

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146332.10 - phase: 0
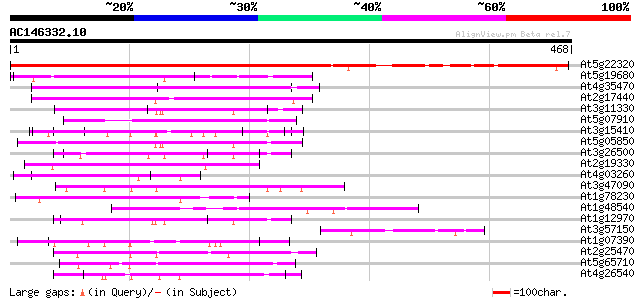
(468 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g22320 unknown protein 381 e-106
At5g19680 unknown protein 94 2e-19
At4g35470 unknown protein 79 4e-15
At2g17440 unknown protein 68 9e-12
At3g11330 unknown protein 68 1e-11
At5g07910 putative protein 65 1e-10
At3g15410 unknown protein 64 1e-10
At5g05850 unknown protein 63 4e-10
At3g26500 unknown protein 60 2e-09
At2g19330 putative leucine-rich-repeat protein 60 2e-09
At4g03260 protein phosphatase regulatory subunit like 60 3e-09
At3g47090 receptor kinase-like protein 58 1e-08
At1g78230 unknown protein; similar to ESTs gb|H36831.1, gb|N9684... 57 2e-08
At1g48540 unknown protein 57 2e-08
At1g12970 unknown protein 57 2e-08
At3g57150 putative pseudouridine synthase (NAP57) 57 3e-08
At1g07390 disease resistance protein, putative 57 3e-08
At2g25470 putative disease resistance protein 56 4e-08
At5g65710 receptor protein kinase-like protein 56 5e-08
At4g26540 receptor protein kinase like protein 55 1e-07
>At5g22320 unknown protein
Length = 452
Score = 381 bits (979), Expect = e-106
Identities = 219/476 (46%), Positives = 310/476 (65%), Gaps = 35/476 (7%)
Query: 1 MTRLSSEQVLKDNNAVNPSSISSLHLTHKALSDVSCLASFNKLEKLDLKFNNLTSLEGLR 60
M L+ EQVLK+ +P S+ L+L HKAL+DVSCL+ F LEKLDL+FNNLT L+GL+
Sbjct: 1 MNSLTVEQVLKEKKTNDPDSVKELNLGHKALTDVSCLSKFKNLEKLDLRFNNLTDLQGLK 60
Query: 61 ACVTLKWLSVVENKLESLEGIQGLTKLTVLNAGKNKLKSMDEIGSLSTIRALILNDNEIV 120
+CV LKWLSVVENKL+SL GI+ LTKLTVLNAGKNKLKSM+EI SL +RALILNDNEI
Sbjct: 61 SCVNLKWLSVVENKLQSLNGIEALTKLTVLNAGKNKLKSMNEISSLVNLRALILNDNEIS 120
Query: 121 SICNLDQMKELNTLVLSKNPIRKIGEALKKVKSITKLSLSHCQLEGIDTSLKFCVELTEL 180
SIC LD +K+LN+LVLS+NPI +IG++L K+K+++K+SLS C+++ I +SLK C +L EL
Sbjct: 121 SICKLDLLKDLNSLVLSRNPISEIGDSLSKLKNLSKISLSDCRIKAIGSSLKSCSDLKEL 180
Query: 181 RLAHNDIKSLPEELMHNSKLRNLDLGNNVIAKWSDIKVLKSLTKLRNLNLQGNPVATNEK 240
RLA+N+IK+LP EL N +L NLD+GNNVI + S ++VL +L+ LRNLN++GNP++ N+K
Sbjct: 181 RLANNEIKALPAELAVNKRLLNLDVGNNVITQLSGLEVLGTLSCLRNLNIRGNPISDNDK 240
Query: 241 VIRKIKN-ALPKLQVFNAKPIDKDTKNEKGHMTDDAHDFSFD--HVDQNEDDHLEAADKR 297
+K++ LP + VFNA+P++K ++N K H+ D D +FD H E++ + KR
Sbjct: 241 SAKKVRTLLLPSVNVFNAQPLEKSSRNAK-HIRLDTDDETFDAYHNKSAEEEQSKEDRKR 299
Query: 298 KSNKKRKETADASEKEAGVYDKENTGHNKDNGNKKKDKLTGTVDPDTKNKSTKKKLKKDD 357
K + KR ++ +E +N+D+ +KKK + T + K KK+ K+
Sbjct: 300 KKSSKRNKS------------EEEEVNNEDHKSKKKKSKSNTNVDQVETK--KKEEHKEK 345
Query: 358 NKPSEKALALEENVNRTEKKKKNRKNKEQSEFDIIDDAEASFAEIFNIKDQENLNHGGEM 417
PS ++ + EKK+K KE E D IDDAE SFAEIF+ +EN+ G E
Sbjct: 346 TIPSN-----NDDDDDAEKKQKRATPKE--ELDAIDDAETSFAEIFS---RENVPKGSED 395
Query: 418 KLQDQVPKDLKLVSSIETLPVKHKSAKMHNVESLSSP-------GTEIGMGGPSTW 466
++ + ++ ++ + K K + + S EIG+GG S W
Sbjct: 396 GIEKKKKSSVQETGLVKVIDTKANKKKKKSEKKQSKSVVIDLPMEVEIGLGGESKW 451
>At5g19680 unknown protein
Length = 328
Score = 94.0 bits (232), Expect = 2e-19
Identities = 82/260 (31%), Positives = 125/260 (47%), Gaps = 37/260 (14%)
Query: 1 MTRLSSEQVLKDNNAVNP-------SSISSLHLTHKALSDVSCLASFNKLEKLDLKFNNL 53
+ +LS Q L D++AV P S + L L L+ V ++ F KL D+ FN +
Sbjct: 62 LKKLSLRQNLIDDSAVEPLSHWDALSDLEELVLRDNKLAKVPDVSIFTKLLVYDISFNEI 121
Query: 54 TSLEGL-RACVTLKWLSVVENKLESLEGIQGLTKLTVLNAGKNKLKSMDEIGSLSTIRAL 112
TSLEG+ +A TLK L V +N++ + I+ L L +L G N+L+ M+
Sbjct: 122 TSLEGISKASSTLKELYVSKNEVNKIMEIEHLHNLQILELGSNRLRVME----------- 170
Query: 113 ILNDNEIVSICNLDQMKELNTLVLSKNPIRKIGEALKKVKSITKLSLSHCQLEGIDTSLK 172
NL+ +L L L +N I+ + L +K I K+SL +L + +
Sbjct: 171 -----------NLENFTKLEELWLGRNRIKVVN--LCGLKCIKKISLQSNRLTSM-KGFE 216
Query: 173 FCVELTELRLAHNDIKSLPEELMHNSKLRNLDLGNNVIAKWSDIKVLKSLTKLRNLNLQG 232
CV L EL L+HN I + E L LR LD+ NN + DI ++LTKL +L L
Sbjct: 217 ECVALEELYLSHNGISKM-EGLSALVNLRVLDVSNNKLTSVDDI---QNLTKLEDLWLND 272
Query: 233 NPVATNEKVIRKIKNALPKL 252
N + + E + + + KL
Sbjct: 273 NQIESLEAITEAVTGSKEKL 292
Score = 76.3 bits (186), Expect = 3e-14
Identities = 48/156 (30%), Positives = 81/156 (51%), Gaps = 6/156 (3%)
Query: 4 LSSEQVLKDNNAVNPSSISSLHLTHKALSDVSCLASFNKLEKLDLKFNNLTSLEGLRACV 63
L S ++ N N + + L L + V+ L ++K+ L+ N LTS++G CV
Sbjct: 161 LGSNRLRVMENLENFTKLEELWLGRNRIKVVN-LCGLKCIKKISLQSNRLTSMKGFEECV 219
Query: 64 TLKWLSVVENKLESLEGIQGLTKLTVLNAGKNKLKSMDEIGSLSTIRALILNDNEIVSIC 123
L+ L + N + +EG+ L L VL+ NKL S+D+I +L+ + L LNDN+I S+
Sbjct: 220 ALEELYLSHNGISKMEGLSALVNLRVLDVSNNKLTSVDDIQNLTKLEDLWLNDNQIESLE 279
Query: 124 NLDQ-----MKELNTLVLSKNPIRKIGEALKKVKSI 154
+ + ++L T+ L NP K + + V+ I
Sbjct: 280 AITEAVTGSKEKLTTIYLENNPCAKSSDYVAAVRQI 315
>At4g35470 unknown protein
Length = 549
Score = 79.3 bits (194), Expect = 4e-15
Identities = 67/245 (27%), Positives = 124/245 (50%), Gaps = 9/245 (3%)
Query: 19 SSISSLHLTHKALSDV-SCLASFNKLEKLDLKFNNLTSL-EGLRACVTLKWLSVVENKLE 76
SS++SL L+ + + + + + L KLDL N + L E + + L +L++ N+L
Sbjct: 246 SSLTSLDLSENHIVVLPNTIGGLSSLTKLDLHSNRIGQLPESIGELLNLVYLNLGSNQLS 305
Query: 77 SL-EGIQGLTKLTVLNAGKNKLKSMDE-IGSLSTIRALILNDNEIVSI-CNLDQMKELNT 133
SL L +L L+ N L + E IGSL +++ L + N+I I ++ L
Sbjct: 306 SLPSAFSRLVRLEELDLSCNNLPILPESIGSLVSLKKLDVETNDIEEIPYSIGGCSSLIE 365
Query: 134 LVLSKNPIRKIGEALKKVKSITKLSLSHCQLEGIDTSLKFCVELTELRLAHNDIKSLPEE 193
L N ++ + EA+ K+ ++ LS+ + + + T++ L EL ++ N+++S+PE
Sbjct: 366 LRADYNKLKALPEAIGKITTLEILSVRYNNIRQLPTTMSSLASLKELDVSFNELESVPES 425
Query: 194 LMHNSKLRNLDLGNNVIAKWSDIKVLKSLTKLRNLNLQGNPVATNEKVIRKIKNALPKLQ 253
L + L L++GNN S + + +L L L++ N + +V+ L KL+
Sbjct: 426 LCFATTLVKLNIGNNFADMVSLPRSIGNLEMLEELDISNNQI----RVLPDSFKMLTKLR 481
Query: 254 VFNAK 258
VF A+
Sbjct: 482 VFRAQ 486
Score = 42.4 bits (98), Expect = 6e-04
Identities = 30/112 (26%), Positives = 54/112 (47%), Gaps = 2/112 (1%)
Query: 124 NLDQMKELNTLVLSKNPIRKIGEALKKVKSITKLSLSHCQLEGIDTSLKFCVELTELRLA 183
+L ++ L +L LS+N I + + + S+TKL L ++ + S+ + L L L
Sbjct: 241 SLGKLSSLTSLDLSENHIVVLPNTIGGLSSLTKLDLHSNRIGQLPESIGELLNLVYLNLG 300
Query: 184 HNDIKSLPEELMHNSKLRNLDLGNNVIAKWSDIKVLKSLTKLRNLNLQGNPV 235
N + SLP +L LDL N + + + SL L+ L+++ N +
Sbjct: 301 SNQLSSLPSAFSRLVRLEELDLSCNNLPILPE--SIGSLVSLKKLDVETNDI 350
Score = 37.4 bits (85), Expect = 0.018
Identities = 34/128 (26%), Positives = 57/128 (43%), Gaps = 11/128 (8%)
Query: 115 NDNEIVSICNLDQM---------KELNTLVLSKNPIRKIGEALKKVKSITKLSLSHCQLE 165
ND E +S+ L + +E+N + + ++L K+ S+T L LS +
Sbjct: 200 NDGEKLSLIKLASLIEVSAKKATQEINLQNKLTEQLEWLPDSLGKLSSLTSLDLSENHIV 259
Query: 166 GIDTSLKFCVELTELRLAHNDIKSLPEELMHNSKLRNLDLGNNVIAKWSDIKVLKSLTKL 225
+ ++ LT+L L N I LPE + L L+LG+N ++ S L +L
Sbjct: 260 VLPNTIGGLSSLTKLDLHSNRIGQLPESIGELLNLVYLNLGSNQLS--SLPSAFSRLVRL 317
Query: 226 RNLNLQGN 233
L+L N
Sbjct: 318 EELDLSCN 325
>At2g17440 unknown protein
Length = 526
Score = 68.2 bits (165), Expect = 9e-12
Identities = 65/251 (25%), Positives = 120/251 (46%), Gaps = 20/251 (7%)
Query: 19 SSISSLHLTHKALSDV-SCLASFNKLEKLDLKFNNLTSL-EGLRACVTLKWLSVVENKLE 76
SS+ L L+ + + + + L +LDL N + L E + + L L++ N+L
Sbjct: 230 SSLVRLDLSENCIMVLPATIGGLISLTRLDLHSNRIGQLPESIGDLLNLVNLNLSGNQLS 289
Query: 77 SL-EGIQGLTKLTVLNAGKNKLKSMDE-IGSLSTIRALILNDNEIV----SICNLDQMKE 130
SL L L L+ N L + E IGSL +++ L + N I SI M+E
Sbjct: 290 SLPSSFNRLIHLEELDLSSNSLSILPESIGSLVSLKKLDVETNNIEEIPHSISGCSSMEE 349
Query: 131 LNTLVLSKNPIRKIGEALKKVKSITKLSLSHCQLEGIDTSLKFCVELTELRLAHNDIKSL 190
L N ++ + EA+ K+ ++ L++ + + + T++ L EL ++ N+++S+
Sbjct: 350 LRA---DYNRLKALPEAVGKLSTLEILTVRYNNIRQLPTTMSSMANLKELDVSFNELESV 406
Query: 191 PEELMHNSKLRNLDLGNNVIAKWSDIKVLKSLTKLRNLNLQGNPV---------ATNEKV 241
PE L + L L++GNN S ++ +L KL L++ N + +N +V
Sbjct: 407 PESLCYAKTLVKLNIGNNFANLRSLPGLIGNLEKLEELDMSNNQIRFLPYSFKTLSNLRV 466
Query: 242 IRKIKNALPKL 252
++ +N L +L
Sbjct: 467 LQTEQNPLEEL 477
Score = 40.4 bits (93), Expect = 0.002
Identities = 37/144 (25%), Positives = 70/144 (47%), Gaps = 6/144 (4%)
Query: 96 KLKSMDEIGSLSTIRALILNDNEIVSICNL-DQMKELNTLV---LSKNPIRKIGEALKKV 151
KL S+ E+ + + L L + + L D + +L++LV LS+N I + + +
Sbjct: 193 KLASLIEVSAKKATQELNLQHRLMDQLEWLPDSLGKLSSLVRLDLSENCIMVLPATIGGL 252
Query: 152 KSITKLSLSHCQLEGIDTSLKFCVELTELRLAHNDIKSLPEELMHNSKLRNLDLGNNVIA 211
S+T+L L ++ + S+ + L L L+ N + SLP L LDL +N ++
Sbjct: 253 ISLTRLDLHSNRIGQLPESIGDLLNLVNLNLSGNQLSSLPSSFNRLIHLEELDLSSNSLS 312
Query: 212 KWSDIKVLKSLTKLRNLNLQGNPV 235
+ + SL L+ L+++ N +
Sbjct: 313 ILPE--SIGSLVSLKKLDVETNNI 334
>At3g11330 unknown protein
Length = 499
Score = 67.8 bits (164), Expect = 1e-11
Identities = 72/239 (30%), Positives = 110/239 (45%), Gaps = 35/239 (14%)
Query: 38 ASFNKLEKLDLKFNNLTSL-EGLRACVTLKWLSVVENKLESL-EGIQGLTKLTVLNAGKN 95
AS N ++++DL L L E L L++ NKLES+ + I GL L L+ N
Sbjct: 195 ASANPVDRVDLSGRKLRLLPEAFGRIQGLLVLNLSNNKLESIPDSIAGLHSLVELDVSTN 254
Query: 96 KLKSM-DEIGSLSTIRALILNDNEIVS----ICN------LD---------------QMK 129
L+++ D IG LS ++ L ++ N++ S IC LD ++
Sbjct: 255 SLETLPDSIGLLSKLKILNVSTNKLTSLPDSICRCGSLVILDVSFNRLTYLPTNIGPELV 314
Query: 130 ELNTLVLSKNPIRKIGEALKKVKSITKLSLSHCQLEGIDTSLKFCVELTELRLAHN--DI 187
L L++ N IR ++ +++S+ L +L G+ S L L L+ N D+
Sbjct: 315 NLEKLLVQYNKIRSFPTSIGEMRSLKHLDAHFNELNGLPDSFVLLTNLEYLNLSSNFSDL 374
Query: 188 KSLPEELMHNSKLRNLDLGNNVIAKWSD-IKVLKSLTKLRNLNLQGNP-VATNEKVIRK 244
K LP L+ LDL NN I D L SLTK LN+ NP V E+V+++
Sbjct: 375 KDLPFSFGELISLQELDLSNNQIHALPDTFGTLDSLTK---LNVDQNPLVVPPEEVVKE 430
Score = 45.1 bits (105), Expect = 9e-05
Identities = 27/100 (27%), Positives = 53/100 (53%)
Query: 116 DNEIVSICNLDQMKELNTLVLSKNPIRKIGEALKKVKSITKLSLSHCQLEGIDTSLKFCV 175
+ E+V I ++ + LS +R + EA +++ + L+LS+ +LE I S+
Sbjct: 185 NEEVVGILQHASANPVDRVDLSGRKLRLLPEAFGRIQGLLVLNLSNNKLESIPDSIAGLH 244
Query: 176 ELTELRLAHNDIKSLPEELMHNSKLRNLDLGNNVIAKWSD 215
L EL ++ N +++LP+ + SKL+ L++ N + D
Sbjct: 245 SLVELDVSTNSLETLPDSIGLLSKLKILNVSTNKLTSLPD 284
>At5g07910 putative protein
Length = 268
Score = 64.7 bits (156), Expect = 1e-10
Identities = 46/196 (23%), Positives = 98/196 (49%), Gaps = 24/196 (12%)
Query: 46 LDLKFNNLTSLEG-LRACVTLKWLSVVENKLESLEGIQGLTKLTVLNAGKNKLKSMDEIG 104
LDL N + + G + + ++ L + +N +E L G +G
Sbjct: 50 LDLTHNKIADVPGEISKLINMQRLLIADNLVERLPG---------------------NLG 88
Query: 105 SLSTIRALILNDNEIVSICN-LDQMKELNTLVLSKNPIRKIGEALKKVKSITKLSLSHCQ 163
L +++ L+L+ N I + + L Q+ L L +S+N + + + + ++++ L++S+ +
Sbjct: 89 KLQSLKVLMLDGNRISCLPDELGQLVRLEQLSISRNMLIYLPDTIGSLRNLLLLNVSNNR 148
Query: 164 LEGIDTSLKFCVELTELRLAHNDIKSLPEELMHNSKLRNLDLGNNVIAKWSDIKVLKSLT 223
L+ + S+ C L E++ N ++ LP L + +L++L L NN + + D +L
Sbjct: 149 LKSLPESVGSCASLEEVQANDNVVEELPASLCNLIQLKSLSLDNNQVNQIPD-GLLIHCK 207
Query: 224 KLRNLNLQGNPVATNE 239
L+NL+L NP++ ++
Sbjct: 208 SLQNLSLHNNPISMDQ 223
>At3g15410 unknown protein
Length = 562
Score = 64.3 bits (155), Expect = 1e-10
Identities = 59/241 (24%), Positives = 117/241 (48%), Gaps = 15/241 (6%)
Query: 17 NPSSISSLHLTHKALSDV-SCLASFNKLEKLDLKFNNLTSL-EGLRACVTLKWLSVVENK 74
N + + L+++H LS + + + ++ LD+ FN+++ L E + + ++L L N+
Sbjct: 66 NLACLVVLNVSHNKLSQLPAAIGELTAMKSLDVSFNSISELPEQIGSAISLVKLDCSSNR 125
Query: 75 LESL-EGIQGLTKLTVLNAGKNKLKSMDE-IGSLSTIRALILNDNEIVS-----ICNLDQ 127
L+ L + I L+ L A N++ S+ E + + S + L + N++ + I +
Sbjct: 126 LKELPDSIGRCLDLSDLKATNNQISSLPEDMVNCSKLSKLDVEGNKLTALSENHIASWTM 185
Query: 128 MKELNTLVLSKNPIRKIGEALKKVKSITKLSLSHCQLEGIDTSLKFCVELTELRLAHNDI 187
+ ELN KN + + + + + + +L L ++ + S+ C L E L N +
Sbjct: 186 LAELNAC---KNMLGVLPQNIGSLSRLIRLDLHQNKISSVPPSIGGCSSLVEFYLGINSL 242
Query: 188 KSLPEELMHNSKLRNLDLGNNVIAKW--SDIKVLKSLTKLRNLNLQG-NPVATNEKVIRK 244
+LP E+ S+L LDL +N + ++ K+ S L N +L G +P N +RK
Sbjct: 243 STLPAEIGDLSRLGTLDLRSNQLKEYPVGACKLKLSYLDLSNNSLTGLHPELGNMTTLRK 302
Query: 245 I 245
+
Sbjct: 303 L 303
Score = 62.8 bits (151), Expect = 4e-10
Identities = 51/168 (30%), Positives = 82/168 (48%), Gaps = 23/168 (13%)
Query: 63 VTLKWLSVVENKLESL-EGIQGLTKLTVLNAGKNKLKSMDEIGSLSTIRALILNDNEIVS 121
V L+ L + N +E L E ++ L L VLN NKL + +
Sbjct: 45 VDLQKLILAHNDIEVLREDLKNLACLVVLNVSHNKLSQLP------------------AA 86
Query: 122 ICNLDQMKELNTLVLSKNPIRKIGEALKKVKSITKLSLSHCQLEGIDTSLKFCVELTELR 181
I L MK L+ +S N I ++ E + S+ KL S +L+ + S+ C++L++L+
Sbjct: 87 IGELTAMKSLD---VSFNSISELPEQIGSAISLVKLDCSSNRLKELPDSIGRCLDLSDLK 143
Query: 182 LAHNDIKSLPEELMHNSKLRNLDLGNNVIAKWSDIKVLKSLTKLRNLN 229
+N I SLPE++++ SKL LD+ N + S+ + S T L LN
Sbjct: 144 ATNNQISSLPEDMVNCSKLSKLDVEGNKLTALSE-NHIASWTMLAELN 190
Score = 55.8 bits (133), Expect = 5e-08
Identities = 62/239 (25%), Positives = 102/239 (41%), Gaps = 41/239 (17%)
Query: 37 LASFNKLEKLDLKFNNLTSLE-GLRACVTLKWLSVVENKLESLEG-IQGLTKLTVLNAGK 94
+ S ++L +LDL N ++S+ + C +L + N L +L I L++L L+
Sbjct: 203 IGSLSRLIRLDLHQNKISSVPPSIGGCSSLVEFYLGINSLSTLPAEIGDLSRLGTLDLRS 262
Query: 95 NKLKSMDEIGSLSTIRALILNDNEIVSIC-NLDQMKELNTLVLSKNPIRKIGEALKK--- 150
N+LK + L L++N + + L M L LVL NP+R + +L
Sbjct: 263 NQLKEYPVGACKLKLSYLDLSNNSLTGLHPELGNMTTLRKLVLVGNPLRTLRSSLVNGPT 322
Query: 151 ---------------------------VKSITKLSLS--HCQLEGIDTS-----LKFCVE 176
+ S ++S+S LEG++ S + E
Sbjct: 323 AALLKYLRSRLSNSEETSASTPTKENVIASAARMSISSKELSLEGLNLSDVPSEVWESGE 382
Query: 177 LTELRLAHNDIKSLPEELMHNSKLRNLDLGNNVIAKWSDIKVLKSLTKLRNLNLQGNPV 235
+T++ L+ N I+ LP +L + L+ L L N I W +LKSL L L L NP+
Sbjct: 383 ITKVNLSKNSIEELPAQLSTSVSLQTLILSRNKIKDWPG-AILKSLPNLMCLKLDNNPL 440
Score = 52.4 bits (124), Expect = 5e-07
Identities = 53/188 (28%), Positives = 93/188 (49%), Gaps = 16/188 (8%)
Query: 20 SISSLHLTHKAL--SDV-SCLASFNKLEKLDLKFNNLTSLEG-LRACVTLKWLSVVENKL 75
SISS L+ + L SDV S + ++ K++L N++ L L V+L+ L + NK+
Sbjct: 357 SISSKELSLEGLNLSDVPSEVWESGEITKVNLSKNSIEELPAQLSTSVSLQTLILSRNKI 416
Query: 76 ESLEG--IQGLTKLTVLNAGKNKLKS--MDEIGSLSTIRALILNDNEIV-----SICNLD 126
+ G ++ L L L N L +D +S ++ L L+ N + C+L
Sbjct: 417 KDWPGAILKSLPNLMCLKLDNNPLNQIPLDGFQVVSGLQILDLSVNAVSFREHPKFCHLP 476
Query: 127 QMKELNTLVLSKNPIRKIGEALKKVKSITKLSLSHCQLEGIDTSLKFCVELTELRLAHND 186
Q++EL LS+ + ++ E + + ++ L L+ L+ I +K L L +++N+
Sbjct: 477 QLREL---YLSRIQLSEVPEDILNLSNLIILDLNQNSLQSIPKGIKNMTSLKHLDISNNN 533
Query: 187 IKSLPEEL 194
I SLP EL
Sbjct: 534 ISSLPPEL 541
Score = 33.9 bits (76), Expect = 0.20
Identities = 19/68 (27%), Positives = 35/68 (50%)
Query: 145 GEALKKVKSITKLSLSHCQLEGIDTSLKFCVELTELRLAHNDIKSLPEELMHNSKLRNLD 204
GE + + KL L+H +E + LK L L ++HN + LP + + +++LD
Sbjct: 38 GENWWEAVDLQKLILAHNDIEVLREDLKNLACLVVLNVSHNKLSQLPAAIGELTAMKSLD 97
Query: 205 LGNNVIAK 212
+ N I++
Sbjct: 98 VSFNSISE 105
>At5g05850 unknown protein
Length = 506
Score = 62.8 bits (151), Expect = 4e-10
Identities = 70/269 (26%), Positives = 122/269 (45%), Gaps = 34/269 (12%)
Query: 7 EQVLKDNNAVNPSSISSLHLTHKALSDVSCLASFNKLEKLDLKFNNLTSL-EGLRACVTL 65
E K+ AV + + + + +S + A N L+++DL L L E L
Sbjct: 171 ESAEKNAAAVAEEEAAEVEVNEEVVSILQQAAE-NPLDRVDLSGRKLKLLPEAFGKIQGL 229
Query: 66 KWLSVVENKLESL-EGIQGLTKLTVLNAGKNKLKSM-DEIGSLSTIRALILNDNEIV--- 120
L++ N+L+++ + I GL L L+ N L+++ D IG LS ++ L ++ N++
Sbjct: 230 LVLNLYNNQLQAIPDSIAGLHNLLELDVSTNFLETLPDSIGLLSKLKILNVSCNKLTTLP 289
Query: 121 -SICN------LD---------------QMKELNTLVLSKNPIRKIGEALKKVKSITKLS 158
SIC+ LD ++ +L L++ N IR + ++ +++S+ L
Sbjct: 290 DSICHCGSLVVLDASYNNLTYLPTNIGFELVKLEKLLIHLNKIRSLPTSIGEMRSLRYLD 349
Query: 159 LSHCQLEGIDTSLKFCVELTELRLAHN--DIKSLPEELMHNSKLRNLDLGNNVIAKWSDI 216
+L G+ S L L L+ N D++ LP L+ LDL NN I D
Sbjct: 350 AHFNELNGLPNSFGLLTNLEYLNLSSNFSDLQDLPASFGDLISLQELDLSNNQIHSLPD- 408
Query: 217 KVLKSLTKLRNLNLQGNP-VATNEKVIRK 244
+L L LNL NP V ++V+++
Sbjct: 409 -AFGTLVNLTKLNLDQNPLVVPPDEVVKQ 436
>At3g26500 unknown protein
Length = 471
Score = 60.5 bits (145), Expect = 2e-09
Identities = 56/199 (28%), Positives = 95/199 (47%), Gaps = 13/199 (6%)
Query: 46 LDLKFNNLTSL-EGLRACVTLKWLSVVENKLESL-EGIQGLTKLTVLNAGKNKLKSMDEI 103
L+L N+LT + + + L+ L V N LESL + I L L +LN N L ++ E
Sbjct: 188 LNLSGNDLTFIPDAISKLKKLEELDVSSNSLESLPDSIGMLLNLRILNVNANNLTALPE- 246
Query: 104 GSLSTIRALILNDNEIVSICNLDQ-----MKELNTLVLSKNPIRKIGEALKKVKSITKLS 158
S++ R+L+ D ++ +L ++ L L + N +R ++ ++ ++ L
Sbjct: 247 -SIAHCRSLVELDASYNNLTSLPTNIGYGLQNLERLSIQLNKLRYFPGSISEMYNLKYLD 305
Query: 159 LSHCQLEGIDTSLKFCVELTELRLAHN--DIKSLPEELMHNSKLRNLDLGNNVIAKWSDI 216
++ GI S+ +L L L+ N ++ +P+ + + LR LDL NN I D
Sbjct: 306 AHMNEIHGIPNSIGRLTKLEVLNLSSNFNNLMGVPDTITDLTNLRELDLSNNQIQAIPD- 364
Query: 217 KVLKSLTKLRNLNLQGNPV 235
L KL LNL NP+
Sbjct: 365 -SFYRLRKLEKLNLDQNPL 382
Score = 54.7 bits (130), Expect = 1e-07
Identities = 51/179 (28%), Positives = 87/179 (48%), Gaps = 7/179 (3%)
Query: 37 LASFNKLEKLDLKFNNLTSL-EGLRACVTLKWLSVVENKLESL-EGIQGLTKLTVLNAGK 94
++ KLE+LD+ N+L SL + + + L+ L+V N L +L E I L L+A
Sbjct: 202 ISKLKKLEELDVSSNSLESLPDSIGMLLNLRILNVNANNLTALPESIAHCRSLVELDASY 261
Query: 95 NKLKSM-DEIG-SLSTIRALILNDNEIVSI-CNLDQMKELNTLVLSKNPIRKIGEALKKV 151
N L S+ IG L + L + N++ ++ +M L L N I I ++ ++
Sbjct: 262 NNLTSLPTNIGYGLQNLERLSIQLNKLRYFPGSISEMYNLKYLDAHMNEIHGIPNSIGRL 321
Query: 152 KSITKLSLS--HCQLEGIDTSLKFCVELTELRLAHNDIKSLPEELMHNSKLRNLDLGNN 208
+ L+LS L G+ ++ L EL L++N I+++P+ KL L+L N
Sbjct: 322 TKLEVLNLSSNFNNLMGVPDTITDLTNLRELDLSNNQIQAIPDSFYRLRKLEKLNLDQN 380
Score = 46.2 bits (108), Expect = 4e-05
Identities = 43/139 (30%), Positives = 71/139 (50%), Gaps = 13/139 (9%)
Query: 37 LASFNKLEKLDLKFNNLTSLE-----GLRACVTLKWLSVVENKLESLEG-IQGLTKLTVL 90
+A L +LD +NNLTSL GL+ L+ LS+ NKL G I + L L
Sbjct: 248 IAHCRSLVELDASYNNLTSLPTNIGYGLQ---NLERLSIQLNKLRYFPGSISEMYNLKYL 304
Query: 91 NAGKNKLKSM-DEIGSLSTIRALIL--NDNEIVSICN-LDQMKELNTLVLSKNPIRKIGE 146
+A N++ + + IG L+ + L L N N ++ + + + + L L LS N I+ I +
Sbjct: 305 DAHMNEIHGIPNSIGRLTKLEVLNLSSNFNNLMGVPDTITDLTNLRELDLSNNQIQAIPD 364
Query: 147 ALKKVKSITKLSLSHCQLE 165
+ +++ + KL+L LE
Sbjct: 365 SFYRLRKLEKLNLDQNPLE 383
Score = 35.8 bits (81), Expect = 0.052
Identities = 27/94 (28%), Positives = 46/94 (48%), Gaps = 5/94 (5%)
Query: 143 KIGEALKKVKS---ITKLSLSHCQLEGIDTSLKFCVELTELRLAHNDIKSLPEELMHNSK 199
K+ LK+ +S + ++ LS +L+ I + V L L L+ ND+ +P+ + K
Sbjct: 148 KVLAVLKEAESGGTVERIDLSSQELKLIPEAFWKVVGLVYLNLSGNDLTFIPDAISKLKK 207
Query: 200 LRNLDLGNNVIAKWSDIKVLKSLTKLRNLNLQGN 233
L LD+ +N + D + L LR LN+ N
Sbjct: 208 LEELDVSSNSLESLPD--SIGMLLNLRILNVNAN 239
>At2g19330 putative leucine-rich-repeat protein
Length = 380
Score = 60.5 bits (145), Expect = 2e-09
Identities = 59/205 (28%), Positives = 102/205 (48%), Gaps = 9/205 (4%)
Query: 13 NNAVNPSSISSLHLTHKALSDV--SCLASFNKLEKLDLKFNNLTSLEGLRACVT-LKWLS 69
N ++N + I L L++ L + S A L LD+ N + +L C++ LK L+
Sbjct: 77 NPSLNLAQICKLDLSNNHLQTIPESLTARLLNLIALDVHSNQIKALPNSIGCLSKLKTLN 136
Query: 70 VVENKLESL-EGIQGLTKLTVLNAGKNKLKSM-DEIG-SLSTIRALILNDNEIVSI-CNL 125
V N L S + IQ L LNA NKL + D IG L+ +R L +N N+++S+ ++
Sbjct: 137 VSGNFLVSFPKSIQHCRSLEELNANFNKLIRLPDSIGFELTNLRKLSINSNKLISLPISI 196
Query: 126 DQMKELNTLVLSKNPIRKIGEALKKVKSITKLSLSHC--QLEGIDTSLKFCVELTELRLA 183
+ L L N + + + L+ + ++ L++S L + +S+ + L EL ++
Sbjct: 197 THLTSLRVLDARLNCLMILPDDLENLINLEILNVSQNFQYLSALPSSIGLLMNLIELDVS 256
Query: 184 HNDIKSLPEELMHNSKLRNLDLGNN 208
+N I LPE + +LR L + N
Sbjct: 257 YNKITVLPESIGCMRRLRKLSVEGN 281
Score = 35.4 bits (80), Expect = 0.068
Identities = 30/111 (27%), Positives = 50/111 (45%), Gaps = 3/111 (2%)
Query: 124 NLDQMKELNTLVLSKNPIRKIGEALKKVKSITKLSLSHCQLEGIDTSLKF-CVELTELRL 182
N ++ + L + LS + + + I KL LS+ L+ I SL + L L +
Sbjct: 55 NNNEEERLEVVNLSGMALESLPNPSLNLAQICKLDLSNNHLQTIPESLTARLLNLIALDV 114
Query: 183 AHNDIKSLPEELMHNSKLRNLDLGNNVIAKWSDIKVLKSLTKLRNLNLQGN 233
N IK+LP + SKL+ L++ N + + K ++ L LN N
Sbjct: 115 HSNQIKALPNSIGCLSKLKTLNVSGNFLVSFP--KSIQHCRSLEELNANFN 163
Score = 31.6 bits (70), Expect = 0.98
Identities = 25/94 (26%), Positives = 51/94 (53%), Gaps = 7/94 (7%)
Query: 157 LSLSHCQLEGIDTSLKFCVELTELRLAHNDIKSLPEELMHNSKLRNL---DLGNNVIAKW 213
++LS LE + ++ +L L++N ++++PE L ++L NL D+ +N I
Sbjct: 65 VNLSGMALESLPNPSLNLAQICKLDLSNNHLQTIPESL--TARLLNLIALDVHSNQIKAL 122
Query: 214 SDIKVLKSLTKLRNLNLQGNPVATNEKVIRKIKN 247
+ + L+KL+ LN+ GN + + K I+ ++
Sbjct: 123 PNS--IGCLSKLKTLNVSGNFLVSFPKSIQHCRS 154
>At4g03260 protein phosphatase regulatory subunit like
Length = 677
Score = 59.7 bits (143), Expect = 3e-09
Identities = 37/119 (31%), Positives = 63/119 (52%), Gaps = 5/119 (4%)
Query: 4 LSSEQVLKDNNAVNPSSISSLHLTHKALSDVSCLASFNKLEKLDLKFNNLTSL-EGLRAC 62
LS +++ P + +L+L+ ++S + L +L LDL +N + L GL +C
Sbjct: 426 LSGNAIVRITAGALPRGLHALNLSKNSISVIEGLRELTRLRVLDLSYNRILRLGHGLASC 485
Query: 63 VTLKWLSVVENKLESLEGIQGLTKLTVLNAGKNKLKSMDEIGSL----STIRALILNDN 117
+LK L + NK+ +EG+ L KLTVL+ NK + +G L S+++A+ L N
Sbjct: 486 SSLKELYLAGNKISEIEGLHRLLKLTVLDLRFNKFSTTKCLGQLAANYSSLQAISLEGN 544
Score = 43.1 bits (100), Expect = 3e-04
Identities = 36/145 (24%), Positives = 70/145 (47%), Gaps = 4/145 (2%)
Query: 19 SSISSLHLTHKALSDVSCLASFNKLEKLDLKFNNLTSLEGLRACVTLKWLSVVENKLESL 78
+S ++ L L + L++F L L+L N + + L L++ +N + +
Sbjct: 397 ASATTAQLVSHGLVVIPFLSAFVGLRVLNLSGNAIVRITAGALPRGLHALNLSKNSISVI 456
Query: 79 EGIQGLTKLTVLNAGKNKLKSMDE-IGSLSTIRALILNDNEIVSICNLDQMKELNTLVLS 137
EG++ LT+L VL+ N++ + + S S+++ L L N+I I L ++ +L L L
Sbjct: 457 EGLRELTRLRVLDLSYNRILRLGHGLASCSSLKELYLAGNKISEIEGLHRLLKLTVLDLR 516
Query: 138 KNPI---RKIGEALKKVKSITKLSL 159
N + +G+ S+ +SL
Sbjct: 517 FNKFSTTKCLGQLAANYSSLQAISL 541
>At3g47090 receptor kinase-like protein
Length = 1009
Score = 58.2 bits (139), Expect = 1e-08
Identities = 72/276 (26%), Positives = 125/276 (45%), Gaps = 35/276 (12%)
Query: 39 SFNKLEKL-----------DLKFNNLTSLEGLRACVTLKWLSVVENKLES---LEGIQGL 84
+F KLE L F +L L+ L C L LSV N+L +
Sbjct: 302 NFGKLENLHYLELANNSLGSYSFGDLAFLDALTNCSHLHGLSVSYNRLGGALPTSIVNMS 361
Query: 85 TKLTVLNAGKNKLKSM--DEIGSLSTIRALILNDNEIVS--ICNLDQMKELNTLVLSKNP 140
T+LTVLN N + +IG+L +++L+L DN + +L + L L+L N
Sbjct: 362 TELTVLNLKGNLIYGSIPHDIGNLIGLQSLLLADNLLTGPLPTSLGNLVGLGELILFSNR 421
Query: 141 IR-KIGEALKKVKSITKLSLSHCQLEGI-DTSLKFCVELTELRLAHNDIK-SLPEELMHN 197
+I + + + KL LS+ EGI SL C + +L++ +N + ++P+E+M
Sbjct: 422 FSGEIPSFIGNLTQLVKLYLSNNSFEGIVPPSLGDCSHMLDLQIGYNKLNGTIPKEIMQI 481
Query: 198 SKLRNLDLGNNVIAKW--SDIKVLKSLTK--LRNLNLQGNPVATNEKVI---------RK 244
L +L++ +N ++ +DI L++L + L N NL G+ T K +
Sbjct: 482 PTLVHLNMESNSLSGSLPNDIGRLQNLVELLLGNNNLSGHLPQTLGKCLSMEVIYLQENH 541
Query: 245 IKNALPKLQ-VFNAKPIDKDTKNEKGHMTDDAHDFS 279
+P ++ + K +D N G +++ +FS
Sbjct: 542 FDGTIPDIKGLMGVKNVDLSNNNLSGSISEYFENFS 577
>At1g78230 unknown protein; similar to ESTs gb|H36831.1,
gb|N96841.1, and gb|AA651293.1
Length = 413
Score = 57.4 bits (137), Expect = 2e-08
Identities = 49/201 (24%), Positives = 100/201 (49%), Gaps = 15/201 (7%)
Query: 6 SEQVLKDNNAVNPSSISS--LHLTHKALSDVSCLASFNKLEKLDLKFNNLTSLEGLRACV 63
SE ++ N+ + S SS H++ L + ++ F L+ +DL N + +
Sbjct: 146 SEAIVHANSLIQSLSKSSSVAHISSIGLKAIPSISHFTSLKSIDLSNNFIVQITPASLPK 205
Query: 64 TLKWLSVVENKLESLEGIQGLTKLTVLNAGKNKLKSMDE-IGSLSTIRALILNDNEIVSI 122
L L++ +NK+ +EG++ LT+L VL+ N++ + + + + + I+ L L N+I ++
Sbjct: 206 GLHALNLSKNKISVIEGLRDLTRLRVLDLSYNRISRIGQGLSNCTLIKELYLAGNKISNV 265
Query: 123 CNLDQMKELNTLVLSKNPI---RKIGEALKKVKSITKLSLSHCQLEGIDTSLKFCVELTE 179
L ++ +L L LS N I + IG+ + S+ L++ + ++ V +
Sbjct: 266 EGLHRLLKLIVLDLSFNKIATTKAIGQLVANYNSLVALNI-------LGNPIQNNVGEDQ 318
Query: 180 LRLAHNDIKSLPEELMHNSKL 200
LR + + LP+ + HN +L
Sbjct: 319 LRKTVSSL--LPKLVYHNKQL 337
>At1g48540 unknown protein
Length = 1063
Score = 57.4 bits (137), Expect = 2e-08
Identities = 65/263 (24%), Positives = 116/263 (43%), Gaps = 32/263 (12%)
Query: 86 KLTVLNAGKNKLKSMDE-IGSLSTIRALILNDNEIVSICNLDQMKELNTLVLSKNPIRKI 144
KL V++ N+L MDE + L +L L+ N+ V + NL + +L L L N +R
Sbjct: 169 KLAVISCACNRLVLMDESLQLLPAAESLDLSRNKFVKVDNLRRCTKLKHLDLGFNHLR-- 226
Query: 145 GEALKKVKSITKLSLSHCQLEGIDTSLKFCVELTELRLAHNDIKSLPEELMHNSKLRNLD 204
+++ LS C L +L L +N + +L + + L+ LD
Sbjct: 227 --------TVSYLSQVSCHL-------------VKLVLRNNALTTL-RGIENLKSLQGLD 264
Query: 205 LGNNVIAKWSDIKVLKSLTKLRNLNLQGNPVATNEKVIRKIKN--ALPKLQVFNAKPIDK 262
+ N+I+ +S+++ L SL++L+ L L+GNPV + + ALP + K I
Sbjct: 265 VSYNIISNFSELEFLWSLSQLKELWLEGNPVCCARWYRAHVFSYVALPDELKLDGKQIGT 324
Query: 263 DTKNEK----GHMTDDAHDFSFDHVDQNEDDHLEAADKRKSNKKRKETADASEKEAGVYD 318
++ H + + F + E+D+ E + RK K + + SE E+ +
Sbjct: 325 REFWKRQIIVAHRQSEPASYGF-YSPAREEDNEEGSCNRKKKKICRLASIHSEAESTYVN 383
Query: 319 KENTGHNKDNGNKKKDKLTGTVD 341
++ D+ NK+ K D
Sbjct: 384 SDHESATCDHENKENMKFNQEAD 406
Score = 40.4 bits (93), Expect = 0.002
Identities = 36/116 (31%), Positives = 55/116 (47%), Gaps = 6/116 (5%)
Query: 9 VLKDNNAVNPSSISSLHLTHKALSDVSCLASFNKLEKLDLKFNNLTSLEGLR--ACVTLK 66
VL D + + SL L+ V L KL+ LDL FN+L ++ L +C +K
Sbjct: 181 VLMDESLQLLPAAESLDLSRNKFVKVDNLRRCTKLKHLDLGFNHLRTVSYLSQVSCHLVK 240
Query: 67 WLSVVENKLESLEGIQGLTKLTVLNAGKNKLKSMDE---IGSLSTIRALILNDNEI 119
L + N L +L GI+ L L L+ N + + E + SLS ++ L L N +
Sbjct: 241 -LVLRNNALTTLRGIENLKSLQGLDVSYNIISNFSELEFLWSLSQLKELWLEGNPV 295
Score = 40.0 bits (92), Expect = 0.003
Identities = 31/105 (29%), Positives = 50/105 (47%), Gaps = 10/105 (9%)
Query: 44 EKLDLKFNNLTSLEGLRACVTLKWLSVVENKLESLEGIQ----GLTKLTVLNAGKNKLKS 99
E LDL N ++ LR C LK L + N L ++ + L KL + N L+
Sbjct: 194 ESLDLSRNKFVKVDNLRRCTKLKHLDLGFNHLRTVSYLSQVSCHLVKLVLRNNALTTLRG 253
Query: 100 MDEIGSLSTIRA---LILNDNEIVSICNLDQMKELNTLVLSKNPI 141
++ + SL + +I N +E+ + +L Q+KE L L NP+
Sbjct: 254 IENLKSLQGLDVSYNIISNFSELEFLWSLSQLKE---LWLEGNPV 295
>At1g12970 unknown protein
Length = 464
Score = 57.4 bits (137), Expect = 2e-08
Identities = 62/223 (27%), Positives = 97/223 (42%), Gaps = 32/223 (14%)
Query: 43 LEKLDLKFNNLTSL-EGLRACVTLKWLSVVENKLESL-EGIQGLTKLTVLNAGKNKLKSM 100
+E++DL + L L + L V L L+V N L L + I GL KL L+ N+L +
Sbjct: 163 VERIDLSDHELKLLPDALGKIVGLVSLNVSRNNLRFLPDTISGLEKLEELDLSSNRLVFL 222
Query: 101 -DEIGSLSTIRALILNDNEIV----------SICNLDQ---------------MKELNTL 134
D IG L +R L + N++ S+ LD + L L
Sbjct: 223 PDSIGLLLNLRILNVTGNKLTLLPESIAQCRSLVELDASFNNLTSLPANFGYGLLNLERL 282
Query: 135 VLSKNPIRKIGEALKKVKSITKLSLSHCQLEGIDTSLKFCVELTELRLAHN--DIKSLPE 192
+ N IR ++ +++S+ L ++ G+ ++ L + L+ N D+ LP+
Sbjct: 283 SIQLNKIRFFPNSICEMRSLRYLDAHMNEIHGLPIAIGRLTNLEVMNLSSNFSDLIELPD 342
Query: 193 ELMHNSKLRNLDLGNNVIAKWSDIKVLKSLTKLRNLNLQGNPV 235
+ + LR LDL NN I D L KL LNL NP+
Sbjct: 343 TISDLANLRELDLSNNQIRVLPD--SFFRLEKLEKLNLDQNPL 383
Score = 43.9 bits (102), Expect = 2e-04
Identities = 38/136 (27%), Positives = 68/136 (49%), Gaps = 7/136 (5%)
Query: 37 LASFNKLEKLDLKFNNLTSLEGL--RACVTLKWLSVVENKLESL-EGIQGLTKLTVLNAG 93
+A L +LD FNNLTSL + L+ LS+ NK+ I + L L+A
Sbjct: 249 IAQCRSLVELDASFNNLTSLPANFGYGLLNLERLSIQLNKIRFFPNSICEMRSLRYLDAH 308
Query: 94 KNKLKSMD-EIGSLSTIRALILNDN--EIVSICN-LDQMKELNTLVLSKNPIRKIGEALK 149
N++ + IG L+ + + L+ N +++ + + + + L L LS N IR + ++
Sbjct: 309 MNEIHGLPIAIGRLTNLEVMNLSSNFSDLIELPDTISDLANLRELDLSNNQIRVLPDSFF 368
Query: 150 KVKSITKLSLSHCQLE 165
+++ + KL+L LE
Sbjct: 369 RLEKLEKLNLDQNPLE 384
Score = 37.7 bits (86), Expect = 0.014
Identities = 27/103 (26%), Positives = 51/103 (49%), Gaps = 2/103 (1%)
Query: 154 ITKLSLSHCQLEGIDTSLKFCVELTELRLAHNDIKSLPEELMHNSKLRNLDLGNNVIAKW 213
+ ++ LS +L+ + +L V L L ++ N+++ LP+ + KL LDL +N +
Sbjct: 163 VERIDLSDHELKLLPDALGKIVGLVSLNVSRNNLRFLPDTISGLEKLEELDLSSNRLVFL 222
Query: 214 SDIKVLKSLTKLRNLNLQGNPVATNEKVIRKIKNALPKLQVFN 256
D + L LR LN+ GN + + I + ++ + FN
Sbjct: 223 PD--SIGLLLNLRILNVTGNKLTLLPESIAQCRSLVELDASFN 263
>At3g57150 putative pseudouridine synthase (NAP57)
Length = 565
Score = 56.6 bits (135), Expect = 3e-08
Identities = 51/148 (34%), Positives = 69/148 (46%), Gaps = 20/148 (13%)
Query: 260 IDKDTKN-EKGHMTDDAHDFSFDHV--------DQNEDDHLEAADKRKSNKKRKETADAS 310
I D +N E G HD S D E + EA +K KS+KK+K+
Sbjct: 427 IKADAENGEAGEARKRKHDDSSDSPAPVTTKKSKTKEVEGEEAEEKVKSSKKKKKKDKEE 486
Query: 311 EKEAGVYDKENTGHNKDNGNKKKDKLTGTVDPDTKNKSTKKKLKKDDNKPSEKALALEEN 370
EKE E G K KKKDK ++ KS KKK KK +K +E A+ E+
Sbjct: 487 EKE------EEAGSEKKEKKKKKDKKEEVIEEVASPKSEKKKKKK--SKDTEAAVDAEDE 538
Query: 371 --VNRTEKKKKNRKNKEQSEFDIIDDAE 396
++EKKKK +K+K++ D DD E
Sbjct: 539 SAAEKSEKKKK-KKDKKKKNKDSEDDEE 565
Score = 39.3 bits (90), Expect = 0.005
Identities = 33/108 (30%), Positives = 49/108 (44%), Gaps = 11/108 (10%)
Query: 307 ADASEKEAGVYDKENTGHNKDNG-----NKKKDKLTGTVDPDTKNKSTKKKLKKDDNKPS 361
ADA EAG K + D+ K K K + + K KS+KKK KKD K
Sbjct: 429 ADAENGEAGEARKRKHDDSSDSPAPVTTKKSKTKEVEGEEAEEKVKSSKKKKKKD--KEE 486
Query: 362 EKALALEENVNRTEKKKKNRKNKEQSEFDIIDDAEASFAEIFNIKDQE 409
EK EE +K+KK +K+K++ + + ++ + KD E
Sbjct: 487 EK----EEEAGSEKKEKKKKKDKKEEVIEEVASPKSEKKKKKKSKDTE 530
Score = 37.4 bits (85), Expect = 0.018
Identities = 31/117 (26%), Positives = 54/117 (45%), Gaps = 5/117 (4%)
Query: 287 EDDHLEAADKRKSNKKRKETADASEKEAGVYDKENT-GHNKDNGNKKKDKLTGTVDPDTK 345
E+ A KRK + A + K++ + E K +KKK K + + +
Sbjct: 432 ENGEAGEARKRKHDDSSDSPAPVTTKKSKTKEVEGEEAEEKVKSSKKKKKKDKEEEKEEE 491
Query: 346 NKSTKKKLKKDDNKPSEKALALEENVNRTEKKKKNRKNKE-QSEFDIIDDAEASFAE 401
S KK+ KK +K E +EE + +KKK +K+K+ ++ D D++ A +E
Sbjct: 492 AGSEKKEKKKKKDKKEE---VIEEVASPKSEKKKKKKSKDTEAAVDAEDESAAEKSE 545
>At1g07390 disease resistance protein, putative
Length = 976
Score = 56.6 bits (135), Expect = 3e-08
Identities = 68/223 (30%), Positives = 102/223 (45%), Gaps = 26/223 (11%)
Query: 7 EQVLKDNNAVNPSSISSLHLTHKALSDVSC--LASFNKLEKLDLKFNNLTSLEGLRACVT 64
E V N +++ L+L + S +S L F LE LDL FN + E + T
Sbjct: 148 EGVFPPQELSNMTNLRVLNLKDNSFSFLSSQGLTDFRDLEVLDLSFNGVNDSEASHSLST 207
Query: 65 --LKWLSVVENKLES---LEGIQGLTKLTVLNAGKNKLK---SMDEIGSLSTIRALILND 116
LK L + N L L+G++ L +L VL NK S + L ++ L L+D
Sbjct: 208 AKLKTLDLNFNPLSDFSQLKGLESLQELQVLKLRGNKFNHTLSTHVLKDLKMLQELDLSD 267
Query: 117 NEIVSICNLDQMKELNTLVLSKNPIRKIGEALKKVKSIT-KLSLSHCQLE------GIDT 169
N NLD ++++ K E ++KV+S+T L + H L + T
Sbjct: 268 N---GFTNLDHGRDVDESRSEKR--FDFREVVQKVESLTCLLEVEHANLYLFMYPYVLST 322
Query: 170 S--LKFC--VELTELRLAHNDIKSLPEELMHNSKLRNLDLGNN 208
+ L C ++L EL L+ N + SLP L + + LR LDL NN
Sbjct: 323 AGYLGICRLMKLRELDLSSNALTSLPYCLGNLTHLRTLDLSNN 365
Score = 43.5 bits (101), Expect = 2e-04
Identities = 47/204 (23%), Positives = 90/204 (44%), Gaps = 37/204 (18%)
Query: 33 DVSCLASFNKLEKLDLKFNNLTSLEGLRACVTLKWLSVVENKLESLEGIQGLTKLTVLNA 92
++S L SF +L+ L+L +N W + + + + L KLT L+
Sbjct: 74 NLSLLHSFPQLQSLNLSWN---------------WFTNLSDHFLGFKSFGTLDKLTTLDF 118
Query: 93 GKNKLKS--MDEIGSLSTIRALILNDNEIVSICNLDQMKELNTLVLSKNPIRKIGEALKK 150
N + + + + ++IR+L L N + + ++ + L +
Sbjct: 119 SHNMFDNSIVPFLNAATSIRSLHLESNYMEGVFPPQELSNMTNL---------------R 163
Query: 151 VKSITKLSLSHCQLEGIDTSLKFCVELTELRLAHNDIK-SLPEELMHNSKLRNLDLGNNV 209
V ++ S S +G+ T + +L L L+ N + S + +KL+ LDL N
Sbjct: 164 VLNLKDNSFSFLSSQGL-TDFR---DLEVLDLSFNGVNDSEASHSLSTAKLKTLDLNFNP 219
Query: 210 IAKWSDIKVLKSLTKLRNLNLQGN 233
++ +S +K L+SL +L+ L L+GN
Sbjct: 220 LSDFSQLKGLESLQELQVLKLRGN 243
>At2g25470 putative disease resistance protein
Length = 910
Score = 56.2 bits (134), Expect = 4e-08
Identities = 64/235 (27%), Positives = 106/235 (44%), Gaps = 22/235 (9%)
Query: 37 LASFNKLEKLDLKFNNLTS---LEGLRACVTLKWLSVVENKLE-SLEGIQGLTKLTVLNA 92
L + L L L +N + ++GL+ L+ L + NKL S++ +Q L L VL
Sbjct: 144 LNAATSLTTLILTYNEMDGPFPIKGLKDLTNLELLDLRANKLNGSMQELQNLINLEVLGL 203
Query: 93 GKNKLKS---MDEIGSLSTIRALILNDNEIVS---ICNLDQMKELNTLVLSKNPIR-KIG 145
+N + ++ L +R L L N V +C L +K+L L LS N + +
Sbjct: 204 AQNHVDGPIPIEVFCKLKNLRDLDLKGNHFVGQIPLC-LGSLKKLRVLDLSSNQLSGDLP 262
Query: 146 EALKKVKSITKLSLSHCQLEGIDTSLKFCVELTELR----LAHNDIKSLPEELMHNSKLR 201
+ ++S+ LSLS +G SL LT L+ L ++ +P L++ KLR
Sbjct: 263 SSFSSLESLEYLSLSDNNFDG-SFSLNPLTNLTNLKFVVVLRFCSLEKIPSFLLYQKKLR 321
Query: 202 NLDLGNNVIAKWSDIKVLKSLTKLRNLNLQGNPVATNEKVIRKIKNALPKLQVFN 256
+DL +N ++ +L + +L L LQ N I I + LQ+F+
Sbjct: 322 LVDLSSNNLSGNIPTWLLTNNPELEVLQLQNNSF-----TIFPIPTMVHNLQIFD 371
Score = 32.0 bits (71), Expect = 0.75
Identities = 29/109 (26%), Positives = 49/109 (44%), Gaps = 9/109 (8%)
Query: 36 CLASFNKLEKLDLKFNNLTS--LEGLRACVTLKWLSVVENKLESLEGIQGLTKLTVLNAG 93
CL S KL LDL N L+ + +L++LS+ +N + + LT LT L
Sbjct: 240 CLGSLKKLRVLDLSSNQLSGDLPSSFSSLESLEYLSLSDNNFDGSFSLNPLTNLTNL--- 296
Query: 94 KNKLKSMDEIGSLSTIRALILNDNE--IVSICNLDQMKELNTLVLSKNP 140
K + SL I + +L + +V + + + + T +L+ NP
Sbjct: 297 --KFVVVLRFCSLEKIPSFLLYQKKLRLVDLSSNNLSGNIPTWLLTNNP 343
>At5g65710 receptor protein kinase-like protein
Length = 976
Score = 55.8 bits (133), Expect = 5e-08
Identities = 56/211 (26%), Positives = 103/211 (48%), Gaps = 19/211 (9%)
Query: 42 KLEKLDLKFNNLTSL--EGLRACVTLKWLSVVENKLESLEGIQ----GLTKLTVLNAGKN 95
KL+K+ N L+ E C +L ++ + +NKL + LT+L + N N
Sbjct: 388 KLQKIITFSNQLSGEIPESYGDCHSLNYIRMADNKLSGEVPARFWELPLTRLELAN--NN 445
Query: 96 KLK-----SMDEIGSLSTIRALILNDNEIVSICNLDQMKELNTLVLSKNP-IRKIGEALK 149
+L+ S+ + LS + N + ++ + L +++L + LS+N + I +
Sbjct: 446 QLQGSIPPSISKARHLSQLEISANNFSGVIPV-KLCDLRDLRVIDLSRNSFLGSIPSCIN 504
Query: 150 KVKSITKLSLSHCQLEG-IDTSLKFCVELTELRLAHNDIK-SLPEELMHNSKLRNLDLGN 207
K+K++ ++ + L+G I +S+ C ELTEL L++N ++ +P EL L LDL N
Sbjct: 505 KLKNLERVEMQENMLDGEIPSSVSSCTELTELNLSNNRLRGGIPPELGDLPVLNYLDLSN 564
Query: 208 NVIAKWSDIKVLKSLTKLRNLNLQGNPVATN 238
N + ++L+ KL N+ N + N
Sbjct: 565 NQLTGEIPAELLR--LKLNQFNVSDNKLYGN 593
Score = 35.8 bits (81), Expect = 0.052
Identities = 31/91 (34%), Positives = 50/91 (54%), Gaps = 6/91 (6%)
Query: 150 KVKSITKLSLSHCQLEG-IDTS-LKFCVELTELRLAHNDIKS-LPEELMHNSKLRNLDLG 206
+++++ ++LS L G ID++ L C +L L L N+ LPE KLR L+L
Sbjct: 96 RIRTLINITLSQNNLNGTIDSAPLSLCSKLQNLILNQNNFSGKLPEFSPEFRKLRVLELE 155
Query: 207 NNVIAKWSDI-KVLKSLTKLRNLNLQGNPVA 236
+N+ +I + LT L+ LNL GNP++
Sbjct: 156 SNLFT--GEIPQSYGRLTALQVLNLNGNPLS 184
Score = 32.3 bits (72), Expect = 0.57
Identities = 43/184 (23%), Positives = 73/184 (39%), Gaps = 34/184 (18%)
Query: 104 GSLSTIRALILNDNEIVSICN-----LDQMKELNTLVLSKNPIRKIGEALKKVKSITKLS 158
G L+ ++ L LN N + I L ++ L+ +S +P I L + ++T L
Sbjct: 168 GRLTALQVLNLNGNPLSGIVPAFLGYLTELTRLDLAYISFDP-SPIPSTLGNLSNLTDLR 226
Query: 159 LSHCQLEG-IDTSLKFCVELTELRLAHNDIKS-------------------------LPE 192
L+H L G I S+ V L L LA N + LPE
Sbjct: 227 LTHSNLVGEIPDSIMNLVLLENLDLAMNSLTGEIPESIGRLESVYQIELYDNRLSGKLPE 286
Query: 193 ELMHNSKLRNLDLGNNVIAKWSDIKVLKSLTKLRNLNLQGNPVATNEKVIRKIKNALPKL 252
+ + ++LRN D+ N + K+ + +L + NL N + + L +
Sbjct: 287 SIGNLTELRNFDVSQNNLTGELPEKI--AALQLISFNLNDNFFTGGLPDVVALNPNLVEF 344
Query: 253 QVFN 256
++FN
Sbjct: 345 KIFN 348
>At4g26540 receptor protein kinase like protein
Length = 926
Score = 54.7 bits (130), Expect = 1e-07
Identities = 59/217 (27%), Positives = 99/217 (45%), Gaps = 22/217 (10%)
Query: 37 LASFNKLEKLDLKFNNLT-SLEGLRACVTLKWLSVVENKLESL--EGIQGLTKLTVLNAG 93
++ LE LDL N+L+ SL G +LK++ +N L S GI LT+LT LN
Sbjct: 473 ISGCESLEFLDLHTNSLSGSLLGTTLPKSLKFIDFSDNALSSTLPPGIGLLTELTKLNLA 532
Query: 94 KNKLKSM--DEIGSLSTIRALILNDNEIVSICNLDQMKELNTLVLSKNP-----IRKIGE 146
KN+L EI + +++ L L +N+ D++ ++ +L +S N + +I
Sbjct: 533 KNRLSGEIPREISTCRSLQLLNLGENDFSGEIP-DELGQIPSLAISLNLSCNRFVGEIPS 591
Query: 147 ALKKVKSITKLSLSHCQLEGIDTSLKFCVELTELRLAHNDIKSLPEELMHNSKLRNLDLG 206
+K++ L +SH QL G L L L +++ND +L + R L L
Sbjct: 592 RFSDLKNLGVLDVSHNQLTGNLNVLTDLQNLVSLNISYNDFSG---DLPNTPFFRRLPL- 647
Query: 207 NNVIAKWSDIKVLKSLTKLRNLNLQGNPVATNEKVIR 243
SD+ + L ++ + +P N V+R
Sbjct: 648 -------SDLASNRGLYISNAISTRPDPTTRNSSVVR 677
Score = 42.7 bits (99), Expect = 4e-04
Identities = 50/182 (27%), Positives = 83/182 (45%), Gaps = 21/182 (11%)
Query: 62 CVTLKWLSVVENKLE--------SLEGIQGLTKLTVLNAGKNKLKSMDEIGSLSTIRALI 113
C L L + E L +L+ +Q + T L +G DEIG + ++ L
Sbjct: 213 CENLVMLGLAETSLSGKLPASIGNLKRVQTIAIYTSLLSGPIP----DEIGYCTELQNLY 268
Query: 114 LNDNEIVSIC--NLDQMKELNTLVL-SKNPIRKIGEALKKVKSITKLSLSHCQLEG-IDT 169
L N I + +K+L +L+L N + KI L + + S L G I
Sbjct: 269 LYQNSISGSIPTTIGGLKKLQSLLLWQNNLVGKIPTELGNCPELWLIDFSENLLTGTIPR 328
Query: 170 SLKFCVELTELRLAHNDIK-SLPEELMHNSKLRNLDLGNNVIAKWSDIKVLKSLTKLRNL 228
S L EL+L+ N I ++PEEL + +KL +L++ NN+I ++ ++ LR+L
Sbjct: 329 SFGKLENLQELQLSVNQISGTIPEELTNCTKLTHLEIDNNLITG----EIPSLMSNLRSL 384
Query: 229 NL 230
+
Sbjct: 385 TM 386
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.308 0.128 0.345
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,580,394
Number of Sequences: 26719
Number of extensions: 482090
Number of successful extensions: 4818
Number of sequences better than 10.0: 645
Number of HSP's better than 10.0 without gapping: 135
Number of HSP's successfully gapped in prelim test: 515
Number of HSP's that attempted gapping in prelim test: 2947
Number of HSP's gapped (non-prelim): 1442
length of query: 468
length of database: 11,318,596
effective HSP length: 103
effective length of query: 365
effective length of database: 8,566,539
effective search space: 3126786735
effective search space used: 3126786735
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 62 (28.5 bits)
Medicago: description of AC146332.10


