query: 61 SLFVVKRNTQPRFQFIVMNRRNTENLVENLLGDFEYEIQVPYLLYRNAAQEVNGIWFYNA 120
SLFVVKR+TQPRFQFIVMNRRNT+NLVENLLGDFEYE+Q PYLLYRNA+QEVNGIWFYN
Sbjct: 61 SLFVVKRSTQPRFQFIVMNRRNTDNLVENLLGDFEYEVQGPYLLYRNASQEVNGIWFYNK 120
Query: 121 RECEEVANLFSRILNAYAKVPPKLKVSSTKSEFEELEAVPTMAVMDGPLEPSSSITSNVA 180
RECEEVA LF+RIL+AY+KV K K SS+KSEFEELEA PTMAVMDGPLEPSS+
Sbjct: 121 RECEEVATLFNRILSAYSKVNQKPKASSSKSEFEELEAKPTMAVMDGPLEPSST----AR 176
Query: 181 DVPDDPSFINFFSAAMGIGNTSNAPITGQPYQSSATISSSGPTHAATPVVPL-----PTL 235
D PDDP+F+NFFS+ M +GNT++ +G PYQSSA H T P+ P +
Sbjct: 177 DAPDDPAFVNFFSSTMNLGNTASGSASG-PYQSSAIPHQPHQPHQPTIAPPVAAAAPPQI 235
Query: 236 QI-PSLPTSTPFTPQHD-APESINSSSQA-TNLVKPSFFVPPLSSAAMMMPPVSSSIPTA 292
Q P L +S+P D PE I+S+S T+LV PSFF PP A + P S+PTA
Sbjct: 236 QSPPPLQSSSPLMTLFDNNPEVISSNSNIHTDLVTPSFFGPPRMMAQPHLIP-GVSMPTA 294
Query: 293 PPLHPTGT--VQRPYGTPMLQPFPPPTPPPSLAPVSSPLPNPAISREKVRDALLVLVQDN 350
PPL+P QR YGTP+LQPFPPPTPPPSLAP + P ISR+KV++ALL L+Q++
Sbjct: 295 PPLNPNNASHQQRSYGTPVLQPFPPPTPPPSLAPAPT---GPVISRDKVKEALLSLLQED 351
Query: 351 QFIDMVYKALLNAHQ 365
+FID + + L NA Q
Sbjct: 352 EFIDKITRTLQNALQ 366
>At1g23540 putative serine/threonine protein kinase
Length = 731
Score = 54.7 bits (130), Expect = 8e-08
Identities = 52/181 (28%), Positives = 70/181 (37%), Gaps = 21/181 (11%)
Query: 160 PTMAVMDGPLEPSSSITSNVADVPDDPSFINFFSAAMGIGNTSNAPI--TGQPYQSSATI 217
P+ D L P SI + D P PS S+ P+ T P ++
Sbjct: 55 PSTPPPDSQLPPLPSILPPLTDSPPPPS-------------DSSPPVDSTPSPPPPTSNE 101
Query: 218 SSSGPTHAATPVVPLPTLQIPSLPTSTPFTPQHDAPESINSSSQATNLVKPSFFVPPLSS 277
S S P + TP P + P S Q P S + + TN P PP
Sbjct: 102 SPSPPEDSETPPAPPNESNDNNPPPSQDL--QSPPPSSPSPNVGPTNPESPPLQSPPAPP 159
Query: 278 AA--MMMPPVSSSIPT-APPLHPTGTVQRPYGTPMLQPFPPPTPPPSLAPVSSPLPNPAI 334
A+ PP S PT PP+ P+G P P P P PT PP P S P+ +P++
Sbjct: 160 ASDPTNSPPASPLDPTNPPPIQPSGPATSPPANPNAPPSPFPTVPPK-TPSSGPVVSPSL 218
Query: 335 S 335
+
Sbjct: 219 T 219
Score = 53.1 bits (126), Expect = 2e-07
Identities = 53/168 (31%), Positives = 71/168 (41%), Gaps = 26/168 (15%)
Query: 171 PSSSITSNVADV---PDDPSFINFFSAAMGIGNTSNAPITGQPYQSSATISSSGPTHAAT 227
PSSS + AD P+ PS N++ P+ P A SS+ P +
Sbjct: 8 PSSSPPAPPADTAPPPETPS-----------ENSALPPVDSSPPSPPADSSSTPPL--SE 54
Query: 228 PVVPLPTLQIPSLPTSTPFTPQHDAPESINSSSQATNLVKPSFFVPPLSSAAMMMPPVSS 287
P P P Q+P LP+ P P D+P + SS + PS PP +S PP S
Sbjct: 55 PSTPPPDSQLPPLPSILP--PLTDSPPPPSDSSPPVDST-PS--PPPPTSNESPSPPEDS 109
Query: 288 SIPTAPPLHPTGTVQRPYGTPMLQPFPPPTPPPSLAPV---SSPLPNP 332
P APP P + LQ PP +P P++ P S PL +P
Sbjct: 110 ETPPAPPNESND--NNPPPSQDLQSPPPSSPSPNVGPTNPESPPLQSP 155
Score = 35.4 bits (80), Expect = 0.050
Identities = 33/101 (32%), Positives = 44/101 (42%), Gaps = 11/101 (10%)
Query: 241 PTSTPFTPQHDAPESINSSSQATNLVKPSFFVPPLSSAAMMMPPVSSSIPT-APPLHPTG 299
P+S+P P D + S+ + L PP+ S+ P SSS P + P P
Sbjct: 8 PSSSPPAPPADTAPPPETPSENSAL-------PPVDSSPPSPPADSSSTPPLSEPSTPPP 60
Query: 300 TVQRPYGTPMLQPFPPPTPPPSLA--PV-SSPLPNPAISRE 337
Q P +L P PPPS + PV S+P P P S E
Sbjct: 61 DSQLPPLPSILPPLTDSPPPPSDSSPPVDSTPSPPPPTSNE 101
>At1g68690 protein kinase, putative
Length = 708
Score = 53.5 bits (127), Expect = 2e-07
Identities = 41/139 (29%), Positives = 54/139 (38%), Gaps = 7/139 (5%)
Query: 200 NTSNAPITGQPYQSSATISSSGPTHAATPVVPLPTLQIPSLPTSTPFTPQHDAPESINSS 259
N + +P T P S S+ P A P P+ T P + P P PES +
Sbjct: 24 NNATSPATPPPVTSPLPPSAPPPNRAPPPPPPVTTSP-PPVANGAPPPPLPKPPESSSPP 82
Query: 260 SQATNLVKPSFFVPPLSSAAMMMPPVSSSIPTAPPLHPTGTVQRPYGTPMLQPFPPP--- 316
Q P PP PP +S P P P+ P P +P P P
Sbjct: 83 PQPVIPSPPPSTSPPPQPVIPSPPPSASPPPALVPPLPSSPPP-PASVPPPRPSPSPPIL 141
Query: 317 --TPPPSLAPVSSPLPNPA 333
+PPPS+ P+ SP P P+
Sbjct: 142 VRSPPPSVRPIQSPPPPPS 160
Score = 48.1 bits (113), Expect = 7e-06
Identities = 39/134 (29%), Positives = 54/134 (40%), Gaps = 11/134 (8%)
Query: 210 PYQSSATISSSGP--THAATPVVPLPTLQIPSLPTSTP---FTPQHDAPESINSSSQATN 264
P +S ++S P +A +P P P LP S P P P + + A
Sbjct: 9 PVSNSPPVTSPPPPLNNATSPATPPPVTS--PLPPSAPPPNRAPPPPPPVTTSPPPVANG 66
Query: 265 LVKPSFFVPPLSSAAMMMPPVSSSIP-TAPPLHPTGTVQRPYGTP---MLQPFPPPTPPP 320
P PP SS+ P + S P T+PP P P +P ++ P P PPP
Sbjct: 67 APPPPLPKPPESSSPPPQPVIPSPPPSTSPPPQPVIPSPPPSASPPPALVPPLPSSPPPP 126
Query: 321 SLAPVSSPLPNPAI 334
+ P P P+P I
Sbjct: 127 ASVPPPRPSPSPPI 140
Score = 46.2 bits (108), Expect = 3e-05
Identities = 46/140 (32%), Positives = 55/140 (38%), Gaps = 22/140 (15%)
Query: 209 QPYQSSATISSSGPTHAATPVVPL----PTLQIPSLPTSTPFTPQHDAPESINSSSQATN 264
QP S S+S P P P P +P LP+S P P P S S
Sbjct: 84 QPVIPSPPPSTSPPPQPVIPSPPPSASPPPALVPPLPSSPP--PPASVPPPRPSPSPPIL 141
Query: 265 LVKPSFFVPPLSSAAMMMPPVSSSIPTA---PPLHPTGTVQRPYGTPMLQPF-------P 314
+ P V P+ S PP S PT PP P+ +RP +P P P
Sbjct: 142 VRSPPPSVRPIQSP----PPPPSDRPTQSPPPPSPPSPPSERPTQSPPSPPSERPTQSPP 197
Query: 315 PPTP--PPSLAPVSSPLPNP 332
PP+P PPS P SP P P
Sbjct: 198 PPSPPSPPSDRPSQSPPPPP 217
Score = 42.0 bits (97), Expect = 5e-04
Identities = 39/111 (35%), Positives = 50/111 (44%), Gaps = 11/111 (9%)
Query: 225 AATPVVPLPTLQIPSLPTSTPFTPQHDAPESINSSSQATNLVKPSFFVPPLSSAAMMMPP 284
A TPV P P P P ++P P ++A S + T+ + PS PP + A PP
Sbjct: 2 ATTPVQP-PVSNSP--PVTSPPPPLNNAT-SPATPPPVTSPLPPS--APPPNRAPPPPPP 55
Query: 285 VSSSIPTAPPLHPTGTVQRPYGTPMLQPFPPPTPP-PSLAPVSSPLPNPAI 334
V+ T+PP G P P PPP P PS P +SP P P I
Sbjct: 56 VT----TSPPPVANGAPPPPLPKPPESSSPPPQPVIPSPPPSTSPPPQPVI 102
Score = 33.1 bits (74), Expect = 0.25
Identities = 25/96 (26%), Positives = 39/96 (40%), Gaps = 8/96 (8%)
Query: 204 APITGQPYQSSATISSSGPTHAATPVVPLPTLQIPSLPTSTPFTPQHDAPESINSSS--Q 261
+P + +P QS + S PT + P P PS P+ P PE +
Sbjct: 174 SPPSERPTQSPPSPPSERPTQSPPPPSP------PSPPSDRPSQSPPPPPEDTKPQPPRR 227
Query: 262 ATNLVKPSFFVPPLSSAAMMMPPVSSSIPTAPPLHP 297
+ N P+F PP S +++P ++ P L P
Sbjct: 228 SPNSPPPTFSSPPRSPPEILVPGSNNPSQNNPTLRP 263
Score = 33.1 bits (74), Expect = 0.25
Identities = 37/139 (26%), Positives = 49/139 (34%), Gaps = 19/139 (13%)
Query: 205 PITGQPYQSSATISSSGPTHAATPVVPLPTLQIPSLPTSTPFTPQHDAPESINSSSQATN 264
P + +P QS S PT + P PS P S P +P S S +
Sbjct: 146 PPSVRPIQSPPPPPSDRPTQSPPP---------PS-PPSPPSERPTQSPPSPPSERPTQS 195
Query: 265 LVKPSFFVPPLSSAAMMMPPVSSSIPTAPPLHPTGTVQRPYGTPMLQPFPPPTPPPSLAP 324
PS PP + PP PP + P P PP +PP L P
Sbjct: 196 PPPPSPPSPPSDRPSQSPPPPPEDTKPQPPR------RSPNSPPPTFSSPPRSPPEILVP 249
Query: 325 VSSPLPNPAISREKVRDAL 343
S+ NP+ + +R L
Sbjct: 250 GSN---NPSQNNPTLRPPL 265
Score = 31.2 bits (69), Expect = 0.93
Identities = 37/132 (28%), Positives = 45/132 (34%), Gaps = 7/132 (5%)
Query: 202 SNAPITGQPYQSSATISSSGPTHAATPVVPLPTLQIPSLPTSTPFTPQH-DAPESINSSS 260
S P P + SS P A+ P P P+ P L S P + + +P S
Sbjct: 104 SPPPSASPPPALVPPLPSSPPPPASVPP-PRPSPSPPILVRSPPPSVRPIQSPPPPPSDR 162
Query: 261 QATNLVKPSFFVPPLSSAAMMMPPVSSSIPTAPPLHPTGTVQRPYGTPMLQPFPPP---- 316
+ PS PP P S PT P P P P P PPP
Sbjct: 163 PTQSPPPPSPPSPPSERPTQSPPSPPSERPTQSP-PPPSPPSPPSDRPSQSPPPPPEDTK 221
Query: 317 TPPPSLAPVSSP 328
PP +P S P
Sbjct: 222 PQPPRRSPNSPP 233
Score = 30.0 bits (66), Expect = 2.1
Identities = 22/67 (32%), Positives = 31/67 (45%), Gaps = 16/67 (23%)
Query: 273 PPLSSAAMMM--PPVSSSIP-TAPPLHPTGTVQRPYGTPMLQPFPPPTPPPSLAPVSSPL 329
PPL++A PPV+S +P +APP P P PPP S PV++
Sbjct: 21 PPLNNATSPATPPPVTSPLPPSAPP-------------PNRAPPPPPPVTTSPPPVANGA 67
Query: 330 PNPAISR 336
P P + +
Sbjct: 68 PPPPLPK 74
>At4g33970 extensin-like protein
Length = 699
Score = 53.1 bits (126), Expect = 2e-07
Identities = 42/135 (31%), Positives = 63/135 (46%), Gaps = 16/135 (11%)
Query: 204 APITGQPYQSSATISSSGPTHAATPVVPLPTLQ---IPSLPTSTPFTPQHDAPESINSSS 260
+P+ P Q + + ++ P H +PV+ P + +PS P P P+ ++P+ +
Sbjct: 451 SPVPTTPVQKPSPVPTT-PVHEPSPVLATPVDKPSPVPSRPVQKPQPPK-ESPQPDDPYD 508
Query: 261 QATNLVKPSFFVPPLSSAAMMMPPVSSSIPTAPPLH-PTGTVQRPYGTPMLQPFPPP--- 316
Q+ + S PP + PPV S P PP+H P V P P+ P PPP
Sbjct: 509 QSPVTKRRS---PPPAPVNSPPPPVYSPPPPPPPVHSPPPPVHSPPPPPVYSPPPPPPPV 565
Query: 317 -TPPPSLAPVSSPLP 330
+PPP PV SP P
Sbjct: 566 HSPPP---PVFSPPP 577
Score = 47.0 bits (110), Expect = 2e-05
Identities = 41/158 (25%), Positives = 61/158 (37%), Gaps = 6/158 (3%)
Query: 200 NTSNAPITGQPYQSSATISSSGPTHAATPVVPLPTLQIPSLPTSTPFTPQHDAPESINSS 259
N+ P+ P S P H+ P P+ + P P +P P P + S
Sbjct: 524 NSPPPPVYSPPPPPPPVHSPPPPVHSPPPP-PVYSPPPPPPPVHSPPPPVFSPPPPVYSP 582
Query: 260 SQATNLVKPSFFVPPLSSAAMMMPPVSSSIPTAPPLH---PTGTVQRPYGTPMLQPFPPP 316
+ P PP + PP S P PP++ P P +P + PPP
Sbjct: 583 PPPVHSPPPPVHSPPPPAPVHSPPPPVHSPPPPPPVYSPPPPVFSPPPSQSPPVVYSPPP 642
Query: 317 TPPP-SLAPVSSPLPNPAISREKVRDALLVLVQDNQFI 353
PP + PV SP P P + +++ A D++FI
Sbjct: 643 RPPKINSPPVQSPPPAP-VEKKETPPAHAPAPSDDEFI 679
Score = 43.1 bits (100), Expect = 2e-04
Identities = 47/146 (32%), Positives = 59/146 (40%), Gaps = 28/146 (19%)
Query: 214 SATISSSGPTHAATPV--------VPLPT--LQIPSLPTSTPF---TPQHDAPESINSSS 260
S+T S P H TPV P+PT +Q PS +TP +P P S
Sbjct: 416 SSTPSKPSPVHKPTPVPTTPVHKPTPVPTTPVQKPSPVPTTPVQKPSPVPTTPVHEPSPV 475
Query: 261 QATNLVKPSFFVPPLSSAAMM--MPPVSSSIPTAPPLHPTGTVQR-----PYGTPMLQPF 313
AT + KPS P+ S + PP S P P T +R P +P +
Sbjct: 476 LATPVDKPS----PVPSRPVQKPQPPKESPQPDDPYDQSPVTKRRSPPPAPVNSPPPPVY 531
Query: 314 PPPTPPPSL----APVSSPLPNPAIS 335
PP PPP + PV SP P P S
Sbjct: 532 SPPPPPPPVHSPPPPVHSPPPPPVYS 557
>At3g24400 protein kinase, putative
Length = 694
Score = 53.1 bits (126), Expect = 2e-07
Identities = 41/134 (30%), Positives = 53/134 (38%), Gaps = 27/134 (20%)
Query: 205 PITGQPYQSSATISSS----GPTHAATPVVPLPTLQIPSLPTSTPFTPQHDAPESINSSS 260
P+T P S T S PT + P+ P P P++ S P TP P S
Sbjct: 77 PLTPSPLPPSPTTPSPPLTPSPTTPSPPLTPSPP---PAITPSPPLTPSPLPPSPTTPS- 132
Query: 261 QATNLVKPSFFVPPLSSAAMMMPPVSSSIPTAPPLHPTGTVQRPYGTPMLQPFPPPTPPP 320
PP S ++ PP++ S P + PL RP P P P TPP
Sbjct: 133 ------------PPPPSPSIPSPPLTPSPPPSSPL-------RPSSPPPPSPATPSTPPR 173
Query: 321 SLAPVSSPLPNPAI 334
S P S+P P P +
Sbjct: 174 SPPPPSTPTPPPRV 187
Score = 47.8 bits (112), Expect = 1e-05
Identities = 44/139 (31%), Positives = 54/139 (38%), Gaps = 23/139 (16%)
Query: 219 SSGPTHAATPVVPLPTLQIPSLPTSTPFTPQHDA---PESINSSSQATNLVK------PS 269
SS P TP P L IP P P TP P ++ T L P
Sbjct: 2 SSAPPPGGTPSPPPQPLPIPPPPQPLPVTPPPPPTALPPALPPPPPPTALPPALPPPPPP 61
Query: 270 FFVPPLSSAAMMMPPVSSSIPT-------APPLHPTGTVQRPYGTPM----LQPFPPPTP 318
VPP+ + PP + P +PPL P+ T P TP + P PP TP
Sbjct: 62 TTVPPIPPSTPSPPPPLTPSPLPPSPTTPSPPLTPSPTTPSPPLTPSPPPAITPSPPLTP 121
Query: 319 ---PPSLAPVSSPLPNPAI 334
PPS S P P+P+I
Sbjct: 122 SPLPPSPTTPSPPPPSPSI 140
Score = 47.4 bits (111), Expect = 1e-05
Identities = 45/123 (36%), Positives = 57/123 (45%), Gaps = 25/123 (20%)
Query: 226 ATPVVPLPTLQIPSLPTSTPFTPQHDAPESINSSSQATNLVKPSFFVPPLS-SAAMMMPP 284
A P P PT +P +P STP P P + S P+ PPL+ S PP
Sbjct: 54 ALPPPPPPTT-VPPIPPSTPSPPPPLTPSPLPPS--------PTTPSPPLTPSPTTPSPP 104
Query: 285 VSSSIPTA----PPLHPT------GTVQRPYGTPMLQPFPP--PTPPPS--LAPVSSPLP 330
++ S P A PPL P+ T P +P + P PP P+PPPS L P S P P
Sbjct: 105 LTPSPPPAITPSPPLTPSPLPPSPTTPSPPPPSPSI-PSPPLTPSPPPSSPLRPSSPPPP 163
Query: 331 NPA 333
+PA
Sbjct: 164 SPA 166
Score = 36.2 bits (82), Expect = 0.029
Identities = 56/231 (24%), Positives = 73/231 (31%), Gaps = 69/231 (29%)
Query: 141 PPKLKVSSTKSEFEELEAVPTMAVMDGPLEPSSSITSNVADVPDDPSFINFFSAAMGIGN 200
PP L T S P+ + P PS +T P P I
Sbjct: 75 PPPL----TPSPLPPSPTTPSPPLTPSPTTPSPPLT------PSPPPAI----------- 113
Query: 201 TSNAPITGQPYQSSATISSSGPTHAATPVVPLPTLQIPSLPT----------STPFTPQH 250
T + P+T P S T S P + P PL PS P +TP TP
Sbjct: 114 TPSPPLTPSPLPPSPTTPSPPPPSPSIPSPPLTPSPPPSSPLRPSSPPPPSPATPSTPPR 173
Query: 251 DAPESINSSSQATNLVKPSFFVPPLSSAAMMMPPVSSSIPTAPPLHPTGTVQRPYGTPML 310
P PS P PP S+ PP P+G
Sbjct: 174 SPP-------------PPSTPTP---------PPRVGSLSPPPPASPSGG---------R 202
Query: 311 QPFPPPTPPPSLAPVSSPLPNPAISREKVRDALLVLVQDNQFIDMVYKALL 361
P P T P S P S S+E + A++ + F+ +V AL+
Sbjct: 203 SPSTPSTTPGSSPPAQS-------SKELSKGAMVGIAIGGGFVLLVALALI 246
>At1g68725 putative arabinogalactan protein AGP19
Length = 248
Score = 53.1 bits (126), Expect = 2e-07
Identities = 48/145 (33%), Positives = 61/145 (41%), Gaps = 18/145 (12%)
Query: 201 TSNAPITGQPYQSSATISSSGPTHAATPVVPLPTLQIPSLPTSTPFTPQHDAPESINSSS 260
T+ P T P ++ T S A+PV P P + PTS P AP+ S
Sbjct: 43 TAAPPTTAAPPPTTTTPPVSAAQPPASPVTPPPAVT----PTSPP------APKVAPVIS 92
Query: 261 QATNLVKPSFFVPPLSSAAMMMPPVSSS-IPTAPPLHPTGTVQRPYGTPMLQPFPPP--- 316
AT +P PP S+ + PPVS PT+PP P P P PPP
Sbjct: 93 PATPPPQPPQS-PPASAPTVSPPPVSPPPAPTSPPPTPASPPPAPASPPPAPASPPPAPV 151
Query: 317 TPPPSLAPVS---SPLPNPAISREK 338
+PPP AP P P PA ++ K
Sbjct: 152 SPPPVQAPSPISLPPAPAPAPTKHK 176
Score = 45.1 bits (105), Expect = 6e-05
Identities = 30/95 (31%), Positives = 42/95 (43%), Gaps = 6/95 (6%)
Query: 241 PTSTPFTPQHDAPESINSSSQATNLVKPSFFVPPLSSAAMMMPPVS---SSIPTAPPLHP 297
P ++P T AP ++ T P+ PP+S+A PV+ + PT+PP
Sbjct: 27 PAASPVTSTTTAPPPTTAAPPTTAAPPPTTTTPPVSAAQPPASPVTPPPAVTPTSPPAPK 86
Query: 298 TGTVQRPYGTPMLQPFPPPTPPPSLAPVSSPLPNP 332
V P P P PP +PP S VS P +P
Sbjct: 87 VAPVISPATPP---PQPPQSPPASAPTVSPPPVSP 118
Score = 41.2 bits (95), Expect = 0.001
Identities = 36/132 (27%), Positives = 52/132 (39%), Gaps = 14/132 (10%)
Query: 213 SSATISSSGPTHAATPVVPLPTLQIPSL--PTSTPFTPQHDAPESINSSSQATNLVKPSF 270
SS ++++ GP AA+PV T P+ P +T P ++++ + V P
Sbjct: 18 SSFSVNAQGP--AASPVTSTTTAPPPTTAAPPTTAAPPPTTTTPPVSAAQPPASPVTPPP 75
Query: 271 FVPPLSSAAMMMPPVSSSI------PTAPPLHPTGTVQRPYGTPMLQPFPPPTP----PP 320
V P S A + PV S P +PP P P PPPTP P
Sbjct: 76 AVTPTSPPAPKVAPVISPATPPPQPPQSPPASAPTVSPPPVSPPPAPTSPPPTPASPPPA 135
Query: 321 SLAPVSSPLPNP 332
+P +P P
Sbjct: 136 PASPPPAPASPP 147
>At1g49490 hypothetical protein
Length = 847
Score = 52.8 bits (125), Expect = 3e-07
Identities = 42/138 (30%), Positives = 54/138 (38%), Gaps = 25/138 (18%)
Query: 200 NTSNAPITGQPYQSSATISSSGP-------THAATPVVPLPTLQIPSLPTSTPFTPQHDA 252
N + P PY +S + P T P P+P+ PS P +P P H
Sbjct: 501 NEAQGPTPDDPYDASPVKNRRSPPPPKVEDTRVPPPQPPMPSPSPPS-PIYSPPPPVHSP 559
Query: 253 PESINSSSQATNLVKPSFFVPPLSSAAMMMPPVSSSIPTAPPLHPTGTVQRPYGTPMLQP 312
P + SS P + PP PPV+S P +PP V P P+ P
Sbjct: 560 PPPVYSSPPP-----PHVYSPP--------PPVASPPPPSPP----PPVHSPPPPPVFSP 602
Query: 313 FPPPTPPPSLAPVSSPLP 330
PP PP +PV SP P
Sbjct: 603 PPPVFSPPPPSPVYSPPP 620
Score = 45.4 bits (106), Expect = 5e-05
Identities = 40/132 (30%), Positives = 50/132 (37%), Gaps = 9/132 (6%)
Query: 200 NTSNAPITGQPYQSSATISSSGPTHAATPVVPLPTLQIPSLPTSTPFTPQHDAPESINSS 259
+ +N P G P + A SS T TP QI S P P Q P S
Sbjct: 643 HNTNQPPMGAPTPTQAPTPSSETTQVPTPSSESDQSQILS-PVQAPTPVQSSTP-----S 696
Query: 260 SQATNLVKPSFFVPPLSSAAMMMPPVSSSIPTAPPLHPTGTVQRPYGTPMLQPFPPPTPP 319
S+ T + PS S A + PV + P P + T Q P + P P
Sbjct: 697 SEPTQVPTPS---SSESYQAPNLSPVQAPTPVQAPTTSSETSQVPTPSSESNQSPSQAPT 753
Query: 320 PSLAPVSSPLPN 331
P L PV +P PN
Sbjct: 754 PILEPVHAPTPN 765
Score = 40.4 bits (93), Expect = 0.002
Identities = 46/201 (22%), Positives = 69/201 (33%), Gaps = 4/201 (1%)
Query: 137 YAKVPPKLKVSSTKSEFEELEAVPTMAVMDGPLEPSSSITSNVADVPDDPSFINFFSAAM 196
Y P K + S + E+ P M P P S I S V P +
Sbjct: 512 YDASPVKNRRSPPPPKVEDTRVPPPQPPMPSP-SPPSPIYSPPPPVHSPPPPVYSSPPPP 570
Query: 197 GIGNTSNAPITGQPYQSSATISSSGPTHAATPVVPLPTLQIPSLPTSTPFTPQHDAPESI 256
+ + + P + S P +P P+ + PS P +P P H P +
Sbjct: 571 HVYSPPPPVASPPPPSPPPPVHSPPPPPVFSPPPPVFSPPPPS-PVYSPPPPSHSPPPPV 629
Query: 257 NSSSQATNLVKPSFFV--PPLSSAAMMMPPVSSSIPTAPPLHPTGTVQRPYGTPMLQPFP 314
S T P+ PP+ + P SS T P + + Q +P+ P P
Sbjct: 630 YSPPPPTFSPPPTHNTNQPPMGAPTPTQAPTPSSETTQVPTPSSESDQSQILSPVQAPTP 689
Query: 315 PPTPPPSLAPVSSPLPNPAIS 335
+ PS P P P+ + S
Sbjct: 690 VQSSTPSSEPTQVPTPSSSES 710
Score = 39.3 bits (90), Expect = 0.003
Identities = 40/150 (26%), Positives = 53/150 (34%), Gaps = 22/150 (14%)
Query: 205 PITGQPYQSSATISSSGPTHAATPVV----PLPTLQIPSLPTSTPFTPQHDAPESINSSS 260
P P S S P H+ P V P P + P P ++P P P ++S
Sbjct: 538 PPMPSPSPPSPIYSPPPPVHSPPPPVYSSPPPPHVYSPPPPVASP--PPPSPPPPVHSPP 595
Query: 261 QATNLVKPS-FFVPPLSSAAMMMPPVSSSIPTAPPLH----------PTGTVQRPYGTPM 309
P F PP S PP S S P PP++ PT +P PM
Sbjct: 596 PPPVFSPPPPVFSPPPPSPVYSPPPPSHSPP--PPVYSPPPPTFSPPPTHNTNQP---PM 650
Query: 310 LQPFPPPTPPPSLAPVSSPLPNPAISREKV 339
P P P PS P P+ + ++
Sbjct: 651 GAPTPTQAPTPSSETTQVPTPSSESDQSQI 680
Score = 38.9 bits (89), Expect = 0.004
Identities = 31/128 (24%), Positives = 46/128 (35%), Gaps = 8/128 (6%)
Query: 209 QPYQSSATISSSGPTHAATPVVPLPTLQIPSLPTSTPFTPQHDAPESINSSSQATNLVKP 268
QP + + P +P P + P+ P S P Q P+ +S N P
Sbjct: 464 QPSKPEDSPKPEQPKPEESPKPEQPQIPEPTKPVSPPNEAQGPTPDDPYDASPVKNRRSP 523
Query: 269 SFFVPPLSSAAMMMPPVSSSIPTAPPLHPTGTVQRPYGTPMLQPFPPPTPPPSLAP---- 324
PP +PP +P+ P P + P +P + P PP +P
Sbjct: 524 ----PPPKVEDTRVPPPQPPMPSPSPPSPIYSPPPPVHSPPPPVYSSPPPPHVYSPPPPV 579
Query: 325 VSSPLPNP 332
S P P+P
Sbjct: 580 ASPPPPSP 587
Score = 30.4 bits (67), Expect = 1.6
Identities = 31/122 (25%), Positives = 50/122 (40%), Gaps = 7/122 (5%)
Query: 201 TSNAPITGQPYQSSATISSSGPTHAATPVVPLPTLQIPSLPTSTPFTPQHDAPESINSSS 260
T+++ + P SS S+ P+ A TP+ L + P+ P S P + E ++S
Sbjct: 729 TTSSETSQVPTPSSE--SNQSPSQAPTPI--LEPVHAPT-PNSKPVQSPTPSSEPVSSPE 783
Query: 261 QATNLVKPSFFVPPLSSAAMMMPPVSSSIPTAPPLHPTGTVQRPYGTPMLQPFPPPTPPP 320
Q+ + P SS P +SIP PP + + P F +PPP
Sbjct: 784 QSEEVEAPEPTPVNPSSVPSSSPSTDTSIP--PPENNDDDDDGDFVLPPHIGFQYASPPP 841
Query: 321 SL 322
+
Sbjct: 842 PM 843
>At3g19430 putative late embryogenesis abundant protein
Length = 550
Score = 52.4 bits (124), Expect = 4e-07
Identities = 44/140 (31%), Positives = 55/140 (38%), Gaps = 13/140 (9%)
Query: 202 SNAPITGQPYQSSATISSSGPTHAATPVVPLPTLQI-PSLPTSTPFTPQHDAPESINSSS 260
S P T P S T S P TP VP PT + P PT TP P P +
Sbjct: 116 SPPPPTPTPSVPSPTPPVSPPPPTPTPSVPSPTPPVSPPPPTPTPSVPSPTPPVPTDPMP 175
Query: 261 QATNLVKPSFFVPPLSSAAMMMPP------VSSSIPTAP---PLHPTGTVQRPYGTPMLQ 311
V P PP + ++ PP + S+P+ P P PT +V P
Sbjct: 176 SPPPPVSPP---PPTPTPSVPSPPDVTPTPPTPSVPSPPDVTPTPPTPSVPSPPDVTPTP 232
Query: 312 PFPPPTPPPSLAPVSSPLPN 331
P PP P PS +P P P+
Sbjct: 233 PTPPSVPTPSGSPPYVPPPS 252
Score = 52.0 bits (123), Expect = 5e-07
Identities = 42/129 (32%), Positives = 53/129 (40%), Gaps = 10/129 (7%)
Query: 205 PITGQPY--QSSATISSSGPTHAATPVVPLPTLQIPSLPTSTPFTPQHDAPESINSSSQA 262
PI G P S G TP P+P + S P TP P P S +
Sbjct: 49 PICGPPSPGDDGGGDDSGGDDGGYTPPAPVPPV---SPPPPTPSVPSPTPPVSPPPPTPT 105
Query: 263 TNLVKPSFFVPPLS-SAAMMMPPVSSSIPTAPPLHPTGTVQRPYGTPMLQPFPPPTPPPS 321
++ P+ PP+S P V S P P PT T P TP + P PPPTP PS
Sbjct: 106 PSVPSPT---PPVSPPPPTPTPSVPSPTPPVSPPPPTPTPSVPSPTPPVSP-PPPTPTPS 161
Query: 322 LAPVSSPLP 330
+ + P+P
Sbjct: 162 VPSPTPPVP 170
Score = 52.0 bits (123), Expect = 5e-07
Identities = 41/117 (35%), Positives = 51/117 (43%), Gaps = 20/117 (17%)
Query: 227 TPVVPLPTLQI-PSLPTSTPFTPQHDAPESINSSSQATNLVKPSFFVPPLSSAAMMMPPV 285
TP VP PT + P PT TP P P S + ++ P+ PP+S PP
Sbjct: 87 TPSVPSPTPPVSPPPPTPTPSVPSPTPPVSPPPPTPTPSVPSPT---PPVS------PPP 137
Query: 286 SSSIPTAP-------PLHPTGTVQRPYGTPMLQPFPPPTPPPSLAPVSSPLPNPAIS 335
+ P+ P P PT T P TP + P P+PPP PVS P P P S
Sbjct: 138 PTPTPSVPSPTPPVSPPPPTPTPSVPSPTPPVPTDPMPSPPP---PVSPPPPTPTPS 191
Score = 52.0 bits (123), Expect = 5e-07
Identities = 42/130 (32%), Positives = 55/130 (42%), Gaps = 10/130 (7%)
Query: 204 APITGQPYQSSATISSSGPTHAATPVVPLPTLQI-PSLPTSTPFTPQHDAPESINSSSQA 262
+P P S T S P TP VP PT + P PT TP P P S +
Sbjct: 82 SPPPPTPSVPSPTPPVSPPPPTPTPSVPSPTPPVSPPPPTPTPSVPSPTPPVSPPPPTPT 141
Query: 263 TNLVKPSFFVPPLSSAAMMMPPVSSSIPTAPPLHPTGTVQRPYGTPMLQPFPPPTPPPSL 322
++ P+ PP+S P + S+P+ P PT + P P + P PPPTP PS+
Sbjct: 142 PSVPSPT---PPVSPPP---PTPTPSVPSPTPPVPTDPMPSP--PPPVSP-PPPTPTPSV 192
Query: 323 APVSSPLPNP 332
P P
Sbjct: 193 PSPPDVTPTP 202
Score = 51.6 bits (122), Expect = 7e-07
Identities = 52/181 (28%), Positives = 68/181 (36%), Gaps = 20/181 (11%)
Query: 160 PTMAVMDGPLEPSSSITSNVADVPDDPSFINFFSAAMGIGNTSNAPITGQPYQSSATISS 219
PT V P P+ S+ S V P S S P T P S T
Sbjct: 93 PTPPVSPPPPTPTPSVPSPTPPVSPPPPTPTP-SVPSPTPPVSPPPPTPTPSVPSPTPPV 151
Query: 220 SGPTHAATPVVPLPTLQIPSLPTSTPFTPQHDAPESINSSSQATNLVKPSFFVPPLSSAA 279
S P TP VP PT +P+ P +P P P + S + V P+ P + S
Sbjct: 152 SPPPPTPTPSVPSPTPPVPTDPMPSPPPPVSPPPPTPTPSVPSPPDVTPTPPTPSVPSPP 211
Query: 280 MMMP-PVSSSIPTAPPLHPTGTVQRPYGTPMLQPFPPPTPPPSLAPVSSP--LPNPAISR 336
+ P P + S+P+ P + PT PPTPP P SP +P P+
Sbjct: 212 DVTPTPPTPSVPSPPDVTPT----------------PPTPPSVPTPSGSPPYVPPPSDEE 255
Query: 337 E 337
E
Sbjct: 256 E 256
Score = 43.5 bits (101), Expect = 2e-04
Identities = 32/78 (41%), Positives = 35/78 (44%), Gaps = 16/78 (20%)
Query: 273 PPLSSAAMMMPPVSSSIPT--------APPLH---PTGTVQRPYGTPMLQPFPPPTPPPS 321
PP S PPVS PT PP+ PT T P TP + P PPPTP PS
Sbjct: 85 PPTPSVPSPTPPVSPPPPTPTPSVPSPTPPVSPPPPTPTPSVPSPTPPVSP-PPPTPTPS 143
Query: 322 L----APVSSPLPNPAIS 335
+ PVS P P P S
Sbjct: 144 VPSPTPPVSPPPPTPTPS 161
Score = 41.6 bits (96), Expect = 7e-04
Identities = 28/74 (37%), Positives = 38/74 (50%), Gaps = 11/74 (14%)
Query: 271 FVPPLSSAAMMMPPVSSSIPT-APPLHP---TGTVQRPYGTPMLQPFPPPTPPPSLA--- 323
+ PP + PP + S+P+ PP+ P T T P TP + P PPPTP PS+
Sbjct: 72 YTPPAPVPPVSPPPPTPSVPSPTPPVSPPPPTPTPSVPSPTPPVSP-PPPTPTPSVPSPT 130
Query: 324 -PVSSP--LPNPAI 334
PVS P P P++
Sbjct: 131 PPVSPPPPTPTPSV 144
Score = 37.4 bits (85), Expect = 0.013
Identities = 25/57 (43%), Positives = 30/57 (51%), Gaps = 9/57 (15%)
Query: 283 PPVSSSIPTAPPLHPTGTVQRPYGTPMLQPFPPPTPPPSL----APVSSPLPNPAIS 335
PP + +P P PT +V P TP + P PPPTP PS+ PVS P P P S
Sbjct: 74 PP--APVPPVSPPPPTPSVPSP--TPPVSP-PPPTPTPSVPSPTPPVSPPPPTPTPS 125
>At3g22120 unknown protein
Length = 351
Score = 51.6 bits (122), Expect = 7e-07
Identities = 40/130 (30%), Positives = 51/130 (38%), Gaps = 10/130 (7%)
Query: 201 TSNAPITGQPYQSSATISSSGPTHAATPVVPLPTLQIPSLPTSTP--FTPQHDAPESINS 258
T P T +P + S+ P H P P P P PT TP TP P I
Sbjct: 149 TKPPPSTPKPPTTKPPPSTPKPPHHKPPPTPCP----PPTPTPTPPVVTPPTPTPPVITP 204
Query: 259 SSQATNLVKPSFFVPPLSSAAMMMPPVSSSIPTAPPLHPTGTVQRPYGTPMLQPFP-PPT 317
+ +V P PP+ + PPV + PP+ T P TP P P PPT
Sbjct: 205 PTPTPPVVTPPTPTPPVITPPTPTPPVITPPTPTPPVVTPPTPTPPVVTP---PTPTPPT 261
Query: 318 PPPSLAPVSS 327
P P P+ +
Sbjct: 262 PIPETCPIDT 271
Score = 47.8 bits (112), Expect = 1e-05
Identities = 40/132 (30%), Positives = 52/132 (39%), Gaps = 16/132 (12%)
Query: 205 PITGQPYQSSATISSSGPTHAATPVV-PLPTLQIPSLPTSTPFTPQHDAPESINSSSQAT 263
P T +P+ T+ H P V P P P P T H P+
Sbjct: 84 PPTVKPHPKPPTVKP----HPKPPTVKPHPKPPTVKPPHPKPPTKPHPHPKP-------- 131
Query: 264 NLVKPSFFVPPLSSAAMMMPPVSS-SIPTAPPLHPTGTVQRPYGTPMLQPFPPPTPPPSL 322
+VKP PP + PP S+ PT P P T + P+ P P PPPTP P+
Sbjct: 132 PIVKPPTKPPPSTPKPPTKPPPSTPKPPTTKP--PPSTPKPPHHKPPPTPCPPPTPTPTP 189
Query: 323 APVSSPLPNPAI 334
V+ P P P +
Sbjct: 190 PVVTPPTPTPPV 201
Score = 42.4 bits (98), Expect = 4e-04
Identities = 38/120 (31%), Positives = 46/120 (37%), Gaps = 14/120 (11%)
Query: 218 SSSGPTHAATPVVPLPTLQIPSLPTSTPFTPQHDAPESINSSSQATNLVKPSFFVPPLSS 277
S+ P P P P P P STP P H P + T P PP +
Sbjct: 143 STPKPPTKPPPSTPKPPTTKP--PPSTPKPPHHKPPPT--PCPPPTPTPTPPVVTPPTPT 198
Query: 278 AAMMMPPVSSSIPTAP---PLHPTGTVQRPYGTPMLQPFPPPTPPPSLAPVSSPLPNPAI 334
++ PP PT P P PT V P TP PPTP P + V+ P P P +
Sbjct: 199 PPVITPP----TPTPPVVTPPTPTPPVITP-PTPTPPVITPPTPTPPV--VTPPTPTPPV 251
Score = 40.0 bits (92), Expect = 0.002
Identities = 38/137 (27%), Positives = 49/137 (35%), Gaps = 14/137 (10%)
Query: 205 PITGQPYQSSATISSSGPTHAATPVVPLPTLQIPSLPTST---PFTPQHDAPESINSSSQ 261
P T +P+ T+ P H P P P + P + T P TP+ ++
Sbjct: 102 PPTVKPHPKPPTVK---PPHPKPPTKPHPHPKPPIVKPPTKPPPSTPKPPTKPPPSTPKP 158
Query: 262 ATNLVKPSFFVPPLSSAAMMMPPVSSSIPTAPPLHPTGTVQRPYGTPMLQPFPPP---TP 318
T PS PP PP PT P P T P + P P P TP
Sbjct: 159 PTTKPPPSTPKPPHHKP----PPTPCPPPTPTPTPPVVTPPTPTPPVITPPTPTPPVVTP 214
Query: 319 PPSLAPV-SSPLPNPAI 334
P PV + P P P +
Sbjct: 215 PTPTPPVITPPTPTPPV 231
Score = 34.3 bits (77), Expect = 0.11
Identities = 21/60 (35%), Positives = 25/60 (41%), Gaps = 6/60 (10%)
Query: 275 LSSAAMMMPPVSSSIPTAPPLHPTGTVQRPYGTPMLQPFPPPT--PPPSLAPVSSPLPNP 332
+S A PP S P PP HP +P P ++P PP PP P P P P
Sbjct: 38 VSYACDCTPPKPSPAPHKPPKHPV----KPPKPPAVKPPKPPAVKPPTPKPPTVKPHPKP 93
Score = 33.5 bits (75), Expect = 0.19
Identities = 25/89 (28%), Positives = 32/89 (35%), Gaps = 7/89 (7%)
Query: 247 TPQHDAPESINSSSQATNLVKPSFFVPPLSSAAMMMPPVSSSIPTAPPLHPTGTVQRPYG 306
TP +P KP PP A + P + PT P HP +P+
Sbjct: 45 TPPKPSPAPHKPPKHPVKPPKPPAVKPPKPPA---VKPPTPKPPTVKP-HPKPPTVKPHP 100
Query: 307 TPML---QPFPPPTPPPSLAPVSSPLPNP 332
P P PP PP P + P P+P
Sbjct: 101 KPPTVKPHPKPPTVKPPHPKPPTKPHPHP 129
>At1g26150 Pto kinase interactor, putative
Length = 760
Score = 51.6 bits (122), Expect = 7e-07
Identities = 60/218 (27%), Positives = 83/218 (37%), Gaps = 36/218 (16%)
Query: 136 AYAKVPPKLKVSSTKSEFEELEAVPTMAVMDGPLEPSSSITSNVADVPDDPSFINFFSAA 195
A + PP+ +SS E P + GP P ++I + P P +
Sbjct: 58 AQSSPPPETPLSSPPPE-----PSPPSPSLTGP--PPTTIPVSPPPEPSPPPPL------ 104
Query: 196 MGIGNTSNAPITGQPYQSSATISSSGPTHAATPVVPLPTLQIPSLPTSTPFTPQHDAPES 255
+ AP P SS SS P T P + PS PT+ P P ++P S
Sbjct: 105 -----PTEAPPPANPV-SSPPPESSPPPPPPTEAPPTTPITSPSPPTNPP--PPPESPPS 156
Query: 256 INSSSQATNLVKPSFFVPPLSSAAMMMP-PVSSSIPTAPPLHPTGTVQRPYGTP---MLQ 311
+ + +N + P VPP S +P P +S IP P P+ TP
Sbjct: 157 LPAPDPPSNPLPPPKLVPPSHSPPRHLPSPPASEIPPPPRHLPSPPASERPSTPPSDSEH 216
Query: 312 PFPP----------PTPPPSLAPVSSPLPNPAISREKV 339
P PP P PP S P SP P+P+ S+ V
Sbjct: 217 PSPPPPGHPKRREQPPPPGSKRPTPSP-PSPSDSKRPV 253
Score = 50.4 bits (119), Expect = 1e-06
Identities = 58/179 (32%), Positives = 72/179 (39%), Gaps = 15/179 (8%)
Query: 169 LEPSSSITSNVADVPDDPSFI--NFFSAAMGIGNTSNAPITGQPYQSSATISSSGPTHAA 226
L PS + +A P PSF N S N + T P QSS + +
Sbjct: 14 LSPSLASPPLMALPPPQPSFPGDNATSPTREPTNGNPPETTNTPAQSSPPPETPLSSPPP 73
Query: 227 TPVVPLPTLQIPSLPTSTPFTPQHDAPESINSSSQATNLVKPSFFVPPLSSAAMMMPPVS 286
P P P+L P PT+ P +P + ++A P PP SS PP +
Sbjct: 74 EPSPPSPSLTGPP-PTTIPVSPPPEPSPPPPLPTEAPPPANPVSSPPPESSPPP--PPPT 130
Query: 287 SSIPTAP---PLHPTGTVQRPYGTPMLQ-PFPP--PTPPPSLAPVS-SP---LPNPAIS 335
+ PT P P PT P P L P PP P PPP L P S SP LP+P S
Sbjct: 131 EAPPTTPITSPSPPTNPPPPPESPPSLPAPDPPSNPLPPPKLVPPSHSPPRHLPSPPAS 189
Score = 45.8 bits (107), Expect = 4e-05
Identities = 53/208 (25%), Positives = 73/208 (34%), Gaps = 24/208 (11%)
Query: 141 PPKLKVSSTKSEFEELEAVPTMAVMDGPLEPSSSITSNVADVPDDPSFINFFSAAMGIGN 200
PP VSS E PT A P+ S S +N P+ P +
Sbjct: 110 PPANPVSSPPPESSPPPPPPTEAPPTTPIT-SPSPPTNPPPPPESPPSLP---------- 158
Query: 201 TSNAPITGQPYQSSATISSSGPTHAATPVVPLPTLQIPSLPTSTPFTPQHDAPESINSSS 260
+ P P S S P H +P P +IP P P P + P + S S
Sbjct: 159 APDPPSNPLPPPKLVPPSHSPPRHLPSP----PASEIPPPPRHLPSPPASERPSTPPSDS 214
Query: 261 QATNLVKPSF----FVPPLSSAAMMMPPVSSSIPTAPPLHPTGTVQRPYGTPMLQPFPPP 316
+ + P PP + P S + P+HP+ P +P P P
Sbjct: 215 EHPSPPPPGHPKRREQPPPPGSKRPTPSPPSPSDSKRPVHPSPPSPPEETLPPPKPSPDP 274
Query: 317 TP-----PPSLAPVSSPLPNPAISREKV 339
P PP+L P SS + P+ R+ V
Sbjct: 275 LPSNSSSPPTLLPPSSVVSPPSPPRKSV 302
Score = 43.9 bits (102), Expect = 1e-04
Identities = 42/158 (26%), Positives = 55/158 (34%), Gaps = 16/158 (10%)
Query: 202 SNAPITGQPYQSSATISSSGPTHAATPVVPLPTLQIPSLPTSTPFTPQHDAPESINSSSQ 261
+ AP T P S + ++ P + P +P P LP P H P + S
Sbjct: 130 TEAPPT-TPITSPSPPTNPPPPPESPPSLPAPDPPSNPLPPPKLVPPSHSPPRHLPSPPA 188
Query: 262 ATNLVKPSFFVPPLSSAAMMMPPVSSSIPTAPPL-HPTGTVQ-RPYGTPMLQPFP----- 314
+ P P +S PP S P+ PP HP Q P G+ P P
Sbjct: 189 SEIPPPPRHLPSPPASERPSTPPSDSEHPSPPPPGHPKRREQPPPPGSKRPTPSPPSPSD 248
Query: 315 --------PPTPPPSLAPVSSPLPNPAISREKVRDALL 344
PP+PP P P P+P S LL
Sbjct: 249 SKRPVHPSPPSPPEETLPPPKPSPDPLPSNSSSPPTLL 286
>At4g18670 extensin-like protein
Length = 839
Score = 50.8 bits (120), Expect = 1e-06
Identities = 53/181 (29%), Positives = 71/181 (38%), Gaps = 26/181 (14%)
Query: 171 PSSSITSNVADVPDDPSFINFFSAAMGIGNTSNAPITGQPYQSSATISSSGPTHAATPVV 230
PS T + P PS + S P G P +S T + G + ++P
Sbjct: 437 PSPPTTPSPGGSPPSPSIVP--------SPPSTTPSPGSP-PTSPTTPTPGGSPPSSPTT 487
Query: 231 PLPTLQIPSLPT---------STPFTPQ---HDAPESINSSSQATNLVKPSFFVPPLSSA 278
P P PS PT S+P TP SI+ S T PS P S
Sbjct: 488 PTPGGSPPSSPTTPTPGGSPPSSPTTPSPGGSPPSPSISPSPPITVPSPPSTPTSPGSPP 547
Query: 279 AMMMPPVSSSIPTAP-PLHPTGTVQRPYGTPMLQPFPP---PTPPPSLAPVSSP-LPNPA 333
+ P SS IP+ P P P + +P + P PP P+PP S +P P +P+P
Sbjct: 548 SPSSPTPSSPIPSPPTPSTPPTPISPGQNSPPIIPSPPFTGPSPPSSPSPPLPPVIPSPP 607
Query: 334 I 334
I
Sbjct: 608 I 608
Score = 47.8 bits (112), Expect = 1e-05
Identities = 47/152 (30%), Positives = 68/152 (43%), Gaps = 33/152 (21%)
Query: 195 AMGIGNTSNAPITGQPYQSSATISSSGPTHAATPVVPLPTLQIPSLPTSTPFTPQHDAPE 254
+ G G ++ P+ S T S G + + + P P + +PS PT TP
Sbjct: 397 SFGCGRSTRPPVV---VPSPPTTPSPGGSPPSPSISPSPPITVPSPPT-TP--------- 443
Query: 255 SINSSSQATNLVKPSFFVPPLSSAAMMMPPVSSSIPT---APPLHPTGTVQRPYGTPMLQ 311
S S + ++V PS PP ++ + PP S + PT +PP P T P G+P
Sbjct: 444 SPGGSPPSPSIV-PS---PPSTTPSPGSPPTSPTTPTPGGSPPSSP--TTPTPGGSP--- 494
Query: 312 PFPPPTPPPSLAPVSSPL--------PNPAIS 335
P P TP P +P SSP P+P+IS
Sbjct: 495 PSSPTTPTPGGSPPSSPTTPSPGGSPPSPSIS 526
Score = 46.2 bits (108), Expect = 3e-05
Identities = 40/132 (30%), Positives = 56/132 (42%), Gaps = 5/132 (3%)
Query: 200 NTSNAPITGQPYQSSATISSSGPTHAATPVVPLPTLQIPSLPTSTPFTP-QHDAPESINS 258
++ P G SS T S G + + + P P + +PS P STP +P +P S
Sbjct: 496 SSPTTPTPGGSPPSSPTTPSPGGSPPSPSISPSPPITVPS-PPSTPTSPGSPPSPSSPTP 554
Query: 259 SSQATNLVKPSFFVPPLSSAAMMMPPVSSSIPTAPPLHPTGTVQRPYGTPMLQPFPPPTP 318
SS + PS P+ S PP+ S P P P+ P P++ P P
Sbjct: 555 SSPIPSPPTPSTPPTPI-SPGQNSPPIIPSPPFTGPSPPSS--PSPPLPPVIPSPPIVGP 611
Query: 319 PPSLAPVSSPLP 330
PS P S+P P
Sbjct: 612 TPSSPPPSTPTP 623
Score = 30.8 bits (68), Expect = 1.2
Identities = 34/131 (25%), Positives = 48/131 (35%), Gaps = 24/131 (18%)
Query: 204 APITGQPYQSSATISSSGPTHAATPVVPLPTLQIPSLPTSTPFTPQHDAPESINSSSQAT 263
+P PY S+ P ++ P P PT S P P TP H P +
Sbjct: 701 SPPPPAPYYYSSPQPPPPPHYSLPP--PTPTYHYISPPP--PPTPIHSPPPQSH------ 750
Query: 264 NLVKPSFFVPPLSSAAMMMPPVSSSIPTAPPLHPTGTVQRPYGTPMLQPFPPPTPPPSLA 323
PP + PP ++ PP P+ P +P + + P PPP++
Sbjct: 751 ---------PPCIEYS---PPPPPTVHYNPPPPPSPAHYSPPPSPPVYYYNSPPPPPAVH 798
Query: 324 PVSSPLPNPAI 334
SP P P I
Sbjct: 799 --YSPPPPPVI 807
>At4g32710 putative protein kinase
Length = 731
Score = 50.1 bits (118), Expect = 2e-06
Identities = 42/135 (31%), Positives = 56/135 (41%), Gaps = 16/135 (11%)
Query: 213 SSATISSSGPTHAATPVVPLPTLQIPSLPTSTPFTPQHDAPESINSSSQATNLVKPSFFV 272
SS ++S P + P +P P P +P P +P + S L P+
Sbjct: 7 SSPAPATSPPAMSLPPADSVPDTSSPPAPPLSPLPPPLSSPPPLPSPPP---LSAPTASP 63
Query: 273 PPLSSAAMMMPPVSS------SIPTAPPLH----PTGTVQRPYGTPMLQPFPP--PTPPP 320
PPL + PP+ S P PPL P+ V P G+P L PF P P+PPP
Sbjct: 64 PPLPVESPPSPPIESPPPPLLESPPPPPLESPSPPSPHVSAPSGSPPL-PFLPAKPSPPP 122
Query: 321 SLAPVSSPLPNPAIS 335
S P + P IS
Sbjct: 123 SSPPSETVPPGNTIS 137
Score = 48.1 bits (113), Expect = 7e-06
Identities = 44/174 (25%), Positives = 67/174 (38%), Gaps = 13/174 (7%)
Query: 168 PLEPSSSITSNVADVPDDP--SFINFFSAAMGIGNTSNAPITGQPYQSSATISSSGPTHA 225
PL P S+ +P P S + + + + PI P + S P
Sbjct: 36 PLSPLPPPLSSPPPLPSPPPLSAPTASPPPLPVESPPSPPIESPP---PPLLESPPPPPL 92
Query: 226 ATPVVPLPTLQIPSLPTSTPFTPQHDAPESINSSSQAT---NLVKPSFFVPPLSSAAMMM 282
+P P P + PS PF P +P + S+ N + P PP S +
Sbjct: 93 ESPSPPSPHVSAPSGSPPLPFLPAKPSPPPSSPPSETVPPGNTISP----PPRSLPSEST 148
Query: 283 PPVSSSIPTAPPLHPTGTVQRPYGTPMLQPFPP-PTPPPSLAPVSSPLPNPAIS 335
PPV+++ P P + +P P + PP P+P P +SP P P +S
Sbjct: 149 PPVNTASPPPPSPPRRRSGPKPSFPPPINSSPPNPSPNTPSLPETSPPPKPPLS 202
Score = 43.9 bits (102), Expect = 1e-04
Identities = 31/78 (39%), Positives = 37/78 (46%), Gaps = 13/78 (16%)
Query: 258 SSSQATNLVKPSFFVPPLSSAAMMMPPVSSSIPTAPPLHPTGTVQRPYGTPMLQPFPPPT 317
SSS A P+ +PP S +P SS P APPL P P+ P P P+
Sbjct: 6 SSSPAPATSPPAMSLPPADS----VPDTSS--PPAPPL-------SPLPPPLSSPPPLPS 52
Query: 318 PPPSLAPVSSPLPNPAIS 335
PPP AP +SP P P S
Sbjct: 53 PPPLSAPTASPPPLPVES 70
Score = 39.7 bits (91), Expect = 0.003
Identities = 57/224 (25%), Positives = 85/224 (37%), Gaps = 37/224 (16%)
Query: 134 LNAYAKVPPKLKVSSTKSEFEELEAVPTMAV-----MDGPLEPSSSITSNVADVP----- 183
L+A PP L V S S E P + ++ P PS +++ P
Sbjct: 56 LSAPTASPPPLPVESPPSPPIESPPPPLLESPPPPPLESPSPPSPHVSAPSGSPPLPFLP 115
Query: 184 --DDPSFINFFSAAMGIGNTSNAPITGQPYQSSATISSSGPTHAATPVVPLPTLQI---- 237
P + S + GNT + P P +S+ P + A+P P P +
Sbjct: 116 AKPSPPPSSPPSETVPPGNTISPPPRSLPSESTP------PVNTASPPPPSPPRRRSGPK 169
Query: 238 PSLPTSTPFTPQHDAPESINSSSQATNLVKPSFFVPPLSSAAMMMPPVSSSIPTAPPLHP 297
PS P +P + +P + S + + KP P S++ P S + T P P
Sbjct: 170 PSFPPPINSSPPNPSPNT-PSLPETSPPPKPPLSTTPFPSSSTPPPKKSPAAVTLPFFGP 228
Query: 298 T-----GTVQRPYGTPMLQP-------FPPPTPPPSLAPVSSPL 329
GTV P G P+++P P TP P L P S P+
Sbjct: 229 AGQLPDGTVAPPIG-PVIEPKTSPAESISPGTPQP-LVPKSLPV 270
Score = 38.9 bits (89), Expect = 0.004
Identities = 44/164 (26%), Positives = 61/164 (36%), Gaps = 27/164 (16%)
Query: 195 AMGIGNTSNAPITGQPYQSSATISSSGPTHAATPVVPLPTLQIPSLPTSTPFTPQHDAPE 254
A + +TS+ P P SS P + P + PT P LP +P +P ++P
Sbjct: 23 ADSVPDTSSPP--APPLSPLPPPLSSPPPLPSPPPLSAPTASPPPLPVESPPSPPIESPP 80
Query: 255 S----------INSSSQATNLVKPSFFVPPLS--SAAMMMPPVSSSIPTAPPLHPTGTVQ 302
+ S S + V PPL A PP S T PP +
Sbjct: 81 PPLLESPPPPPLESPSPPSPHVSAPSGSPPLPFLPAKPSPPPSSPPSETVPPGNTISPPP 140
Query: 303 RPYGTPMLQPF-----PPPTPP-------PSL-APVSSPLPNPA 333
R + P PPP+PP PS P++S PNP+
Sbjct: 141 RSLPSESTPPVNTASPPPPSPPRRRSGPKPSFPPPINSSPPNPS 184
Score = 37.0 bits (84), Expect = 0.017
Identities = 20/59 (33%), Positives = 29/59 (48%)
Query: 274 PLSSAAMMMPPVSSSIPTAPPLHPTGTVQRPYGTPMLQPFPPPTPPPSLAPVSSPLPNP 332
P SS A P + S+P A + T + P +P+ P P P PS P+S+P +P
Sbjct: 5 PSSSPAPATSPPAMSLPPADSVPDTSSPPAPPLSPLPPPLSSPPPLPSPPPLSAPTASP 63
>At5g38560 putative protein
Length = 681
Score = 49.7 bits (117), Expect = 3e-06
Identities = 41/135 (30%), Positives = 54/135 (39%), Gaps = 6/135 (4%)
Query: 205 PITGQPYQSSATISSSG-PTHAATPVVPLPTLQIPSLPTSTPFTPQHDAPESINSSSQAT 263
PI P +S+T + T TP P P PS P S P P + SS +
Sbjct: 8 PILSPPSSNSSTTAPPPLQTQPTTPSAPPPVTPPPSPPQSPPPVVSSSPPPPVVSSPPPS 67
Query: 264 NLVKPS---FFVPPLSSAAMMMPPVSSSIPTAPPLHPTGTVQRPYGTPMLQPFPPPTPPP 320
+ PS PP + A+ PPV + P PP P T P T P P +P P
Sbjct: 68 SSPPPSPPVITSPPPTVASSPPPPVVIASP--PPSTPATTPPAPPQTVSPPPPPDASPSP 125
Query: 321 SLAPVSSPLPNPAIS 335
++P P P+ S
Sbjct: 126 PAPTTTNPPPKPSPS 140
Score = 48.9 bits (115), Expect = 4e-06
Identities = 45/147 (30%), Positives = 56/147 (37%), Gaps = 12/147 (8%)
Query: 202 SNAPITGQPYQSSATISSSGPTHAATPVVPLPTLQIP-SLPTSTPFTPQHDA----PESI 256
S+ P + P S I+S PT A++P P+ P S P +TP P P
Sbjct: 62 SSPPPSSSPPPSPPVITSPPPTVASSPPPPVVIASPPPSTPATTPPAPPQTVSPPPPPDA 121
Query: 257 NSSSQATNLVKPSFFVPPLSSAAMMMPPVSSSIPTAP-PLHPTGTVQR----PYGTPMLQ 311
+ S A P P PP + P P P PT T P T
Sbjct: 122 SPSPPAPTTTNPPPKPSPSPPGETPSPPGETPSPPKPSPSTPTPTTTTSPPPPPATSASP 181
Query: 312 PFPPPTPPPSLAPVSSPLPNPAISREK 338
P PT P +LAP P P P + REK
Sbjct: 182 PSSNPTDPSTLAP--PPTPLPVVPREK 206
Score = 44.7 bits (104), Expect = 8e-05
Identities = 36/118 (30%), Positives = 49/118 (41%), Gaps = 4/118 (3%)
Query: 229 VVPLPTLQIPSLPTSTPFTPQ-HDAPESINSSSQATNLVKPSFFVPPLSSAAMMMPPVSS 287
V PLP L PS +ST P P + ++ T P PP+ S++ P VSS
Sbjct: 4 VPPLPILSPPSSNSSTTAPPPLQTQPTTPSAPPPVTPPPSPPQSPPPVVSSSPPPPVVSS 63
Query: 288 SIPTAPPLHPTGTVQRPYGTPMLQPFPP---PTPPPSLAPVSSPLPNPAISREKVRDA 342
P++ P + P T P PP +PPPS + P P +S DA
Sbjct: 64 PPPSSSPPPSPPVITSPPPTVASSPPPPVVIASPPPSTPATTPPAPPQTVSPPPPPDA 121
>At5g14920 unknown protein
Length = 275
Score = 49.3 bits (116), Expect = 3e-06
Identities = 42/137 (30%), Positives = 57/137 (40%), Gaps = 15/137 (10%)
Query: 210 PYQSSATISSSGPTHAATPVVPLPTLQIPSLPTSTPFTPQHDAPESINSSSQATNLVKPS 269
P S + ++ P+ A P P P+ + P+LPT TP P P + + VKP
Sbjct: 36 PTLPSPSPATKPPSPALKP--PTPSYKPPTLPT-TPIKPPTTKPP-VKPPTIPVTPVKPP 91
Query: 270 FFVPPLSSAAMMMPPVSSSIPTA------PPLH--PTGTVQRPYGTPMLQPFPPPTPPPS 321
PP+ + P PT PP + PT TV+ P +P+ PP TPP
Sbjct: 92 VSTPPIKLPPVQPPTYKPPTPTVKPPSVQPPTYKPPTPTVKPPTTSPVK---PPTTPPVQ 148
Query: 322 LAPVSSPLPNPAISREK 338
PV P P S K
Sbjct: 149 SPPVQPPTYKPPTSPVK 165
Score = 46.6 bits (109), Expect = 2e-05
Identities = 44/126 (34%), Positives = 52/126 (40%), Gaps = 24/126 (19%)
Query: 222 PTHAATPVVPLPTLQIPSL--PTSTPFTPQHDAPESINSSSQATNLVKPSFFVPPLSSAA 279
P T P PT++ PS+ PT P TP P T+ VKP PP+ S
Sbjct: 101 PVQPPTYKPPTPTVKPPSVQPPTYKPPTPTVKPP--------TTSPVKPPT-TPPVQSPP 151
Query: 280 MM----MPPVSSSIP--TAPPLHPTGT---VQRPYGTPMLQPFPPPTPPPSLAPVSSPLP 330
+ PP S P T PP+ P T VQ P P P PPT P PV P P
Sbjct: 152 VQPPTYKPPTSPVKPPTTTPPVKPPTTTPPVQPPTYNPPTTPVKPPTAP----PVKPPTP 207
Query: 331 NPAISR 336
P +R
Sbjct: 208 PPVRTR 213
Score = 45.1 bits (105), Expect = 6e-05
Identities = 31/122 (25%), Positives = 46/122 (37%), Gaps = 6/122 (4%)
Query: 213 SSATISSSGPTHAATPVVPLPTLQIPSLPTSTPFTPQHDAPESINSSSQATNLVKPSFFV 272
++ ++S A +P PTL PS T P S + T +KP
Sbjct: 16 TNVVFAASNEESNALVSLPTPTLPSPSPATKPPSPALKPPTPSYKPPTLPTTPIKPPTTK 75
Query: 273 PPLSSAAMMMPPVSSSIPTAPPLHPTGTVQRPYGTPMLQPFPPPTPPPSLAPVSSPLPNP 332
PP+ + + PV + T P P P P +P P PPS+ P + P P
Sbjct: 76 PPVKPPTIPVTPVKPPVSTPPIKLP------PVQPPTYKPPTPTVKPPSVQPPTYKPPTP 129
Query: 333 AI 334
+
Sbjct: 130 TV 131
>At2g14890 arabinogalactan-protein AGP9
Length = 191
Score = 49.3 bits (116), Expect = 3e-06
Identities = 40/124 (32%), Positives = 52/124 (41%), Gaps = 13/124 (10%)
Query: 215 ATISSSGPTH--AATPVVPLPTLQIPSLPTSTPFTPQHDAPESINSSSQ-ATNLVKPSFF 271
A ++ PT ATP P PT P+ +TP P AP + +S T P+
Sbjct: 16 AGVTGQAPTSPPTATPAPPTPTTPPPA---ATP--PPVSAPPPVTTSPPPVTTAPPPANP 70
Query: 272 VPPLSSAAMMMPPVSSSIPTAPPLHPTGTVQRPYGTPMLQPFPPPTP---PPSLAPVSSP 328
PP+SS PP ++ P A P P P TP PPP P PP+ P +P
Sbjct: 71 PPPVSSPPPASPPPATPPPVASP--PPPVASPPPATPPPVATPPPAPLASPPAQVPAPAP 128
Query: 329 LPNP 332
P
Sbjct: 129 TTKP 132
Score = 48.9 bits (115), Expect = 4e-06
Identities = 43/136 (31%), Positives = 55/136 (39%), Gaps = 13/136 (9%)
Query: 204 APITGQPYQSSATISSSGPTHAATPVVPLPTLQIPSLPTSTPFTPQHDAPESINSSSQAT 263
A +TGQ S T + + PT P P P P P S P P +P + ++
Sbjct: 16 AGVTGQAPTSPPTATPAPPT----PTTPPPAATPP--PVSAP-PPVTTSPPPVTTAPPPA 68
Query: 264 NLVKPSFFVPPLSSAAMMMPPVSSSIPTAPPLHPTGTVQRPYGTPMLQPFP-PPTPPPSL 322
N P PP S PPV+S P P P P TP P PP P+
Sbjct: 69 NPPPPVSSPPPASPPPATPPPVAS--PPPPVASPPPATPPPVATPPPAPLASPPAQVPAP 126
Query: 323 APVS---SPLPNPAIS 335
AP + SP P+P+ S
Sbjct: 127 APTTKPDSPSPSPSSS 142
Score = 32.0 bits (71), Expect = 0.55
Identities = 31/117 (26%), Positives = 45/117 (37%), Gaps = 7/117 (5%)
Query: 201 TSNAPITGQPYQSSATISSSGPTHAATPVVPLPTLQIPSLPTSTPFTPQHDAPESINSSS 260
TS P+T P ++ S P A+ P P + P P ++P P P ++
Sbjct: 56 TSPPPVTTAPPPANPPPPVSSPPPASPPPATPPPVASPPPPVASP-PPATPPPV---ATP 111
Query: 261 QATNLVKPSFFVPPLSSAAMMMPPVSSSIPTAPPLHPTGTVQRPYGTPMLQPFPPPT 317
L P VP + A P S P++ P P+ P T + P P PT
Sbjct: 112 PPAPLASPPAQVP--APAPTTKPDSPSPSPSSSPPLPSSDAPGP-STDSISPAPSPT 165
>At4g13340 extensin-like protein
Length = 760
Score = 48.9 bits (115), Expect = 4e-06
Identities = 45/152 (29%), Positives = 54/152 (34%), Gaps = 27/152 (17%)
Query: 202 SNAPITGQPYQSSATISSSGPTHAATPVVPLPTLQIPSLPTSTPFTPQHDAPESINS--- 258
S P+ + ++S P V PLP PSLP+ P P P ++ S
Sbjct: 377 SRPPVNCGSFSCGRSVSPRPPV-----VTPLPP---PSLPSPPPPAPIFSTPPTLTSPPP 428
Query: 259 --------SSQATNLVKPSFFVPPLSSAAMMMPPVSSSIPTAPPLHPTGTVQRPYGT--- 307
S P + PP PPV S P PP P V P
Sbjct: 429 PSPPPPVYSPPPPPPPPPPVYSPPPPPPPPPPPPVYSPPPPPPPPPPPPPVYSPPPPSPP 488
Query: 308 ----PMLQPFPPPTPPPSLAPVSSPLPNPAIS 335
P+ P PPP PPP PV SP P P S
Sbjct: 489 PPPPPVYSP-PPPPPPPPPPPVYSPPPPPVYS 519
Score = 43.5 bits (101), Expect = 2e-04
Identities = 36/121 (29%), Positives = 49/121 (39%), Gaps = 11/121 (9%)
Query: 217 ISSSGPTHAATPVVPLPTLQIPSLPTSTPFTPQHDAPESINSSSQATNLVKPSFFVPPLS 276
+SS PT +P P P ++ P P ++P P SS + S PP+
Sbjct: 598 VSSPPPTPVYSPPPPPPCIEPPPPPPCIEYSPPPPPPVVHYSSPPPPPVYYSSPPPPPVY 657
Query: 277 -SAAMMMPPVSSSIPTAPPLHPTGTVQRPYGTPMLQP--FPPPTPPPSLAPVSSPLPNPA 333
S+ PPV S P P +H Y +P P + P PPPS SP P P
Sbjct: 658 YSSPPPPPPVHYSSPPPPEVH--------YHSPPPSPVHYSSPPPPPSAPCEESPPPAPV 709
Query: 334 I 334
+
Sbjct: 710 V 710
Score = 42.4 bits (98), Expect = 4e-04
Identities = 36/114 (31%), Positives = 42/114 (36%), Gaps = 21/114 (18%)
Query: 222 PTHAATPVVPLPTLQIPSLPTSTPFTPQHDAPESINSSSQATNLVKPSFFVPPLSSAAMM 281
P +++ P P P P T P P H P P F PP
Sbjct: 516 PVYSSPPPPPSPA-PTPVYCTRPPPPPPHSPPP-------------PQFSPPPPEPYYYS 561
Query: 282 MPPVSSSIPTAPPLHPTGTVQRPYGTPMLQPFPPPTPPPSLAPVSSPLPNPAIS 335
PP S P PP P P P + P+ P PPP+ PVSSP P P S
Sbjct: 562 SPPPPHSSP--PPHSPPPPHSPP---PPIYPYLSPPPPPT--PVSSPPPTPVYS 608
Score = 41.2 bits (95), Expect = 0.001
Identities = 39/132 (29%), Positives = 46/132 (34%), Gaps = 7/132 (5%)
Query: 204 APITGQPYQSSATISSSGPTHAATPVVPLPTLQIPSLPT-STPFTPQHDAPESINSSSQA 262
API P T++S P PV P P P S P P P + S
Sbjct: 414 APIFSTP----PTLTSPPPPSPPPPVYSPPPPPPPPPPVYSPPPPPPPPPPPPVYSPPPP 469
Query: 263 TNLVKPS--FFVPPLSSAAMMMPPVSSSIPTAPPLHPTGTVQRPYGTPMLQPFPPPTPPP 320
P + PP S PPV S P PP P P P PPP+P P
Sbjct: 470 PPPPPPPPPVYSPPPPSPPPPPPPVYSPPPPPPPPPPPPVYSPPPPPVYSSPPPPPSPAP 529
Query: 321 SLAPVSSPLPNP 332
+ + P P P
Sbjct: 530 TPVYCTRPPPPP 541
Score = 41.2 bits (95), Expect = 0.001
Identities = 37/125 (29%), Positives = 52/125 (41%), Gaps = 18/125 (14%)
Query: 222 PTHAATPVV----PLPTLQIPSLPTSTPFTPQ---HDAPESINSSSQATNLVKPSFFVPP 274
P+ A TPV P P P P +P P+ + +P +SS + P PP
Sbjct: 525 PSPAPTPVYCTRPPPPPPHSPPPPQFSPPPPEPYYYSSPPPPHSSPPPHSPPPPHSPPPP 584
Query: 275 LSSAAMMMPPVSSSIPTAPPLHPTGTVQRPYGTPMLQPFPPPT-------PPPSLAPVSS 327
+ + PP + ++PP PT P P ++P PPP PPP + SS
Sbjct: 585 IYP--YLSPPPPPTPVSSPP--PTPVYSPPPPPPCIEPPPPPPCIEYSPPPPPPVVHYSS 640
Query: 328 PLPNP 332
P P P
Sbjct: 641 PPPPP 645
Score = 34.7 bits (78), Expect = 0.085
Identities = 33/124 (26%), Positives = 41/124 (32%), Gaps = 16/124 (12%)
Query: 211 YQSSATISSSGPTHAATPVVPLPTLQIPSLPTSTPFTPQHDAPESINSSSQATNLVKPSF 270
Y S SS P H+ P P P L P TP P +
Sbjct: 560 YSSPPPPHSSPPPHSPPPPHSPPPPIYPYLSPPPPPTPVSSPPPT-------------PV 606
Query: 271 FVPPLSSAAMMMPPVSSSIPTAPPLHPTGTVQRPYGTPMLQPFPPPTPPPSLAPVSSPLP 330
+ PP + PP I +PP P Y +P P +PPP SSP P
Sbjct: 607 YSPPPPPPCIEPPPPPPCIEYSPPPPPPVV---HYSSPPPPPVYYSSPPPPPVYYSSPPP 663
Query: 331 NPAI 334
P +
Sbjct: 664 PPPV 667
Score = 32.0 bits (71), Expect = 0.55
Identities = 33/112 (29%), Positives = 39/112 (34%), Gaps = 10/112 (8%)
Query: 222 PTHAATPVVPLPTLQIPSLPTSTPFTPQHDAPESINSSSQATNLVKPSFFVPPLSSAAMM 281
P + P P P P + + P +P P + P + PP
Sbjct: 461 PPVYSPPPPPPPPPPPPPVYSPPPPSPPPPPPPVYSPPPPPPPPPPPPVYSPP------- 513
Query: 282 MPPVSSSIPTAPPLHPTGTV-QRPYGTPMLQPFPPPTPPPSLAP--VSSPLP 330
PPV SS P P PT RP P P PP PP P SSP P
Sbjct: 514 PPPVYSSPPPPPSPAPTPVYCTRPPPPPPHSPPPPQFSPPPPEPYYYSSPPP 565
>At3g19020 hypothetical protein
Length = 951
Score = 48.9 bits (115), Expect = 4e-06
Identities = 47/160 (29%), Positives = 60/160 (37%), Gaps = 16/160 (10%)
Query: 182 VPDDPSFINFFSAAMGIGNTSNAPITGQPYQSSATISSSGPTHAATPVVPLPTLQIPSLP 241
VP+DP A I P + ++ T S P H+ P P+ + P P
Sbjct: 604 VPNDPY------DASPIKKRRPQPPSPSTEETKTTSPQSPPVHSPPPPPPVHS---PPPP 654
Query: 242 TSTPFTPQHDAPESINSSSQATNLVKPSFFVPPLSSAAMMMPPVSSSIPTA----PPLH- 296
+P P H P + S + P P PPV S P PP+H
Sbjct: 655 VFSPPPPMHSPPPPVYSPPPPVHSPPPPPVHSPPPPVHSPPPPVHSPPPPVHSPPPPVHS 714
Query: 297 PTGTVQRPYGTPMLQPFPPPT-PPPSLAPVSSPLPNPAIS 335
P V P P+ P PPP PP AP+ SP P P S
Sbjct: 715 PPPPVHSP-PPPVQSPPPPPVFSPPPPAPIYSPPPPPVHS 753
Score = 48.5 bits (114), Expect = 6e-06
Identities = 36/114 (31%), Positives = 46/114 (39%), Gaps = 8/114 (7%)
Query: 222 PTHAATPVV---PLPTLQIPSLPTSTPFTPQHDAPESINSSSQATNLVKPSFFVPPLSSA 278
P ++ P V P P + P P +P P H P ++S + P PP
Sbjct: 668 PVYSPPPPVHSPPPPPVHSPPPPVHSPPPPVHSPPPPVHSPPPPVHSPPPPVHSPPPPVQ 727
Query: 279 AMMMPPVSSSIPTAPPLHPTGTVQRPYGTPMLQPFPPP--TPPPSLAPVSSPLP 330
+ PPV S P AP P P+ P PPP +PPP PV SP P
Sbjct: 728 SPPPPPVFSPPPPAPIYSPPPPPVHSPPPPVHSPPPPPVHSPPP---PVHSPPP 778
Score = 45.8 bits (107), Expect = 4e-05
Identities = 43/133 (32%), Positives = 51/133 (38%), Gaps = 30/133 (22%)
Query: 222 PTHAATPVV--PLPTLQIPSLPT--------STPFTPQHDAPESINSSSQATNLVKPSFF 271
P H+ P V P P + P P S P P H P ++S + P
Sbjct: 647 PVHSPPPPVFSPPPPMHSPPPPVYSPPPPVHSPPPPPVHSPPPPVHSPPPPVHSPPPPVH 706
Query: 272 VPPLSSAAMMMPPVSSSIPTAPPLH-PTGTVQRPYGTPMLQPFPP-----PTPPPSLA-- 323
PP PPV S PP+H P VQ P P+ P PP P PPP +
Sbjct: 707 SPP--------PPVHS---PPPPVHSPPPPVQSPPPPPVFSPPPPAPIYSPPPPPVHSPP 755
Query: 324 -PVSSPLPNPAIS 335
PV SP P P S
Sbjct: 756 PPVHSPPPPPVHS 768
Score = 45.8 bits (107), Expect = 4e-05
Identities = 36/121 (29%), Positives = 45/121 (36%), Gaps = 15/121 (12%)
Query: 222 PTHAATPVV---PLPTLQIPSLPTSTPFTPQHDAPESINSSSQATNLVKPSFFVPPLSSA 278
P H+ P P P + P P +P P H P ++S + P PP
Sbjct: 675 PVHSPPPPPVHSPPPPVHSPPPPVHSPPPPVHSPPPPVHSPPPPVHSPPPPVQSPPPPPV 734
Query: 279 AMMMPPVSSSIPTAPPLH-PTGTVQRPYGTPMLQPFPPP--------TPPPSLAPVSSPL 329
PP P PP+H P V P P+ P PP +PPP PV SP
Sbjct: 735 FSPPPPAPIYSPPPPPVHSPPPPVHSPPPPPVHSPPPPVHSPPPPVHSPPP---PVHSPP 791
Query: 330 P 330
P
Sbjct: 792 P 792
Score = 43.1 bits (100), Expect = 2e-04
Identities = 33/116 (28%), Positives = 42/116 (35%), Gaps = 12/116 (10%)
Query: 217 ISSSGPTHAATPVVPLPTLQIPSLPTSTPFTPQHD-APESINSSSQATNLVKPSFFVPPL 275
+ S P +P P P P P +P P H P ++S + P PP
Sbjct: 726 VQSPPPPPVFSPPPPAPIYSPPPPPVHSPPPPVHSPPPPPVHSPPPPVHSPPPPVHSPP- 784
Query: 276 SSAAMMMPPVSSSIPTAPPLHPTGTVQRPYGTPMLQPFPPPTPPPSLAPVSSPLPN 331
PPV S PP+H Y P PPP P L P +SP+ N
Sbjct: 785 -------PPVHS---PPPPVHSPPPPSPIYSPPPPVFSPPPKPVTPLPPATSPMAN 830
Score = 40.4 bits (93), Expect = 0.002
Identities = 35/114 (30%), Positives = 43/114 (37%), Gaps = 20/114 (17%)
Query: 222 PTHAATPVVPLPTLQIPSLPTSTPFTPQHDAPESINSSSQATNLVKPSFFVPPLSSAAMM 281
P H+ P V P P P +P P H P ++S P PP
Sbjct: 750 PVHSPPPPVHSP----PPPPVHSPPPPVHSPPPPVHSP-------PPPVHSPP------- 791
Query: 282 MPPVSSSIPTAPPLHPTGTVQRPYGTPMLQPFPPPTPPPSLAPVSSPLPNPAIS 335
PPV S P +P P V P P + P PP T P + AP S + IS
Sbjct: 792 -PPVHSPPPPSPIYSPPPPVFSPPPKP-VTPLPPATSPMANAPTPSSSESGEIS 843
Score = 40.0 bits (92), Expect = 0.002
Identities = 35/119 (29%), Positives = 41/119 (34%), Gaps = 18/119 (15%)
Query: 222 PTHAATPVV---PLPTLQIPSLPTSTPFTPQHDAPESINSSSQATNLVKPSFFVPPLSSA 278
P H+ P V P P P P +P P H P ++S + P PP
Sbjct: 661 PMHSPPPPVYSPPPPVHSPPPPPVHSPPPPVHSPPPPVHSPPPPVHSPPPPVHSPP---- 716
Query: 279 AMMMPPVSSSIPTAPPLHPTGTVQRPYGTPMLQPFPPPT-----PP--PSLAPVSSPLP 330
PPV S P P P P+ P PPP P P PV SP P
Sbjct: 717 ----PPVHSPPPPVQSPPPPPVFSPPPPAPIYSPPPPPVHSPPPPVHSPPPPPVHSPPP 771
Score = 37.7 bits (86), Expect = 0.010
Identities = 32/115 (27%), Positives = 37/115 (31%), Gaps = 9/115 (7%)
Query: 218 SSSGPTHAATPVVPLPTLQIPSLPTSTPFTPQHDAPESINSSSQATNLVKPSFFVPPLSS 277
S P + P P P P P+ P P+ PES S T KP P S
Sbjct: 428 SKPKPEESPKPQQPSPK---PETPSHEPSNPKEPKPESPKQESPKTEQPKPKPESPKQES 484
Query: 278 AAMMMPPVSSSIPTAPPLHPTGTVQRPYGTPMLQPFPPPTPPPSLAPVSSPLPNP 332
P P + Q P P + P P PP P SP P P
Sbjct: 485 PKQEAPKPEQPKPKPESPKQESSKQEP---PKPEESPKPEPP---KPEESPKPQP 533
Score = 30.8 bits (68), Expect = 1.2
Identities = 34/140 (24%), Positives = 42/140 (29%), Gaps = 12/140 (8%)
Query: 204 APITGQPYQSSATISSSGPTHAAT-PVVPLP---------TLQIPSLPTSTPFTPQHDAP 253
+P T QP + P A P P P + Q P P +P
Sbjct: 467 SPKTEQPKPKPESPKQESPKQEAPKPEQPKPKPESPKQESSKQEPPKPEESPKPEPPKPE 526
Query: 254 ESINSSSQATNLVKPSFFVPPLSSAAMMMPPVSSSIPTAPPLHPTGTVQRPYGTPMLQPF 313
ES KP P P S P P + P P Q
Sbjct: 527 ESPKPQPPKQETPKPEESPKPQPPKQETPKPEESPKPQPPKQETPKPEESPKPQPPKQEQ 586
Query: 314 PPPTPPPSLA--PVSSPLPN 331
PP T P + P+ SP+PN
Sbjct: 587 PPKTEAPKMGSPPLESPVPN 606
>At5g15780 proline-rich protein
Length = 401
Score = 48.5 bits (114), Expect = 6e-06
Identities = 41/117 (35%), Positives = 56/117 (47%), Gaps = 17/117 (14%)
Query: 228 PVVPLPTLQIPSLPTS-----TPFTPQHDA----PESINSSSQATNLVKPSFFVP--PLS 276
P +P+P L +P LP P PQ A +S + + T +KP+FF P PL+
Sbjct: 208 PNLPVPKLPVPDLPLPLVPPLLPPGPQKSASLHNKKSDSLKDKKTEALKPNFFFPPNPLN 267
Query: 277 SAAMMMP-PVSSSIPTAPPLHPTGTVQRPYGTPMLQPFPPPTPPPSLAPVSSPLPNP 332
+++ P P+ SIPT P L P + P P L P P PP+L P LP P
Sbjct: 268 PPSIIPPNPLIPSIPT-PTLPPNPLIPSP---PSLPPIPLIPTPPTL-PTIPLLPTP 319
Score = 46.6 bits (109), Expect = 2e-05
Identities = 44/152 (28%), Positives = 62/152 (39%), Gaps = 45/152 (29%)
Query: 228 PVVPLPTLQIPSLPTSTPFTPQHDAP----ESINSSSQATNLVKPSFFVPP--LSSAAMM 281
P +P+P L +P +P P PQ A +S + + T +KP+FF PP L+ +++
Sbjct: 213 PKLPVPDLPLPLVPPLLPPGPQKSASLHNKKSDSLKDKKTEALKPNFFFPPNPLNPPSII 272
Query: 282 MP------------------------------PVSSSIPTAP--PLHPTGTVQRPYGTPM 309
P P ++PT P P PT T+ P
Sbjct: 273 PPNPLIPSIPTPTLPPNPLIPSPPSLPPIPLIPTPPTLPTIPLLPTPPTPTLPPIPTIPT 332
Query: 310 LQPFP--PPTP---PPSL--APVSSPLPNPAI 334
L P P PP P PPSL P S P+P P +
Sbjct: 333 LPPLPVLPPVPIVNPPSLPPPPPSFPVPLPPV 364
Score = 38.5 bits (88), Expect = 0.006
Identities = 37/116 (31%), Positives = 47/116 (39%), Gaps = 21/116 (18%)
Query: 222 PTHAATPVVPLPTLQ----IPSLPTSTPFTPQHDAPESINSSSQATNLVKPSFFVPPLSS 277
P + P +P PTL IPS P S P P P ++ + P PP +
Sbjct: 273 PPNPLIPSIPTPTLPPNPLIPS-PPSLPPIPLIPTPPTLPTI--------PLLPTPPTPT 323
Query: 278 AAMMMPPVSSSIPTAPPLH---PTGTVQRPYGTPMLQPFPPPTPPPSLAPVSSPLP 330
+PP+ + IPT PPL P V P P FP P PP P P+P
Sbjct: 324 ----LPPIPT-IPTLPPLPVLPPVPIVNPPSLPPPPPSFPVPLPPVPGLPGIPPVP 374
Score = 30.8 bits (68), Expect = 1.2
Identities = 34/114 (29%), Positives = 45/114 (38%), Gaps = 25/114 (21%)
Query: 239 SLPTSTPFTPQHDAPESINSSSQATNLVKPSF-FVPPLSSAAMMMPPVSS-SIPTAPPLH 296
+L S P P P + + L P+ VPPL + + PV +P PPL
Sbjct: 172 NLRGSKPLLPDPSFPPPLQDPPNPSPL--PNLPIVPPLPNLPVPKLPVPDLPLPLVPPLL 229
Query: 297 PTGTVQ------------RPYGTPMLQP---FPP-PTPPPSLAPVSSPLPNPAI 334
P G + + T L+P FPP P PPS+ P PNP I
Sbjct: 230 PPGPQKSASLHNKKSDSLKDKKTEALKPNFFFPPNPLNPPSIIP-----PNPLI 278
Score = 30.8 bits (68), Expect = 1.2
Identities = 19/44 (43%), Positives = 24/44 (54%), Gaps = 1/44 (2%)
Query: 304 PYGTPMLQPFPPPTPPPSLAPVSSPLPNPAISREKVRDALLVLV 347
P P LQ P P+P P+L P+ PLPN + + V D L LV
Sbjct: 183 PSFPPPLQDPPNPSPLPNL-PIVPPLPNLPVPKLPVPDLPLPLV 225
>At4g22470 extensin - like protein
Length = 375
Score = 48.5 bits (114), Expect = 6e-06
Identities = 36/108 (33%), Positives = 46/108 (42%), Gaps = 21/108 (19%)
Query: 228 PVVPLPTLQIPSLPTSTPFTPQHDAPESINSSSQATNLVKPSFFVPPLSSAAMMMPPVSS 287
P P P Q P+ PT P P +D P S+ PP+++ +PP
Sbjct: 44 PPPPQPDPQPPTPPTFQPAPPANDQPPPPPQSTSP----------PPVATTPPALPPK-- 91
Query: 288 SIPTAPPLHPTGTVQRPYGTPMLQPFPPPTPPPSLAPVSSPLPNPAIS 335
P PPL P P TP P P PPP++ P SP P PAI+
Sbjct: 92 --PLPPPLSP------PQTTPPPPPAITPPPPPAITPPLSP-PPPAIT 130
Score = 43.9 bits (102), Expect = 1e-04
Identities = 34/114 (29%), Positives = 46/114 (39%), Gaps = 21/114 (18%)
Query: 218 SSSGPTHAATPVVPLPTLQIPSLPTSTPFTPQHDAPESINSSSQATNLVKPSFFVPPLSS 277
S+S P A TP P+LP P P P++ A P PPLS
Sbjct: 75 STSPPPVATTP---------PALPPK-PLPPPLSPPQTTPPPPPAITPPPPPAITPPLSP 124
Query: 278 ---AAMMMPPVSSSIPTAPPLHPTGTVQRPYGTPMLQPFPPPTPPPSLAPVSSP 328
A PP++++ P PP +P P+ P P PPP++ P SP
Sbjct: 125 PPPAITPPPPLATTPPALPP--------KPLPPPLSPPQTTPPPPPAITPPLSP 170
Score = 41.6 bits (96), Expect = 7e-04
Identities = 34/114 (29%), Positives = 41/114 (35%), Gaps = 2/114 (1%)
Query: 222 PTHAATPVVPLPTLQIPSLPTSTPFTPQHDAPESINSSSQATNLVKPSFFVPPLSSAAMM 281
PT P Q P P ST P P ++ L P PP A+
Sbjct: 54 PTPPTFQPAPPANDQPPPPPQSTSPPPVATTPPALPPKPLPPPLSPPQTTPPP--PPAIT 111
Query: 282 MPPVSSSIPTAPPLHPTGTVQRPYGTPMLQPFPPPTPPPSLAPVSSPLPNPAIS 335
PP + P P P T P T P P PPP P ++P P PAI+
Sbjct: 112 PPPPPAITPPLSPPPPAITPPPPLATTPPALPPKPLPPPLSPPQTTPPPPPAIT 165
>At2g25050 hypothetical protein
Length = 742
Score = 48.5 bits (114), Expect = 6e-06
Identities = 48/168 (28%), Positives = 71/168 (41%), Gaps = 35/168 (20%)
Query: 201 TSNAPITGQPY--QSSATISSSGPTHA-----ATPVVPLPTLQIPSLPTSTPFTPQHDAP 253
TS+ P P QS + S+GP A ++P+ PL L+I S P P P +
Sbjct: 524 TSSQPKKASPQCPQSPTPVHSNGPPSAEAAVTSSPLPPLKPLRILSRPPPPPPPPPISSL 583
Query: 254 ESINSSSQATNLVK-----PSFFVPPLSS------AAMMMPPV---------SSSIPTAP 293
S S S +N + P PPL S ++ + PP+ + P P
Sbjct: 584 RSTPSPSSTSNSIATQGPPPPPPPPPLQSHRSALSSSPLPPPLPPKKLLATTNPPPPPPP 643
Query: 294 PLHPTGTVQRPYGTPMLQPFPPPTPPPSLAPVS-------SPLPNPAI 334
PLH + P + +L+ PP PPP+ AP+S P+P P +
Sbjct: 644 PLHSNSRMGAPTSSLVLKS-PPVPPPPAPAPLSRSHNGNIPPVPGPPL 690
Score = 41.6 bits (96), Expect = 7e-04
Identities = 53/174 (30%), Positives = 69/174 (39%), Gaps = 19/174 (10%)
Query: 171 PSSSITSNVADVPDDPSFIN-FFSAAMGIGNTSNAPITGQPYQSSATISSSGPTHAATPV 229
P +S N + P+ I+ F S+ +G TS G +T SS A+ +
Sbjct: 459 PVTSPLPNRSPTQGSPASISRFHSSPSSLGITSILHDHGSCKDEESTSSSP----ASPSI 514
Query: 230 VPLPTLQ--IPSLPT-STPFTPQHDAPESINSSSQATNLVKPSFFVPPLSSAAMMMPPVS 286
LPTL S P ++P PQ P N A V S +PPL ++ P
Sbjct: 515 SFLPTLHPLTSSQPKKASPQCPQSPTPVHSNGPPSAEAAVTSSP-LPPLKPLRILSRPPP 573
Query: 287 SSIPTAPPLHPTGTVQRPYGTP---MLQPFPPPTPPPSL-----APVSSPLPNP 332
P PP+ + P T Q PPP PPP L A SSPLP P
Sbjct: 574 P--PPPPPISSLRSTPSPSSTSNSIATQGPPPPPPPPPLQSHRSALSSSPLPPP 625
Score = 35.4 bits (80), Expect = 0.050
Identities = 38/140 (27%), Positives = 55/140 (39%), Gaps = 16/140 (11%)
Query: 210 PYQSSATISSSGPTHAATPVVPLPTLQIPSLPTSTPFTP-QHDAPESINSSSQATNLVKP 268
P + ++S P + T P + S P+S T HD + S +++ P
Sbjct: 453 PRMVQSPVTSPLPNRSPTQGSPASISRFHSSPSSLGITSILHDHGSCKDEESTSSSPASP 512
Query: 269 SF-FVPPLSSAAMMMPPVSS-SIPTAP-PLHPTGTVQRPYGT-----PMLQPF-----PP 315
S F+P L P +S P +P P+H G P L+P PP
Sbjct: 513 SISFLPTLHPLTSSQPKKASPQCPQSPTPVHSNGPPSAEAAVTSSPLPPLKPLRILSRPP 572
Query: 316 PTPPPSLAPVSSPLPNPAIS 335
P PPP P+SS P+ S
Sbjct: 573 PPPPP--PPISSLRSTPSPS 590
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.315 0.131 0.380
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,698,515
Number of Sequences: 26719
Number of extensions: 426828
Number of successful extensions: 5753
Number of sequences better than 10.0: 341
Number of HSP's better than 10.0 without gapping: 139
Number of HSP's successfully gapped in prelim test: 213
Number of HSP's that attempted gapping in prelim test: 2182
Number of HSP's gapped (non-prelim): 1355
length of query: 366
length of database: 11,318,596
effective HSP length: 101
effective length of query: 265
effective length of database: 8,619,977
effective search space: 2284293905
effective search space used: 2284293905
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 61 (28.1 bits)
Medicago: description of AC146332.11


