

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146331.7 - phase: 0
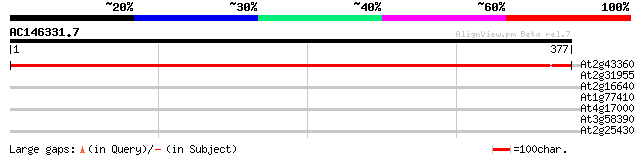
(377 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g43360 biotin synthase (Bio B) 615 e-177
At2g31955 molybdopterin synthase (CNX2) 38 0.008
At2g16640 putative chloroplast outer membrane protein 29 3.7
At1g77410 unknown protein 29 3.7
At4g17000 hypothetical protein 28 8.2
At3g58390 pelota-like protein 28 8.2
At2g25430 unknown protein 28 8.2
>At2g43360 biotin synthase (Bio B)
Length = 378
Score = 615 bits (1587), Expect = e-177
Identities = 297/378 (78%), Positives = 342/378 (89%), Gaps = 2/378 (0%)
Query: 1 MFWLRPILRSQSRSSI-WVLQHCNSFSTSSAAAIQAEKTIQNGPRNDWTKDEVKSIYDSP 59
M +R + RSQ R S+ LQ + +S+ SAA+ +AE+TI+ GPRNDW++DE+KS+YDSP
Sbjct: 1 MMLVRSVFRSQLRPSVSGGLQSASCYSSLSAASAEAERTIREGPRNDWSRDEIKSVYDSP 60
Query: 60 ILDLLFHGAQVHRHAHNFREVQQCTLLSVKTGGCSEDCSYCPQSSRYDTGLKGQKLLNKD 119
+LDLLFHGAQVHRH HNFREVQQCTLLS+KTGGCSEDCSYCPQSSRY TG+K Q+L++KD
Sbjct: 61 LLDLLFHGAQVHRHVHNFREVQQCTLLSIKTGGCSEDCSYCPQSSRYSTGVKAQRLMSKD 120
Query: 120 AVLQAAVKAKEAGSTRFCMGAAWRDTIGRKTNFNQILEYVKEIKGMGMEVCCTLGMLDKD 179
AV+ AA KAKEAGSTRFCMGAAWRDTIGRKTNF+QILEY+KEI+GMGMEVCCTLGM++K
Sbjct: 121 AVIDAAKKAKEAGSTRFCMGAAWRDTIGRKTNFSQILEYIKEIRGMGMEVCCTLGMIEKQ 180
Query: 180 QAGELKKAGLTAYNHNLDTSREYYPNIITTRTYDERLKTLEFVRDAGINVCSGGIIGLGE 239
QA ELKKAGLTAYNHNLDTSREYYPN+ITTR+YD+RL+TL VRDAGINVCSGGIIGLGE
Sbjct: 181 QALELKKAGLTAYNHNLDTSREYYPNVITTRSYDDRLETLSHVRDAGINVCSGGIIGLGE 240
Query: 240 AEDDRVGLLHTLSTLPTHPESVPINALIAVKGTPLQDQKPVEIWEMIRMIATARITMPKA 299
AE+DR+GLLHTL+TLP+HPESVPINAL+AVKGTPL+DQKPVEIWEMIRMI TARI MPKA
Sbjct: 241 AEEDRIGLLHTLATLPSHPESVPINALLAVKGTPLEDQKPVEIWEMIRMIGTARIVMPKA 300
Query: 300 MVRLSAGRVRFSVPEQALCFLAGANSIFAGEKLLTTANNDFDADQLMFKVLGLLPKAPSL 359
MVRLSAGRVRFS+ EQALCFLAGANSIF GEKLLTT NNDFDADQLMFK LGL+PK PS
Sbjct: 301 MVRLSAGRVRFSMSEQALCFLAGANSIFTGEKLLTTPNNDFDADQLMFKTLGLIPKPPSF 360
Query: 360 DDDETNEAENYKEAASSS 377
+D+ +E+EN ++ AS+S
Sbjct: 361 SEDD-SESENCEKVASAS 377
>At2g31955 molybdopterin synthase (CNX2)
Length = 390
Score = 38.1 bits (87), Expect = 0.008
Identities = 46/205 (22%), Positives = 83/205 (40%), Gaps = 21/205 (10%)
Query: 45 NDWTKDEVKSIYDSPILDLLFHGAQVHRHAHNFREVQQCTLLSVKTGGCSEDCSYCPQSS 104
+ + +V I D+P+ D+L F + +S+ T C+ C YC S
Sbjct: 47 SSYAAHQVDQIKDNPVSDMLID---------KFGRLHTYLRISL-TERCNLRCQYCMPSE 96
Query: 105 RYDTGLKGQKLLNKDAVLQAAVKAKEAGSTRFCMGAAWRDTIGRKT---NFNQILEYVKE 161
+ K Q LL++ +++ A AG + R T G T + +I +
Sbjct: 97 GVELTPKPQ-LLSQSEIVRLAGLFVSAGVNKI------RLTGGEPTVRKDIEEICLQLSS 149
Query: 162 IKGMGMEVCCTLGMLDKDQAGELKKAGLTAYNHNLDTSREYYPNIITTRT-YDERLKTLE 220
+KG+ T G+ + LK+ GL + N +LDT +T R +D +K+++
Sbjct: 150 LKGLKNLAITTNGITLAKKLPRLKECGLDSLNISLDTLVPAKFEFLTRRKGHDRVMKSID 209
Query: 221 FVRDAGINVCSGGIIGLGEAEDDRV 245
+ G N + + DD +
Sbjct: 210 TAIELGYNPVKVNCVIMRGLNDDEI 234
>At2g16640 putative chloroplast outer membrane protein
Length = 1206
Score = 29.3 bits (64), Expect = 3.7
Identities = 24/90 (26%), Positives = 40/90 (43%), Gaps = 7/90 (7%)
Query: 195 NLDTSREYYPNIITTRTYDERLKTLEFVRDAGINVCSGGIIGLGEAEDDRVGLL---HTL 251
N +T Y NI+T + D + + V + + S G GE E D L H
Sbjct: 303 NGETGAAYTSNIVTNASGDNEVSSA--VTSSPLEESSSG--EKGETEGDSTCLKPEQHLA 358
Query: 252 STLPTHPESVPINALIAVKGTPLQDQKPVE 281
S+ ++PES +++ G ++ KPV+
Sbjct: 359 SSPHSYPESTEVHSNSGSPGVTSREHKPVQ 388
>At1g77410 unknown protein
Length = 815
Score = 29.3 bits (64), Expect = 3.7
Identities = 24/98 (24%), Positives = 43/98 (43%), Gaps = 20/98 (20%)
Query: 155 ILEYVKEIKGMGMEVCCTLGMLDKDQAGELKKAGLTAYNHNLDTSREYYPNIITTRTYDE 214
I++++KE+K G+ VC +G + GE GL + HN+ I RT +E
Sbjct: 95 IVKFIKEVKNHGLYVCLRIGPFIQ---GEWSYGGLPFWLHNVQG--------IVFRTDNE 143
Query: 215 RLK---------TLEFVRDAGINVCSGGIIGLGEAEDD 243
K ++ ++ + GG I L + E++
Sbjct: 144 PFKYHMKRYAKMIVKLMKSENLYASQGGPIILSQIENE 181
>At4g17000 hypothetical protein
Length = 674
Score = 28.1 bits (61), Expect = 8.2
Identities = 16/66 (24%), Positives = 32/66 (48%), Gaps = 1/66 (1%)
Query: 252 STLPTHPESVPINALIAVKGTPLQDQKPVEIWEMIRMIATARITMPKAMVRLSAGRVRFS 311
+T+P P ++ L + P + + E +++++T + PKA + V S
Sbjct: 98 NTVPAKPSRSRVSKLAMISSIPQKGNGNIRSKE-VKVVSTNKNVTPKAKAKGKESAVISS 156
Query: 312 VPEQAL 317
VP++AL
Sbjct: 157 VPQKAL 162
>At3g58390 pelota-like protein
Length = 395
Score = 28.1 bits (61), Expect = 8.2
Identities = 17/54 (31%), Positives = 29/54 (53%), Gaps = 3/54 (5%)
Query: 4 LRPILRSQSRSSIWVLQHCNSFSTSSAAAIQAEKTIQNGPRNDWTKDEVKSIYD 57
LRPIL ++SR ++L H NS S + + + + N ++ EVK++ D
Sbjct: 247 LRPILENKSR---FILVHTNSGYKHSLSEVLHDPNVMNMIKDTKAAKEVKALND 297
>At2g25430 unknown protein
Length = 653
Score = 28.1 bits (61), Expect = 8.2
Identities = 17/41 (41%), Positives = 22/41 (53%), Gaps = 4/41 (9%)
Query: 210 RTY----DERLKTLEFVRDAGINVCSGGIIGLGEAEDDRVG 246
RTY D+RL+ F R +G++V SGG DDR G
Sbjct: 139 RTYAGYLDQRLELALFERKSGVSVNSGGNSSHHSNNDDRYG 179
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.133 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,289,467
Number of Sequences: 26719
Number of extensions: 343492
Number of successful extensions: 745
Number of sequences better than 10.0: 7
Number of HSP's better than 10.0 without gapping: 1
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 744
Number of HSP's gapped (non-prelim): 7
length of query: 377
length of database: 11,318,596
effective HSP length: 101
effective length of query: 276
effective length of database: 8,619,977
effective search space: 2379113652
effective search space used: 2379113652
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC146331.7


