

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146331.9 + phase: 0
(75 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
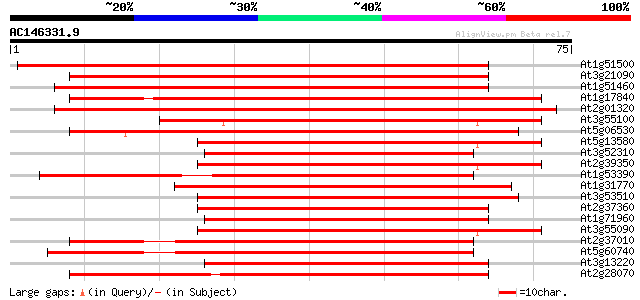
Score E
Sequences producing significant alignments: (bits) Value
At1g51500 ATP-dependent transmembrane transporter, putative 99 5e-22
At3g21090 ABC transporter, putative 94 9e-21
At1g51460 ATP-dependent transmembrane transporter like protein 93 3e-20
At1g17840 putative ABC transporter (At1g17840) 72 5e-14
At2g01320 putative ABC transporter 60 2e-10
At3g55100 ABC transporter - like protein 54 2e-08
At5g06530 ABC transporter like protein 53 2e-08
At5g13580 ABC transporter-like protein 52 5e-08
At3g52310 ABC transporter-like protein 52 7e-08
At2g39350 ABC transporter like protein 51 9e-08
At1g53390 putative ABC transporter gb|AAD31586.1 50 2e-07
At1g31770 unknown protein 50 2e-07
At3g53510 ABC transporter -like protein 50 2e-07
At2g37360 ABC transporter like protein 50 3e-07
At1g71960 putative ABC transporter 50 3e-07
At3g55090 ABC transporter - like protein 49 6e-07
At2g37010 putative ABC transporter 48 8e-07
At5g60740 xABC transporter - like protein 48 1e-06
At3g13220 ABC transporter 48 1e-06
At2g28070 putative ABC transporter 48 1e-06
>At1g51500 ATP-dependent transmembrane transporter, putative
Length = 687
Score = 98.6 bits (244), Expect = 5e-22
Identities = 48/63 (76%), Positives = 54/63 (85%)
Query: 2 EIGYLMVLAWEDVRVMLPNFGKGPTKRLLNGLNGFAEPGRIMAIMGPSGSGKSTLLDTLA 61
EIG LAWED+ V++PNF GPT+RLL+GLNG AEPGRIMAIMGPSGSGKSTLLD+LA
Sbjct: 17 EIGRGAYLAWEDLTVVIPNFSGGPTRRLLDGLNGHAEPGRIMAIMGPSGSGKSTLLDSLA 76
Query: 62 GSL 64
G L
Sbjct: 77 GRL 79
>At3g21090 ABC transporter, putative
Length = 680
Score = 94.4 bits (233), Expect = 9e-21
Identities = 44/56 (78%), Positives = 50/56 (88%)
Query: 9 LAWEDVRVMLPNFGKGPTKRLLNGLNGFAEPGRIMAIMGPSGSGKSTLLDTLAGSL 64
LAWED+ V++PNF GPT+RLL LNG+AEPGRIMAIMGPSGSGKSTLLD+LAG L
Sbjct: 25 LAWEDLTVVIPNFSDGPTRRLLQRLNGYAEPGRIMAIMGPSGSGKSTLLDSLAGRL 80
>At1g51460 ATP-dependent transmembrane transporter like protein
Length = 678
Score = 92.8 bits (229), Expect = 3e-20
Identities = 43/58 (74%), Positives = 50/58 (86%)
Query: 7 MVLAWEDVRVMLPNFGKGPTKRLLNGLNGFAEPGRIMAIMGPSGSGKSTLLDTLAGSL 64
M +AWED+ V++PNFG+G TKRLLNG+NG EP RI+AIMGPSGSGKSTLLD LAG L
Sbjct: 8 MYVAWEDLTVVIPNFGEGATKRLLNGVNGCGEPNRILAIMGPSGSGKSTLLDALAGRL 65
>At1g17840 putative ABC transporter (At1g17840)
Length = 703
Score = 72.0 bits (175), Expect = 5e-14
Identities = 34/63 (53%), Positives = 45/63 (70%), Gaps = 1/63 (1%)
Query: 9 LAWEDVRVMLPNFGKGPTKRLLNGLNGFAEPGRIMAIMGPSGSGKSTLLDTLAGSLKVTS 68
L W+D+ VM+ G G T+ +L GL G+AEPG + A+MGPSGSGKST+LD LA L +
Sbjct: 50 LTWQDLTVMV-TMGDGETQNVLEGLTGYAEPGSLTALMGPSGSGKSTMLDALASRLAANA 108
Query: 69 FLN 71
FL+
Sbjct: 109 FLS 111
>At2g01320 putative ABC transporter
Length = 725
Score = 60.1 bits (144), Expect = 2e-10
Identities = 28/67 (41%), Positives = 45/67 (66%)
Query: 7 MVLAWEDVRVMLPNFGKGPTKRLLNGLNGFAEPGRIMAIMGPSGSGKSTLLDTLAGSLKV 66
+ + W ++ L + + LL ++G A+PGR++AIMGPSGSGK+TLL+ LAG L +
Sbjct: 68 VTIRWRNITCSLSDKSSKSVRFLLKNVSGEAKPGRLLAIMGPSGSGKTTLLNVLAGQLSL 127
Query: 67 TSFLNMS 73
+ L++S
Sbjct: 128 SPRLHLS 134
>At3g55100 ABC transporter - like protein
Length = 662
Score = 53.5 bits (127), Expect = 2e-08
Identities = 32/58 (55%), Positives = 38/58 (65%), Gaps = 7/58 (12%)
Query: 21 FGKGPTK--RLLNGLNGFAEPGRIMAIMGPSGSGKSTLLDTLA-----GSLKVTSFLN 71
FG P K LLNG+ G A+ G I+AI+G SG+GKSTL+D LA GSLK T LN
Sbjct: 41 FGHSPAKIKTLLNGITGEAKEGEILAILGASGAGKSTLIDALAGQIAEGSLKGTVTLN 98
>At5g06530 ABC transporter like protein
Length = 627
Score = 53.1 bits (126), Expect = 2e-08
Identities = 26/62 (41%), Positives = 39/62 (61%), Gaps = 2/62 (3%)
Query: 9 LAWEDV--RVMLPNFGKGPTKRLLNGLNGFAEPGRIMAIMGPSGSGKSTLLDTLAGSLKV 66
L + DV +V++ K +L G++G PG ++A+MGPSGSGK+TLL LAG +
Sbjct: 33 LKFRDVTYKVVIKKLTSSVEKEILTGISGSVNPGEVLALMGPSGSGKTTLLSLLAGRISQ 92
Query: 67 TS 68
+S
Sbjct: 93 SS 94
>At5g13580 ABC transporter-like protein
Length = 727
Score = 52.0 bits (123), Expect = 5e-08
Identities = 29/51 (56%), Positives = 34/51 (65%), Gaps = 5/51 (9%)
Query: 26 TKRLLNGLNGFAEPGRIMAIMGPSGSGKSTLLDTLA-----GSLKVTSFLN 71
TK LLNG+ G A G I+A++G SGSGKSTL+D LA GSLK LN
Sbjct: 105 TKTLLNGITGEARDGEILAVLGASGSGKSTLIDALANRIAKGSLKGNVTLN 155
>At3g52310 ABC transporter-like protein
Length = 737
Score = 51.6 bits (122), Expect = 7e-08
Identities = 22/36 (61%), Positives = 30/36 (83%)
Query: 27 KRLLNGLNGFAEPGRIMAIMGPSGSGKSTLLDTLAG 62
K +LNG++G A PG ++A+MGPSGSGK+TLL+ L G
Sbjct: 165 KSILNGISGSAYPGELLALMGPSGSGKTTLLNALGG 200
>At2g39350 ABC transporter like protein
Length = 740
Score = 51.2 bits (121), Expect = 9e-08
Identities = 29/51 (56%), Positives = 34/51 (65%), Gaps = 5/51 (9%)
Query: 26 TKRLLNGLNGFAEPGRIMAIMGPSGSGKSTLLDTLA-----GSLKVTSFLN 71
TK LLN ++G G IMA++G SGSGKSTL+D LA GSLK T LN
Sbjct: 106 TKTLLNNISGETRDGEIMAVLGASGSGKSTLIDALANRIAKGSLKGTVKLN 156
>At1g53390 putative ABC transporter gb|AAD31586.1
Length = 1096
Score = 50.4 bits (119), Expect = 2e-07
Identities = 26/58 (44%), Positives = 41/58 (69%), Gaps = 4/58 (6%)
Query: 5 YLMVLAWEDVRVMLPNFGKGPTKRLLNGLNGFAEPGRIMAIMGPSGSGKSTLLDTLAG 62
+LM L+++D+ + L + GK ++L + G +PGRI A+MGPSG+GK++LL LAG
Sbjct: 473 HLMELSFKDLTLTLKSNGK----QVLRCVTGSMKPGRITAVMGPSGAGKTSLLSALAG 526
>At1g31770 unknown protein
Length = 648
Score = 50.4 bits (119), Expect = 2e-07
Identities = 23/45 (51%), Positives = 29/45 (64%)
Query: 23 KGPTKRLLNGLNGFAEPGRIMAIMGPSGSGKSTLLDTLAGSLKVT 67
K K +LNG+ G PG +A++GPSGSGK+TLL L G L T
Sbjct: 75 KSKEKTILNGITGMVCPGEFLAMLGPSGSGKTTLLSALGGRLSKT 119
>At3g53510 ABC transporter -like protein
Length = 739
Score = 50.1 bits (118), Expect = 2e-07
Identities = 24/43 (55%), Positives = 31/43 (71%)
Query: 26 TKRLLNGLNGFAEPGRIMAIMGPSGSGKSTLLDTLAGSLKVTS 68
TK LLNG++G A G +MA++G SGSGKSTL+D LA + S
Sbjct: 123 TKVLLNGISGEAREGEMMAVLGASGSGKSTLIDALANRISKES 165
>At2g37360 ABC transporter like protein
Length = 755
Score = 49.7 bits (117), Expect = 3e-07
Identities = 23/39 (58%), Positives = 30/39 (75%)
Query: 26 TKRLLNGLNGFAEPGRIMAIMGPSGSGKSTLLDTLAGSL 64
TK LLNG++G A G +MA++G SGSGKSTL+D LA +
Sbjct: 130 TKILLNGISGEAREGEMMAVLGASGSGKSTLIDALANRI 168
>At1g71960 putative ABC transporter
Length = 662
Score = 49.7 bits (117), Expect = 3e-07
Identities = 21/38 (55%), Positives = 29/38 (76%)
Query: 27 KRLLNGLNGFAEPGRIMAIMGPSGSGKSTLLDTLAGSL 64
+ +L+G+ G PG MA++GPSGSGKSTLL+ +AG L
Sbjct: 81 RTILSGVTGMISPGEFMAVLGPSGSGKSTLLNAVAGRL 118
>At3g55090 ABC transporter - like protein
Length = 720
Score = 48.5 bits (114), Expect = 6e-07
Identities = 27/51 (52%), Positives = 34/51 (65%), Gaps = 5/51 (9%)
Query: 26 TKRLLNGLNGFAEPGRIMAIMGPSGSGKSTLLDTLA-----GSLKVTSFLN 71
TK LL+ ++G G I+A++G SGSGKSTL+D LA GSLK T LN
Sbjct: 88 TKTLLDNISGETRDGEILAVLGASGSGKSTLIDALANRIAKGSLKGTVTLN 138
>At2g37010 putative ABC transporter
Length = 386
Score = 48.1 bits (113), Expect = 8e-07
Identities = 24/54 (44%), Positives = 35/54 (64%), Gaps = 4/54 (7%)
Query: 9 LAWEDVRVMLPNFGKGPTKRLLNGLNGFAEPGRIMAIMGPSGSGKSTLLDTLAG 62
+A++D+ + L KG K +L + G PGR+ A+MGPSG+GK+T L LAG
Sbjct: 9 VAFKDLTLTL----KGKHKHILRSVTGKIMPGRVSAVMGPSGAGKTTFLSALAG 58
>At5g60740 xABC transporter - like protein
Length = 1061
Score = 47.8 bits (112), Expect = 1e-06
Identities = 23/57 (40%), Positives = 36/57 (62%), Gaps = 4/57 (7%)
Query: 6 LMVLAWEDVRVMLPNFGKGPTKRLLNGLNGFAEPGRIMAIMGPSGSGKSTLLDTLAG 62
++ +A++D+ + L KG K L+ + G PGR+ A+MGPSG+GK+T L L G
Sbjct: 496 MIEVAFKDLSITL----KGKNKHLMRCVTGKLSPGRVSAVMGPSGAGKTTFLTALTG 548
>At3g13220 ABC transporter
Length = 647
Score = 47.8 bits (112), Expect = 1e-06
Identities = 21/38 (55%), Positives = 27/38 (70%)
Query: 27 KRLLNGLNGFAEPGRIMAIMGPSGSGKSTLLDTLAGSL 64
K +L G+ G PG I+A+MGPSGSGK+TLL + G L
Sbjct: 66 KHILKGITGSTGPGEILALMGPSGSGKTTLLKIMGGRL 103
>At2g28070 putative ABC transporter
Length = 705
Score = 47.8 bits (112), Expect = 1e-06
Identities = 26/56 (46%), Positives = 35/56 (62%), Gaps = 1/56 (1%)
Query: 9 LAWEDVRVMLPNFGKGPTKRLLNGLNGFAEPGRIMAIMGPSGSGKSTLLDTLAGSL 64
+AW+D+ V + K K ++ NG+A PG + IMGP+ SGKSTLL LAG L
Sbjct: 114 IAWKDLTVTMKGKRKYSDK-VVKSSNGYAFPGTMTVIMGPAKSGKSTLLRALAGRL 168
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.140 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,639,048
Number of Sequences: 26719
Number of extensions: 56752
Number of successful extensions: 578
Number of sequences better than 10.0: 209
Number of HSP's better than 10.0 without gapping: 187
Number of HSP's successfully gapped in prelim test: 22
Number of HSP's that attempted gapping in prelim test: 338
Number of HSP's gapped (non-prelim): 262
length of query: 75
length of database: 11,318,596
effective HSP length: 51
effective length of query: 24
effective length of database: 9,955,927
effective search space: 238942248
effective search space used: 238942248
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 52 (24.6 bits)
Medicago: description of AC146331.9


