

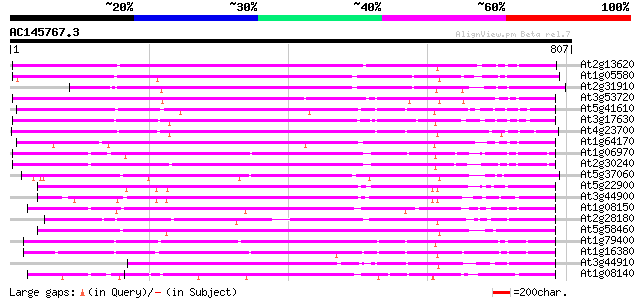
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC145767.3 - phase: 0
(807 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g13620 putative Na/H antiporter 514 e-146
At1g05580 unknown protein 454 e-128
At2g31910 putative Na+/H+ antiporter 404 e-113
At3g53720 unknown protein 404 e-112
At5g41610 Na+/H+ antiporter-like protein 391 e-109
At3g17630 unknown protein 383 e-106
At4g23700 putative Na+/H+-exchanging protein 376 e-104
At1g64170 hypothetical protein 318 1e-86
At1g06970 hypothetical protein 273 4e-73
At2g30240 putative Na/H antiporter 263 4e-70
At5g37060 putative transporter protein 256 4e-68
At5g22900 Na+/H+ antiporter-like protein 253 3e-67
At3g44900 putative protein 251 1e-66
At1g08150 hypothetical protein 249 4e-66
At2g28180 hypothetical protein 243 2e-64
At5g58460 putative protein 235 6e-62
At1g79400 hypothetical protein 228 8e-60
At1g16380 putative Na/H antiporter 215 9e-56
At3g44910 putative protein 207 2e-53
At1g08140 hypothetical protein 205 9e-53
>At2g13620 putative Na/H antiporter
Length = 821
Score = 514 bits (1324), Expect = e-146
Identities = 289/794 (36%), Positives = 474/794 (59%), Gaps = 29/794 (3%)
Query: 4 CYNVTLSASNNIWMTDDVMIKRVPLLCLQIAYNLLVSRFLFFILKPLHVPLIVAQMLTGF 63
CY ++ +N +W D+ + +PL LQ+ ++V+RF FILKP P +++++L G
Sbjct: 15 CYAPSMITTNGVWQGDNPLDFSLPLFVLQLTLVVVVTRFFVFILKPFRQPRVISEILGGI 74
Query: 64 SVSRSLLGYFEGVITLFYNPRGILAVETISNLGIMYYVFLSGLEMNSDTILRSRKKGTSM 123
+ S+LG + R ++ +ET++N+G++Y++FL G+EM+ + ++ K+ ++
Sbjct: 75 VLGPSVLGRSTKFAHTIFPQRSVMVLETMANVGLLYFLFLVGVEMDIMVVRKTGKRALTI 134
Query: 124 AIAGIVTSMLFGVGFLTLQQKLLDKK-EKTHIKAYLFWCLTLSVTGFPVLARILAKLKLL 182
AI G+V L G F + D + T+I LF + LSVT FPVLARILA+LKL+
Sbjct: 135 AIGGMVLPFLIGAAFSFSMHRSEDHLGQGTYI---LFLGVALSVTAFPVLARILAELKLI 191
Query: 183 YTKLGKDTLTAAMLTDAYGWVLFTLLIPAANNWGEGY--YLSVITTFLFIAFCFTVVRPI 240
T++G+ +++AA++ D + W+L L I A + + +I++ +FIA C VVRP
Sbjct: 192 NTEIGRISMSAALVNDMFAWILLALAIALAESDKTSFASLWVMISSAVFIAVCVFVVRPG 251
Query: 241 LTPIIENRTNKNMWRKSHMLDVFIGLFICSYITDFLGTHPIVGAFVFGLILPHGKFADMV 300
+ II + + H+ + G+ I +ITD +GTH + GAFVFGL++P+G +
Sbjct: 252 IAWIIRKTPEGENFSEFHICLILTGVMISGFITDAIGTHSVFGAFVFGLVIPNGPLGLTL 311
Query: 301 MEMSDDFVTGILCPVYFSGFGFKLDLPVLWNTPNSVLMMLIMVLLCIPKVLSSLIVTFFF 360
+E +DFV+G+L P++F+ G K ++ + + + L++ L C KV+ ++IV FF
Sbjct: 312 IEKLEDFVSGLLLPLFFAISGLKTNIAAIQGPATWLTLFLVIFLACAGKVIGTVIVAFFH 371
Query: 361 GMPARDGLSIGLLLNTKGVMAVILQNVAWDKRFLDPCSFMVMTLAIILMTVIVSPLINAI 420
GMP R+G+++GLLLNTKG++ +I+ NV D++ LD +F M L ++MT +++P++ +
Sbjct: 372 GMPVREGITLGLLLNTKGLVEMIVLNVGKDQKVLDDETFATMVLVALVMTGVITPIVTIL 431
Query: 421 YKPKFRFMQSQLRTVQKLRFDMELRVVACVHNAKHATNIIHVLEATNATRISPVHVSAVH 480
YKP + + + RT+Q+ + D ELRV+ CVH ++ II++LEA++ T+ SP+ + +H
Sbjct: 432 YKPVKKSVSYKRRTIQQTKPDSELRVLVCVHTPRNVPTIINLLEASHPTKRSPICIYVLH 491
Query: 481 LLELTRHGTAILVSQMADLNNIAGAETTNYGPKAEFEIITTAFKEFVEQYNAVRFDTSSV 540
L+ELT +A+L+ + T +A+ + I AF+ + + V +
Sbjct: 492 LVELTGRASAMLIVHNTRKSGRPALNRT----QAQSDHIINAFENYEQHAAFVAVQPLTA 547
Query: 541 VSSYTTIHEDIYNVAEEKRASLILLPFHKEYSTVGGSPEISHNEHCEINENVLQHAPCSV 600
+S Y+T+HED+ ++AE+KR S I++PFHK+ TV G E ++ + +N+N+L+++PCSV
Sbjct: 548 ISPYSTMHEDVCSLAEDKRVSFIIIPFHKQ-QTVDGGMESTNPAYRLVNQNLLENSPCSV 606
Query: 601 GIFVDRGLGSLLK-----TKMRIITLFIGGPDDREALSIAWRMAGHSGTQLHVVR-IHLL 654
GI VDRGL + +++ LF GGPDDREAL+ AWRMA H G L V+R IH
Sbjct: 607 GILVDRGLNGATRLNSNTVSLQVAVLFFGGPDDREALAYAWRMAQHPGITLTVLRFIHDE 666
Query: 655 GKA--AEEKVLKKKISKSPHGMLSTVMDGVMQKELDEEYIFSFRHKAVNNNDSIVYLEKE 712
+A A + K P MD Q++LD++YI FR + +SIVY+EK
Sbjct: 667 DEADTASTRATNDSDLKIPK------MDHRKQRQLDDDYINLFRAENA-EYESIVYIEKL 719
Query: 713 VHSDTGEEIPTLLNEIDKPGYDLYIVGQGSGKNKTIFLKLLEWCDHPELGVIGDILASTS 772
V GEE + +D +DL+IVG+G G + + L +W + PELG IGD+LAS+
Sbjct: 720 V--SNGEETVAAVRSMDS-SHDLFIVGRGEGMSSPLTAGLTDWSECPELGAIGDLLASSD 776
Query: 773 FGTHSSVLIVQQYM 786
F SVL+VQQY+
Sbjct: 777 FAATVSVLVVQQYV 790
>At1g05580 unknown protein
Length = 867
Score = 454 bits (1169), Expect = e-128
Identities = 273/804 (33%), Positives = 437/804 (53%), Gaps = 40/804 (4%)
Query: 4 CYNVTL---SASNNIWMTDDVMIKRVPLLCLQIAYNLLVSRFLFFILKPLHVPLIVAQML 60
CY+ +L W + + +P Q+ L R L+++ +PL++P VAQ+L
Sbjct: 26 CYDQSLLFEKREQKGWESGSTLASSLPFFITQLFVANLSYRVLYYLTRPLYLPPFVAQIL 85
Query: 61 TGFSVSRSLLGYFEGVITLFYNPRGILAVETISNLGIMYYVFLSGLEMNSDTILRSRKKG 120
G S S+LG +I + R + +ET +NL ++Y +FL GL M+ + + K
Sbjct: 86 CGLLFSPSVLGNTRFIIAHVFPYRFTMVLETFANLALVYNIFLLGLGMDLRMVRITELKP 145
Query: 121 TSMAIAGIVTSMLFGVGFLTLQQKLLDKKEKTHIKAYLFWCLTLSVTGFPVLARILAKLK 180
+A G++ ++ G L K I +FW + L+ T FP LARILA LK
Sbjct: 146 VIIAFTGLLVALPVGAFLYYLPGNGHPDKI---ISGCVFWSVALACTNFPDLARILADLK 202
Query: 181 LLYTKLGKDTLTAAMLTDAYGWVLFTLLIPA---ANNWGEGYYLSVITTFLFIAFCFTVV 237
LL + +G+ + AA++TD WVL + + W + +ITT +F+ C V+
Sbjct: 203 LLRSDMGRTAMCAAIVTDLCTWVLLVFGFASFSKSGTWNKMMPFVIITTAIFVLLCIFVI 262
Query: 238 RPILTPIIENRTNKNMWRKSHMLDVFIGLFICSYITDFLGTHPIVGAFVFGLILPHGKFA 297
RP + I +H+ + G+ +C ITD G H I GAF+FGL +PH
Sbjct: 263 RPGIAWIFAKTVKAGHVGDTHVWFILGGVVLCGLITDACGVHSITGAFLFGLSIPHDHII 322
Query: 298 -DMVMEMSDDFVTGILCPVYFSGFGFKLDLPVLWNTPNSVLMMLIMVLLCIPKVLSSLIV 356
+M+ E DF++GIL P+++ G + D+ + + +M++++ + K+++++I
Sbjct: 323 RNMIEEKLHDFLSGILMPLFYIICGLRADIGFMLQFTDKFMMVVVICSSFLVKIVTTVIT 382
Query: 357 TFFFGMPARDGLSIGLLLNTKGVMAVILQNVAWDKRFLDPCSFMVMTLAIILMTVIVSPL 416
+ F +P RD +IG L+NTKG +++++ N D + LD + MT+A+++M+++V PL
Sbjct: 383 SLFMHIPMRDAFAIGALMNTKGTLSLVVLNAGRDTKALDSPMYTHMTIALLVMSLVVEPL 442
Query: 417 INAIYKPKFRFMQSQLRTVQKLRFDMELRVVACVHNAKHATNIIHVLEATNATRISPVHV 476
+ YKPK + + RTVQK++ + ELRV+ACVH + + I ++L+ +NAT+ SP+ V
Sbjct: 443 LAFAYKPKKKLAHYKHRTVQKIKGETELRVLACVHVLPNVSGITNLLQVSNATKQSPLSV 502
Query: 477 SAVHLLELTRHGTAILVSQMADLNNIAGAETTNYGP--KAEFEIITTAFKEFVEQYNAVR 534
A+HL+ELT TA L+ + N N+ +AE + I F+ +A+
Sbjct: 503 FAIHLVELTGRTTASLL-----IMNDECKPKANFSDRVRAESDQIAETFEAMEVNNDAMT 557
Query: 535 FDTSSVVSSYTTIHEDIYNVAEEKRASLILLPFHKEYSTVGGSPEISHNEHCEINENVLQ 594
T + VS Y T+HEDI +AE+KR I+LP+HK + G E ++ H EIN+NVL
Sbjct: 558 VQTITAVSPYATMHEDICVLAEDKRVCFIILPYHKHLTPDGRMGE-GNSSHAEINQNVLS 616
Query: 595 HAPCSVGIFVDRGLGSLLKTKMR-------IITLFIGGPDDREALSIAWRMAGHSGTQLH 647
HAPCSVGI VDRG+ + R + LF+GGPDDREALS AWRM G +L
Sbjct: 617 HAPCSVGILVDRGMAMVRSESFRGESMKREVAMLFVGGPDDREALSYAWRMVGQHVIKLT 676
Query: 648 VVRIHLLGKAAEEKVLKKKISKSPHGMLSTVMDGVMQKELDEEYIFSFRHKAVNNNDSIV 707
VVR E + K++ + +K++D+E I+ F K +N++ S+
Sbjct: 677 VVR---FVPGREALISSGKVA----------AEYEREKQVDDECIYEFNFKTMNDS-SVK 722
Query: 708 YLEKEVHSDTGEEIPTLLNEIDKPGYDLYIVGQGSGKNKTIFLKLLEWCDHPELGVIGDI 767
Y+EK V +D + I T+ D YDLY+VG+G + + L +W PELG IGD
Sbjct: 723 YIEKVV-NDGQDTIATIREMEDNNSYDLYVVGRGYNSDSPVTAGLNDWSSSPELGTIGDT 781
Query: 768 LASTSFGTHSSVLIVQQYMVGRKR 791
LAS++F H+SVL++QQY +++
Sbjct: 782 LASSNFTMHASVLVIQQYSATKRQ 805
>At2g31910 putative Na+/H+ antiporter
Length = 735
Score = 404 bits (1039), Expect = e-113
Identities = 258/736 (35%), Positives = 395/736 (53%), Gaps = 55/736 (7%)
Query: 87 LAVETISNLGIMYYVFLSGLEMNSDTILRSRKKGTSMAIAGIVTSMLFGVGFLTLQQKLL 146
+ +ET +NL ++Y VFL GL ++ I K +AI G++ ++L G G L
Sbjct: 1 MLLETFANLALVYNVFLLGLGLDLRMIKIKDIKPVIIAIVGLLAALLAGAGLYYLPSN-- 58
Query: 147 DKKEKTHIKAYLFWCLTLSVTGFPVLARILAKLKLLYTKLGKDTLTAAMLTDAYGWVLFT 206
+ +K + ++W + T FP LARILA LKLL T +G + AA++TD W+LF
Sbjct: 59 GEADKI-LAGCMYWSIAFGCTNFPDLARILADLKLLRTDMGHTAMCAAVVTDLCTWILFI 117
Query: 207 LLIPAANNWG---EGYYLSVITTFLFIAFCFTVVRPILTPIIENRTNKNMWRKSHMLDVF 263
+ + G E S+ +T F+ C+ V++P + I N +H+
Sbjct: 118 FGMAIFSKSGVRNEMLPYSLASTIAFVLLCYFVIQPGVAWIFNNTVEGGQVGDTHVWYTL 177
Query: 264 IGLFICSYITDFLGTHPIVGAFVFGLILPHGKFA-DMVMEMSDDFVTGILCPVYFSGFGF 322
G+ ICS IT+ G H I GAF+FGL +PH M+ E DF++G+L P+++ G
Sbjct: 178 AGVIICSLITEVCGVHSITGAFLFGLSIPHDHIIRKMIEEKLHDFLSGMLMPLFYIICGL 237
Query: 323 KLDLPVLWNTPNSVLMMLIMVLLCIPKVLSSLIVTFFFGMPARDGLSIGLLLNTKGVMAV 382
+ D+ + T + +M ++ + K+LS++ + F +P RDGL+IG L+NTKG MA+
Sbjct: 238 RADIGYMNRTVSVGMMAVVTSASVMVKILSTMFCSIFLRIPLRDGLAIGALMNTKGTMAL 297
Query: 383 ILQNVAWDKRFLDPCSFMVMTLAIILMTVIVSPLINAIYKPKFRFMQSQLRTVQKLRFDM 442
++ N D + LD + +TLA ++M+++V PL+ YKPK + + + RT+QK + +
Sbjct: 298 VILNAGRDTKALDVIMYTHLTLAFLVMSMVVQPLLAIAYKPKKKLIFYKNRTIQKHKGES 357
Query: 443 ELRVVACVHNAKHATNIIHVLEATNATRISPVHVSAVHLLELTRHGTAILVSQMADLNNI 502
EL V+ CVH + + I ++L+ +N T+ SP++V A+HL+ELT TA L+ + N
Sbjct: 358 ELCVLTCVHVLPNVSGITNLLQLSNPTKKSPLNVFAIHLVELTGRTTASLL-----IMND 412
Query: 503 AGAETTNYGP--KAEFEIITTAFKEFVEQYNAVRFDTSSVVSSYTTIHEDIYNVAEEKRA 560
N+ +AE + I F + V T + VS Y T+ EDI +AE+K+A
Sbjct: 413 EAKPKANFADRVRAESDQIAEMFTALEVNNDGVMVQTITAVSPYATMDEDICLLAEDKQA 472
Query: 561 SLILLPFHKEYSTVGGSPEISHNEHCEINENVLQHAPCSVGIFVDRGLGSLL-------- 612
ILLP+HK ++ G E + H EIN+NV+ HAPCSVGI VDRG+ ++
Sbjct: 473 CFILLPYHKNMTSDGRLNE-GNAVHAEINQNVMSHAPCSVGILVDRGMTTVRFESFMFQG 531
Query: 613 -KTKMRIITLFIGGPDDREALSIAWRMAGHSGTQLHVVR-------IHLLGKAAEEKVLK 664
TK I LF+GG DDREAL+ AWRM G QL VVR + G+AA+E
Sbjct: 532 ETTKKEIAMLFLGGRDDREALAYAWRMVGQEMVQLTVVRFVPSQEALVSAGEAADEY--- 588
Query: 665 KKISKSPHGMLSTVMDGVMQKELDEEYIFSFRHKAVNNNDSIVYLEKEVHSDTGEEIPTL 724
K +DEE I+ F K + N+ S+ Y+EK V + G+E T
Sbjct: 589 -----------------EKDKHVDEESIYEFNFKTM-NDPSVTYVEKVVKN--GQETITA 628
Query: 725 LNEI-DKPGYDLYIVGQGSGKNKTIFLKLLEWCDHPELGVIGDILASTSFGTHSSVLIVQ 783
+ E+ D YDLYIVG+G + L +W P+LG+IGD L S++F +SVL+VQ
Sbjct: 629 ILELEDNNSYDLYIVGRGYQVETPVTSGLTDWNSTPDLGIIGDTLISSNFTMQASVLVVQ 688
Query: 784 QYMVGRKRVVYKCHNE 799
QY ++ + E
Sbjct: 689 QYSSANRQTAENNNQE 704
>At3g53720 unknown protein
Length = 842
Score = 404 bits (1038), Expect = e-112
Identities = 257/824 (31%), Positives = 444/824 (53%), Gaps = 54/824 (6%)
Query: 5 YNVTL--SASNNIWMTDDVMIKRVPLLCLQIAYNLLVSRFLFFILKPLHVPLIVAQMLTG 62
+N+T ++SN +W D+ + PLL +Q A + VSRFL + KPL P ++A+++ G
Sbjct: 3 FNITSVKTSSNGVWQGDNPLNFAFPLLIVQTALIIAVSRFLAVLFKPLRQPKVIAEIVGG 62
Query: 63 FSVSRSLLGYFEGVITLFYNPRGILAVETISNLGIMYYVFLSGLEMNSDTILRSRKKGTS 122
+ S LG + + + +E+++++G+++++FL GLE++ +I RS K+
Sbjct: 63 ILLGPSALGRNMAYMDRIFPKWSMPILESVASIGLLFFLFLVGLELDLSSIRRSGKRAFG 122
Query: 123 MAIAGIVTSMLFGVGF-LTLQQKLLDKKEKTHIKAYL-FWCLTLSVTGFPVLARILAKLK 180
+A+AGI + GVG ++ L +K +L F + LS+T FPVLARILA+LK
Sbjct: 123 IAVAGITLPFIAGVGVAFVIRNTLYTAADKPGYAEFLVFMGVALSITAFPVLARILAELK 182
Query: 181 LLYTKLGKDTLTAAMLTDAYGWVLFTLLIPAANNWGEG---------YYLSVITTFLFIA 231
LL T++G+ + AA D W+L L + A N GEG +++ F+
Sbjct: 183 LLTTQIGETAMAAAAFNDVAAWILLALAVALAGNGGEGGGEKKSPLVSLWVLLSGAGFVV 242
Query: 232 FCFTVVRPILTPIIENRTNKN-MWRKSHMLDVFIGLFICSYITDFLGTHPIVGAFVFGLI 290
F V+RP + + + + +N + R+S++ G+ + + TD +G H I GAFVFGL
Sbjct: 243 FMLVVIRPGMKWVAKRGSPENDVVRESYVCLTLAGVMVSGFATDLIGIHSIFGAFVFGLT 302
Query: 291 LPH-GKFADMVMEMSDDFVTGILCPVYFSGFGFKLDLPVLWNTPNSVLMMLIMVLLCIPK 349
+P G+F ++E +DFV+G+L P+YF+ G K D+ + + ++ L++V C K
Sbjct: 303 IPKDGEFGQRLIERIEDFVSGLLLPLYFATSGLKTDVAKIRGAESWGMLGLVVVTACAGK 362
Query: 350 VLSSLIVTFFFGMPARDGLSIGLLLNTKGVMAVILQNVAWDKRFLDPCSFMVMTLAIILM 409
++ + +V +PAR+ L++G L+NTKG++ +I+ N+ +K+ L+ +F ++ L +
Sbjct: 363 IVGTFVVAVMVKVPAREALTLGFLMNTKGLVELIVLNIGKEKKVLNDETFAILVLMALFT 422
Query: 410 TVIVSPLINAIYKPKFRFMQSQLRTVQKLRFDM--ELRVVACVHNAKHATNIIHVLEATN 467
T I +P + AIYKP R +L+ + + ELR++AC+H + +++I ++E+
Sbjct: 423 TFITTPTVMAIYKPA-RGTHRKLKDLSASQDSTKEELRILACLHGPANVSSLISLVESIR 481
Query: 468 ATRISPVHVSAVHLLELTRHGTAILVSQMADLNNIAGAETTNYGPKAEFEIITTAFKEFV 527
T+I + + +HL+ELT ++I++ Q A N + +G + + F+ +
Sbjct: 482 TTKILRLKLFVMHLMELTERSSSIIMVQRARKNGLPFVHRYRHGER--HSNVIGGFEAY- 538
Query: 528 EQYNAVRFDTSSVVSSYTTIHEDIYNVAEEKRASLILLPFHKEYST-------------V 574
Q V + VS T+HEDI ++A+ KR ++I+LPFHK ++
Sbjct: 539 RQLGRVAVRPITAVSPLPTMHEDICHMADTKRVTMIILPFHKRWNADHGHSHHHQDGGGD 598
Query: 575 GGSPEISHNEHCEINENVLQHAPCSVGIFVDRGLGSLLKTKM---------RIITLFIGG 625
G PE + +N+ VL++APCSV + VDRGLGS+ + R+ +F GG
Sbjct: 599 GNVPENVGHGWRLVNQRVLKNAPCSVAVLVDRGLGSIEAQTLSLDGSNVVERVCVIFFGG 658
Query: 626 PDDREALSIAWRMAGHSGTQLHVVRI----HLLGKAAEEKVLKKKISKSPHGMLSTVMDG 681
PDDRE++ + RMA H ++ V+R L A + K + + L+T +D
Sbjct: 659 PDDRESIELGGRMAEHPAVKVTVIRFLVRETLRSTAVTLRPAPSKGKEKNYAFLTTNVDP 718
Query: 682 VMQKELDEEYIFSFRHKAVNNNDSIVYLEKEVHSDTGEEIPTLLNEIDKPGYDLYIVGQG 741
+KELDE + F+ K + + Y EKE ++ EEI ++ D +DL +VG+G
Sbjct: 719 EKEKELDEGALEDFKSKW---KEMVEYKEKEPNNII-EEILSIGQSKD---FDLIVVGRG 771
Query: 742 SGKNKTIFLKLLEWCDHPELGVIGDILASTSFGTHSSVLIVQQY 785
+ + +HPELG IGD+LAS+ S+L+VQQ+
Sbjct: 772 RIPSAEVAALAERQAEHPELGPIGDVLASSINHIIPSILVVQQH 815
>At5g41610 Na+/H+ antiporter-like protein
Length = 810
Score = 391 bits (1004), Expect = e-109
Identities = 247/793 (31%), Positives = 444/793 (55%), Gaps = 39/793 (4%)
Query: 10 SASNNIWMTDDVMIKRVPLLCLQIAYNLLVSRFLFFILKPLHVPLIVAQMLTGFSVSRSL 69
+ SN ++ D+ + +PL LQI ++++R L ++L+PL P ++A+++ G + SL
Sbjct: 15 ATSNGVFQGDNPIDFALPLAILQIVIVIVLTRVLAYLLRPLRQPRVIAEVIGGIMLGPSL 74
Query: 70 LGYFEGVITLFYNPRGILAVETISNLGIMYYVFLSGLEMNSDTILRSRKKGTSMAIAGIV 129
LG + + + + + +ET++NLG+++++FL+GLE+++ + R+ KK +A+AGI
Sbjct: 75 LGRSKAFLDAVFPKKSLTVLETLANLGLLFFLFLAGLEIDTKALRRTGKKALGIALAGIT 134
Query: 130 TSMLFGVGFLTLQQKLLDKKEKTHIKAYL-FWCLTLSVTGFPVLARILAKLKLLYTKLGK 188
G+G + + + K + A+L F + LS+T FPVLARILA+LKLL T++G+
Sbjct: 135 LPFALGIGSSFVLKATISKGVNS--TAFLVFMGVALSITAFPVLARILAELKLLTTEIGR 192
Query: 189 DTLTAAMLTDAYGWVLFTLLIPAANNWGEGYYLSVITTFLFIAFCFTVVRP--ILTPI-- 244
++AA + D W+L L I + + +++ ++F++ C V+ I+ PI
Sbjct: 193 LAMSAAAVNDVAAWILLALAIALSGSNTS----PLVSLWVFLSGCAFVIGASFIIPPIFR 248
Query: 245 -IENRTNKNM-WRKSHMLDVFIGLFICSYITDFLGTHPIVGAFVFGLILP-HGKFADMVM 301
I R ++ ++++ + +C +ITD +G H + GAFV G+++P G FA ++
Sbjct: 249 WISRRCHEGEPIEETYICATLAVVLVCGFITDAIGIHSMFGAFVVGVLIPKEGPFAGALV 308
Query: 302 EMSDDFVTGILCPVYFSGFGFKLDLPVLWNTPNSVLMMLIMVLLCIPKVLSSLIVTFFFG 361
E +D V+G+ P+YF G K ++ + + L++L+ C K+L +L V+ F
Sbjct: 309 EKVEDLVSGLFLPLYFVASGLKTNVATIQGAQSWGLLVLVTATACFGKILGTLGVSLAFK 368
Query: 362 MPARDGLSIGLLLNTKGVMAVILQNVAWDKRFLDPCSFMVMTLAIILMTVIVSPLINAIY 421
+P R+ +++G L+NTKG++ +I+ N+ D++ L+ +F +M L + T I +P++ A+Y
Sbjct: 369 IPMREAITLGFLMNTKGLVELIVLNIGKDRKVLNDQTFAIMVLMALFTTFITTPVVMAVY 428
Query: 422 KPKFRFMQS---QLRTVQKLRFDMELRVVACVHNAKHATNIIHVLEATNA-TRISPVHVS 477
KP R + + R V++ + +LR++ C H A ++I++LEA+ + + V
Sbjct: 429 KPARRAKKEGEYKHRAVERENTNTQLRILTCFHGAGSIPSMINLLEASRGIEKGEGLCVY 488
Query: 478 AVHLLELTRHGTAILVSQMADLNNIAGAETTNYGPKAEFEIITTAFKEFVEQYNAVRFDT 537
A+HL EL+ +AIL+ N + G A+ + + AF+ F +Q + V
Sbjct: 489 ALHLRELSERSSAILMVHKVRKNGM--PFWNRRGVNADADQVVVAFQAF-QQLSRVNVRP 545
Query: 538 SSVVSSYTTIHEDIYNVAEEKRASLILLPFHKEYSTVGGSPEISHNEHCEINENVLQHAP 597
+ +SS + IHEDI A K+A++++LPFHK + + GS E + ++ +N VL AP
Sbjct: 546 MTAISSMSDIHEDICTTAVRKKAAIVILPFHK-HQQLDGSLETTRGDYRWVNRRVLLQAP 604
Query: 598 CSVGIFVDRGLG-----SLLKTKMRIITLFIGGPDDREALSIAWRMAGHSGTQLHVVRIH 652
CSVGIFVDRGLG S ++ LF GGPDDREAL+ RMA H G L V R
Sbjct: 605 CSVGIFVDRGLGGSSQVSAQDVSYSVVVLFFGGPDDREALAYGLRMAEHPGIVLTVFRFV 664
Query: 653 LLGKAAEEKVLKKKISKSPHGMLSTVMDGVMQKELDEEYIFSFRHKAVNNNDSIVYLEKE 712
+ + E ++ ++S + + V + DEE + R K + ++S+ ++EK+
Sbjct: 665 VSPERVGE-IVNVEVSNN-----NNENQSVKNLKSDEEIMSEIR-KISSVDESVKFVEKQ 717
Query: 713 VHSDTGEEIPTLLNEIDKPGYDLYIVGQGSGKNKTIFLKLLEWCDHPELGVIGDILASTS 772
+ + ++ + + E+ + +L++VG+ G I L + E + PELG +G +L S
Sbjct: 718 I-ENAAVDVRSAIEEVRRS--NLFLVGRMPGGE--IALAIRENSECPELGPVGSLLISPE 772
Query: 773 FGTHSSVLIVQQY 785
T +SVL++QQY
Sbjct: 773 SSTKASVLVIQQY 785
>At3g17630 unknown protein
Length = 857
Score = 383 bits (983), Expect = e-106
Identities = 242/792 (30%), Positives = 422/792 (52%), Gaps = 39/792 (4%)
Query: 4 CYNVTLSASNNIWMTDDVMIKRVPLLCLQIAYNLLVSRFLFFILKPLHVPLIVAQMLTGF 63
C + SN + + + +PL+ LQI ++ +R L + LKPL P ++A+++ G
Sbjct: 67 CPGPMKATSNGAFQNESPLDFALPLIILQIVLVVVFTRLLAYFLKPLKQPRVIAEIIGGI 126
Query: 64 SVSRSLLGYFEGVITLFYNPRGILAVETISNLGIMYYVFLSGLEMNSDTILRSRKKGTSM 123
+ S LG + + + + + ++T++N+G+++++FL GLE++ I ++ KK +
Sbjct: 127 LLGPSALGRSKAYLDTIFPKKSLTVLDTLANIGLLFFLFLVGLELDFAAIKKTGKKSLLI 186
Query: 124 AIAGIVTSMLFGVGFLTLQQKLLDKKEKTHIKAYLFWCLTLSVTGFPVLARILAKLKLLY 183
AIAGI + GVG + + K + +F + LS+T FPVLARILA+LKLL
Sbjct: 187 AIAGISLPFIVGVGTSFVLSATISKGVD-QLPFIVFMGVALSITAFPVLARILAELKLLT 245
Query: 184 TKLGKDTLTAAMLTDAYGWVLFTLLIPAANNWGEGYYLSVITTFL---FIAFCFTVVRPI 240
T +G+ ++AA + D W+L L I + + G +SV F+ F ++P+
Sbjct: 246 TDIGRMAMSAAGVNDVAAWILLALAIALSGD-GTSPLVSVWVLLCGTGFVIFAVVAIKPL 304
Query: 241 LTPIIENRTNKNMWRKSHMLDVFIGLFICSYITDFLGTHPIVGAFVFGLILP-HGKFADM 299
L + ++ ++ + S++TD +G H + GAFV G++ P G F +
Sbjct: 305 LAYMARRCPEGEPVKELYVCVTLTVVLAASFVTDTIGIHALFGAFVVGIVAPKEGPFCRI 364
Query: 300 VMEMSDDFVTGILCPVYFSGFGFKLDLPVLWNTPNSVLMMLIMVLLCIPKVLSSLIVTFF 359
+ E +D V+G+L P+YF+ G K D+ + + L++L+++ C K++ ++ +
Sbjct: 365 LTEKIEDLVSGLLLPLYFAASGLKTDVTTIRGAQSWGLLVLVILTTCFGKIVGTVGSSML 424
Query: 360 FGMPARDGLSIGLLLNTKGVMAVILQNVAWDKRFLDPCSFMVMTLAIILMTVIVSPLINA 419
+P R+ +++G L+NTKG++ +I+ N+ D++ L+ +F ++ L + T I +P++
Sbjct: 425 CKVPFREAVTLGFLMNTKGLVELIVLNIGKDRKVLNDQAFAILVLMALFTTFITTPIVML 484
Query: 420 IYKPKFRFMQSQLRTVQKLRFDMELRVVACVHNAKHATNIIHVLEATNAT-RISPVHVSA 478
IYKP + + RT+Q+ D ELR++AC H+ ++ +I+++E++ T + + V A
Sbjct: 485 IYKPARKGAPYKHRTIQRKDHDSELRILACFHSTRNIPTLINLIESSRGTGKKGRLCVYA 544
Query: 479 VHLLELTRHGTAILVSQMADLNNIAGAETTNYGPKAEFEIITTAFKEFVEQYNAVRFDTS 538
+HL+EL+ +AI + A N G N ++ +++ AF+ + + AV
Sbjct: 545 MHLMELSERSSAIAMVHKARNN---GLPIWNKIERSTDQMV-IAFEAY-QHLRAVAVRPM 599
Query: 539 SVVSSYTTIHEDIYNVAEEKRASLILLPFHKEYSTVGGSPEISHNEHCEINENVLQHAPC 598
+ +S ++IHEDI A +KR ++ILLPFHK G I H H E+N+ VLQ APC
Sbjct: 600 TAISGLSSIHEDICTSAHQKRVAMILLPFHKHQRMDGAMESIGHRFH-EVNQRVLQRAPC 658
Query: 599 SVGIFVDRGLGS-----LLKTKMRIITLFIGGPDDREALSIAWRMAGHSGTQLHVVRIHL 653
SVGI VDRGLG + +++ F GG DDREAL+ +M H G L V
Sbjct: 659 SVGILVDRGLGGTSQVVASEVAYKVVIPFFGGLDDREALAYGMKMVEHPGITLTVY---- 714
Query: 654 LGKAAEEKVLKKKISKSPHGMLSTVMDGVMQKELDEEYIFSFRHKAVNNNDSIVYLEKEV 713
K + K+ KS H +KE DEE++ + N+S+ Y E+ V
Sbjct: 715 --KFVAARGTLKRFEKSEHDEKEK-----KEKETDEEFVRELMNDP-RGNESLAYEERVV 766
Query: 714 HSDTGEEIPTLLNEIDKPGYDLYIVGQGSGKNKTIFLKLLEWCDHPELGVIGDILASTSF 773
S ++I L + K +L++VG+ + L++ D PELG +G +L+S+ F
Sbjct: 767 ESK--DDIIATLKSMSK--CNLFVVGRNAA-----VASLVKSTDCPELGPVGRLLSSSEF 817
Query: 774 GTHSSVLIVQQY 785
T +SVL+VQ Y
Sbjct: 818 STTASVLVVQGY 829
>At4g23700 putative Na+/H+-exchanging protein
Length = 820
Score = 376 bits (965), Expect = e-104
Identities = 254/808 (31%), Positives = 436/808 (53%), Gaps = 36/808 (4%)
Query: 3 TCYNVTLSASNNIWMTDDVMIKRVPLLCLQIAYNLLVSRFLFFILKPLHVPLIVAQMLTG 62
TC + SN ++ ++ + +PLL LQI LL++R L F+L+PL P ++A+++ G
Sbjct: 7 TCPGPMKATSNGVFQGENPLEHALPLLILQICIVLLLTRLLAFLLRPLRQPRVIAEIVGG 66
Query: 63 FSVSRSLLGYFEGVITLFYNPRGILAVETISNLGIMYYVFLSGLEMNSDTILRSRKKGTS 122
+ S LG I + P+ + ++T++NLG+++++FL GLE++ ++ R+ K+ S
Sbjct: 67 ILLGPSALGKSTKFINTVFPPKSLTVLDTLANLGLIFFLFLVGLELDPKSLKRTGKRALS 126
Query: 123 MAIAGIVTSMLFGVG-FLTLQQKLLDKKEKTHIKAYLFWCLTLSVTGFPVLARILAKLKL 181
+A+AGI + G+G L+ + D K +F + LS+T FPVLARILA++KL
Sbjct: 127 IALAGITLPFVLGIGTSFALRSSIADGASKAPF--LVFMGVALSITAFPVLARILAEIKL 184
Query: 182 LYTKLGKDTLTAAMLTDAYGWVLFTLLIPAANNWGEGYY-LSVITTFL----FIAFCFTV 236
L T +GK L+AA + D W+L L + + GEG L+ + FL F+ FC V
Sbjct: 185 LTTDIGKIALSAAAVNDVAAWILLALAVALS---GEGSSPLTSLWVFLSGCGFVLFCIFV 241
Query: 237 VRPILTPIIENRTNKNMWRKSHMLDVFIGLFICSYITDFLGTHPIVGAFVFGLILP-HGK 295
V+P + I + + ++ + S++TDF+G H + GAFV G+I P G
Sbjct: 242 VQPGIKLIAKRCPEGEPVNELYVCCTLGIVLAASFVTDFIGIHALFGAFVIGVIFPKEGN 301
Query: 296 FADMVMEMSDDFVTGILCPVYFSGFGFKLDLPVLWNTPNSVLMMLIMVLLCIPKVLSSLI 355
FA+ ++E +D V+G+ P+YF G K ++ + + L++L++ C K++ +++
Sbjct: 302 FANALVEKVEDLVSGLFLPLYFVSSGLKTNVATIQGAQSWGLLVLVIFNACFGKIIGTVL 361
Query: 356 VTFFFGMPARDGLSIGLLLNTKGVMAVILQNVAWDKRFLDPCSFMVMTLAIILMTVIVSP 415
V+ + +P L++G L+NTKG++ +I+ N+ D+ L+ F +M L I T + +P
Sbjct: 362 VSLYCKVPLDQSLALGFLMNTKGLVELIVLNIGKDRGVLNDQIFAIMVLMAIFTTFMTTP 421
Query: 416 LINAIYKPKFRFMQSQL--RTVQKL-RFDMELRVVACVHNAKHATNIIHVLEATNA-TRI 471
L+ A+YKP ++ RTV++ R + L ++ C + + I++++EA+ R
Sbjct: 422 LVLAVYKPGKSLTKADYKNRTVEETNRSNKPLCLMFCFQSIMNIPTIVNLIEASRGINRK 481
Query: 472 SPVHVSAVHLLELTRHGTAILVSQMADLNNIA-GAETTNYGPKAEFEIITTAFKEFVEQY 530
+ V A+HL+EL+ +AIL++ N + + + + +++ AF+ F +
Sbjct: 482 ENLSVYAMHLMELSERSSAILMAHKVRRNGLPFWNKDKSENNSSSSDMVVVAFEAF-RRL 540
Query: 531 NAVRFDTSSVVSSYTTIHEDIYNVAEEKRASLILLPFHKEYSTVGGSPEISHNEHCEINE 590
+ V + +S TIHEDI AE K+ ++++LPFHK + + + E + N++ IN+
Sbjct: 541 SRVSVRPMTAISPMATIHEDICQSAERKKTAMVILPFHK-HVRLDRTWETTRNDYRWINK 599
Query: 591 NVLQHAPCSVGIFVDRGLGSLLKT-----KMRIITLFIGGPDDREALSIAWRMAGHSGTQ 645
V++ +PCSV I VDRGLG + + I LF GG DDREAL+ A RMA H G
Sbjct: 600 KVMEESPCSVAILVDRGLGGTTRVASSDFSLTITVLFFGGNDDREALAFAVRMAEHPGIS 659
Query: 646 LHVVRIHLLGKAAEEKVLKKKISKSPHGMLSTVMDGVMQKELDEEYIFSFRHKAVNNNDS 705
L VVR + E V + + ++D EL + + + +N+DS
Sbjct: 660 LTVVRFIPSDEFKPENVRIEITEDQLCSGATRLIDIEAITELKAK--IKEKESSRSNSDS 717
Query: 706 ---IVYLEKEVHSDTGEEIPTLLNEIDKPGYDLYIVGQGSGKNKTIFLKLLEWCDHPELG 762
I+Y EK V EE+ ++ E K +L++VG+ + + + D PELG
Sbjct: 718 ESHIIYEEKIV--KCYEEVIEVIKEYSKS--NLFLVGKSPEGSVASGINVRS--DTPELG 771
Query: 763 VIGDILA-STSFGTHSSVLIVQQYMVGR 789
IG++L S S T +SVL+VQQY+ R
Sbjct: 772 PIGNLLTESESVSTVASVLVVQQYIASR 799
>At1g64170 hypothetical protein
Length = 847
Score = 318 bits (814), Expect = 1e-86
Identities = 227/837 (27%), Positives = 409/837 (48%), Gaps = 92/837 (10%)
Query: 10 SASNNIWMTDDVMIKRVPLLCLQIAYNLLVSRFLFFILKPLHVPLIVAQMLT-------- 61
+ SN ++ + + PL+ LQI + V+R L F+L+P+ P +VA+++
Sbjct: 23 TTSNGVFDGESPLDFAFPLVILQICLVVAVTRSLAFLLRPMRQPRVVAEIIVSPPSTGLG 82
Query: 62 ----------------------------GFSVSRSLLGYFEGVITLFYNPRGILAVETIS 93
G + S LG + R + ++T++
Sbjct: 83 QSYSFRFNKYPTRLKYELYRDSVQLITGGILLGPSALGRITSYKNSIFPARSLTVLDTLA 142
Query: 94 NLGIMYYVFLSGLEMNSDTILRSRKKGTSMAIAGIVTSMLFGVGFLT---LQQKLLDKKE 150
NLG++ ++FL GLE++ ++ R+ KK S+A AG++ + FG+G +T +
Sbjct: 143 NLGLLLFLFLVGLEIDLTSLRRTGKKAISIAAAGML--LPFGMGIVTSFAFPEASSSGDN 200
Query: 151 KTHIKAYLFWCLTLSVTGFPVLARILAKLKLLYTKLGKDTLTAAMLTDAYGWVLFTLLIP 210
+ +F + LS+T F VLARILA+LKLL T LG+ ++ AA + D WVL L +
Sbjct: 201 SKVLPFIIFMGVALSITAFGVLARILAELKLLTTDLGRISMNAAAINDVAAWVLLALAVS 260
Query: 211 AANNWGEGYY--LSVITTFLFIAFCFTVVRPILTPIIENRTNKNMWRKSHMLDVFIGLFI 268
+ + +++ F+ CF +V I I + ++ + +
Sbjct: 261 LSGDRNSPLVPLWVLLSGIAFVIACFLIVPRIFKFISRRCPEGEPIGEMYVCVALCAVLL 320
Query: 269 CSYITDFLGTHPIVGAFVFGLILPHGKFADMVMEMSDDFVTGILCPVYFSGFGFKLDLPV 328
+ TD +G H I GAFV G++ P G F+D ++E +D V G+L P+YF G K D+
Sbjct: 321 AGFATDAIGIHAIFGAFVMGVLFPKGHFSDAIVEKIEDLVMGLLLPLYFVMSGLKTDITT 380
Query: 329 LWNTPNSVLMMLIMVLLCIPKVLSSLIVTFFFGMPARDGLSIGLLLNTKGVMAVILQNVA 388
+ + + L++V C K++ ++ V + R+ + +G+L+NTKG++ +I+ N+
Sbjct: 381 IQGVKSWGRLALVIVTACFGKIVGTVSVALLCKVRLRESVVLGVLMNTKGLVELIVLNIG 440
Query: 389 WDKRFLDPCSFMVMTLAIILMTVIVSPLINAIYKP------------KFRFMQSQLRTVQ 436
D++ L +F +M L I T I +P++ A+YKP K R + ++ +
Sbjct: 441 KDRKVLSDQTFAIMVLMAIFTTFITTPIVLALYKPSETTQTHSSVSYKNRKHRRKIENDE 500
Query: 437 KLRFDMELRVVACVHNAKHATNIIHVLEATNATRISPVH--VSAVHLLELTRHGTAILVS 494
+ +L+V+ C+ ++K ++ ++EAT + + V +HL +L+ ++I +
Sbjct: 501 EGEKMQQLKVLVCLQSSKDIDPMMKIMEATRGSNETKERFCVYVMHLTQLSERPSSIRMV 560
Query: 495 QMADLNNIAGAETTNYGPKAEFEIITTAFKEFVEQYNAVRFDTSSVVSSYTTIHEDIYNV 554
Q N + + K E T E + ++V + + +S +TIHEDI +
Sbjct: 561 QKVRSNGL-----PFWNKKRENSSAVTVAFEASSKLSSVSVRSVTAISPLSTIHEDICSS 615
Query: 555 AEEKRASLILLPFHKEYSTVGGSPEISHNEHCEINENVLQHAPCSVGIFVDRGLG----- 609
A+ K + ++LPFHK++ ++ E +E+ IN+ VL+++PCSVGI VDRGLG
Sbjct: 616 ADSKCTAFVILPFHKQWRSLEKEFETVRSEYQGINKRVLENSPCSVGILVDRGLGDNNSP 675
Query: 610 -SLLKTKMRIITLFIGGPDDREALSIAWRMAGHSGTQLHVVRIHLLGKAAEEKVLKKKIS 668
+ + + LF GG DDREAL RMA H G L VV I A +++ ++ S
Sbjct: 676 VASSNFSLSVNVLFFGGCDDREALVYGLRMAEHPGVNLTVVVISGPESARFDRLEAQETS 735
Query: 669 KSPHGMLSTVMDGVMQKELDEEYIFSFRHKAVNNNDSIVYLEKEVHSDTGEEIPTLLNEI 728
LDE+++ + + +A ++ + E+ V+S EE+ ++ +
Sbjct: 736 LC---------------SLDEQFLAAIKKRA----NAARFEERTVNST--EEVVEIIRQF 774
Query: 729 DKPGYDLYIVGQGSGKNKTIFLKLLEWCDHPELGVIGDILASTSFGTHSSVLIVQQY 785
+ D+ +VG+ S L +++ + PELG +G+++ S T SVL+VQQY
Sbjct: 775 YE--CDILLVGKSSKGPMVSRLPVMK-IECPELGPVGNLIVSNEISTSVSVLVVQQY 828
>At1g06970 hypothetical protein
Length = 829
Score = 273 bits (697), Expect = 4e-73
Identities = 212/800 (26%), Positives = 394/800 (48%), Gaps = 62/800 (7%)
Query: 4 CYNVTLSASNNIWMTDDVMIKRVPLLCLQIAYNLLVSRFLFFILKPLHVPLIVAQMLTGF 63
C + S +++ D + +PL+ LQ++ ++ SR L+ +LKPL +I AQ+L G
Sbjct: 28 CQKNHMLTSKGVFLGSDPLKYAMPLMLLQMSVIIITSRLLYRLLKPLKQGMISAQVLAGI 87
Query: 64 SVSRSLLGYFEGVITLFYNPRGILAVETISNLGIMYYVFLSGLEMNSDTILRSRKKGTSM 123
+ SL G + +F G + ++T+SNLG ++FL GL +++ I RK G+
Sbjct: 88 ILGPSLFGQSSAYMQMFLPISGKITLQTLSNLGFFIHLFLLGLRIDASII---RKAGSKA 144
Query: 124 AIAGIVTSML-FGVGFLTLQQKLLDKKEKTHIKAYLFWCLTL-----SVTGFPVLARILA 177
+ G + L F +G LT +L K ++ + C++ ++T FPV +LA
Sbjct: 145 ILIGTASYALPFSLGNLT----VLFLKNTYNLPPDVVHCISTVISLNAMTSFPVTTTVLA 200
Query: 178 KLKLLYTKLGKDTLTAAMLTDAYGWVLFTLLIPAANNWGEGYYLSVITTFLFIAFCFTVV 237
+L +L + LG+ +++ +A+ W++ + + S + I F V
Sbjct: 201 ELNILNSDLGRLATNCSIVCEAFSWIVALVFRMFLRDGTLASVWSFVWVTALILVIFFVC 260
Query: 238 RPILTPIIENRT-NKNMWRKSHMLDVFIGLFICSYITDFLGTHPIVGAFVFGLILPHG-K 295
RP + + E R+ + + + + + L S ++ LG H GAF G+ LP G
Sbjct: 261 RPAIIWLTERRSISIDKAGEIPFFPIIMVLLTISLTSEVLGVHAAFGAFWLGVSLPDGPP 320
Query: 296 FADMVMEMSDDFVTGILCPVYFSGFGFKLDLPVLWNTPNSVLMMLIMVLL-CIPKVLSSL 354
+ + F T ++ P + S G + + ++ + ++ +I++ C K L +
Sbjct: 321 LGTGLTTKLEMFATSLMLPCFISISGLQTNFFIIGESHVKIIEAVILITYGC--KFLGTA 378
Query: 355 IVTFFFGMPARDGLSIGLLLNTKGVMAVILQNVAWDKRFLDPCSFMVMTLAIILMTVIVS 414
+ + + D S+ LL+ +GV+ + + D++ L+ F ++ + ++L+T I
Sbjct: 379 AASAYCNIQIGDAFSLALLMCCQGVIEIYTCVMWKDEKVLNTECFNLLIITLLLVTGISR 438
Query: 415 PLINAIYKPKFRFMQSQLRTVQKLR-FDMELRVVACVHNAKHATNIIHVLEATNATRISP 473
L+ +Y P R+ RT+ R +++ R++ CV+N ++ +++++LEA+ +R SP
Sbjct: 439 FLVVCLYDPSKRYRSKSKRTILDTRQRNLQFRLLLCVYNVENVPSMVNLLEASYPSRFSP 498
Query: 474 VHVSAVHLLELTRHGTAILV--SQMADLNNIAGAETTNYGPKAEFEIITTAFKEFVEQ-Y 530
+ V +HL+EL A+LV QM L+ T I F+ F +Q
Sbjct: 499 ISVFTLHLVELKGRAHAVLVPHHQMNKLDPNTVQSTH----------IVNGFQRFEQQNQ 548
Query: 531 NAVRFDTSSVVSSYTTIHEDIYNVAEEKRASLILLPFHKEYSTVGGSPEISHNEHCEINE 590
+ + + +++I++DI +A +K+A+LI++PFHK+Y+ + G+ + + IN
Sbjct: 549 GTLMAQHFTAAAPFSSINDDICTLALDKKATLIVIPFHKQYA-IDGTVDHVNPSIRNINL 607
Query: 591 NVLQHAPCSVGIFVDRGLGS------LLKTKMRIITLFIGGPDDREALSIAWRMAGHSGT 644
NVL+ APCSVGIF+DRG + T + +FI G DD EAL+ + R+A H
Sbjct: 608 NVLEKAPCSVGIFIDRGETEGRRSVLMSYTWRNVAVIFIEGRDDAEALAFSMRIAEH--P 665
Query: 645 QLHVVRIHLLGKAAEEKVLKKKISKSPHGMLSTVMDGVMQKELDEEYIFSFRHKAVNNND 704
++ V IH K++ ++ + V+D ++ EL E Y+ + +
Sbjct: 666 EVSVTMIHFRHKSSLQQ--------------NHVVD--VESELAESYLINDFKNFAMSKP 709
Query: 705 SIVYLEKEVHSDTGEEIPTLLNEIDKPGYDLYIVGQGSGKNKTIFLKLLEWCDHPELGVI 764
I Y E+ V G E +++ + +DL +VG+ ++ L +W + PELGVI
Sbjct: 710 KISYREEIVRD--GVETTQVISSLG-DSFDLVVVGRDHDLESSVLYGLTDWSECPELGVI 766
Query: 765 GDILASTSFGTHSSVLIVQQ 784
GD+ AS+ F H SVL++ Q
Sbjct: 767 GDMFASSDF--HFSVLVIHQ 784
>At2g30240 putative Na/H antiporter
Length = 831
Score = 263 bits (671), Expect = 4e-70
Identities = 222/800 (27%), Positives = 384/800 (47%), Gaps = 63/800 (7%)
Query: 4 CYNVTLSASNNIWMTDDVMIKRVPLLCLQIAYNLLVSRFLFFILKPLHVPLIVAQMLTGF 63
C + S I+M + + +PLL LQ++ ++ SR +F +L+PL +I AQ+LTG
Sbjct: 30 CQAQNMLTSRGIFMKSNPLKYALPLLLLQMSVIIVTSRLIFRVLQPLKQGMISAQVLTGV 89
Query: 64 SVSRSLLGYFEGVITLFYNPRGILAVETISNLGIMYYVFLSGLEMNSDTILRSRKKGTSM 123
+ S LG+ + +F G + ++T+SN+G + ++FL GL+++ I RK G+
Sbjct: 90 VLGPSFLGHNVIYMNMFLPAGGKIIIQTLSNVGFVIHLFLLGLKIDGSII---RKAGSKA 146
Query: 124 AIAGIVT-SMLFGVGFLTLQQKLLDKKEKTHIKAYLFWCLTL-SVTGFPVLARILAKLKL 181
+ G + + F +G LT+ + + + ++L S+T FPV +LA+L +
Sbjct: 147 ILIGTASYAFPFSLGNLTIMFISKTMGLPSDVISCTSSAISLSSMTSFPVTTTVLAELNI 206
Query: 182 LYTKLGKDTLTAAMLTDAYGW--VLFTLLIPAANNWGEGYYLSVITTFLFIAFCFTVVRP 239
L ++LG+ +M+ + W L L Y LS+I L + + V RP
Sbjct: 207 LNSELGRLATHCSMVCEVCSWFVALAFNLYTRDRTMTSLYALSMIIGLLLV--IYFVFRP 264
Query: 240 ILTPIIENRTNKNMWRKS--HMLDVFIGLFICSYITDFLGTHPIVGAFVFGLILPHG-KF 296
I+ + + +T K+M +K V + L I S + +G H GAF G+ LP G
Sbjct: 265 IIVWLTQRKT-KSMDKKDVVPFFPVLLLLSIASLSGEAMGVHAAFGAFWLGVSLPDGPPL 323
Query: 297 ADMVMEMSDDFVTGILCPVYFSGFGFKLDLPVLWNTPNSVLMMLIMVLLCI--PKVLSSL 354
+ + F + + P + + G + + + + ++M+ ++LL K L +
Sbjct: 324 GTELAAKLEMFASNLFLPCFIAISGLQTNFFEITESHEHHVVMIEIILLITYGCKFLGTA 383
Query: 355 IVTFFFGMPARDGLSIGLLLNTKGVMAVILQNVAWDKRFLDPCSFMVMTLAIILMTVIVS 414
+ + D L + L+ +G++ V V D + +D F ++ + I+ +T I
Sbjct: 384 AASAYCQTQIGDALCLAFLMCCQGIIEVYTTIVWKDAQVVDTECFNLVIITILFVTGISR 443
Query: 415 PLINAIYKPKFRFMQSQLRTVQKLR-FDMELRVVACVHNAKHATNIIHVLEATNATRISP 473
L+ +Y P R+ RT+ R +++LR++ ++N ++ +++++LEAT TR +P
Sbjct: 444 FLVVYLYDPSKRYKSKSKRTILNTRQHNLQLRLLLGLYNVENVPSMVNLLEATYPTRFNP 503
Query: 474 VHVSAVHLLELTRHGTAILV--SQMADLNNIAGAETTNYGPKAEFEIITTAFKEFVEQY- 530
+ +HL+EL A+L QM L+ T I AF+ F ++Y
Sbjct: 504 ISFFTLHLVELKGRAHALLTPHHQMNKLDPNTAQSTH----------IVNAFQRFEQKYQ 553
Query: 531 NAVRFDTSSVVSSYTTIHEDIYNVAEEKRASLILLPFHKEYSTVGGSPEISHNEHCEINE 590
A+ + + Y++I+ DI +A +K+A+LI++PFHK+Y+ G ++ + IN
Sbjct: 554 GALMAQHFTAAAPYSSINNDICTLALDKKATLIVIPFHKQYAIDGTVGQV-NGPIRTINL 612
Query: 591 NVLQHAPCSVGIFVDRGLGS------LLKTKMRIITLFIGGPDDREALSIAWRMAGHSGT 644
NVL APCSV IF+DRG + T + LFIGG DD EAL++ RMA
Sbjct: 613 NVLDAAPCSVAIFIDRGETEGRRSVLMTNTWQNVAMLFIGGKDDAEALALCMRMA--EKP 670
Query: 645 QLHVVRIHLLGKAAEEKVLKKKISKSPHGMLSTVMDGVMQKELDEEYIFSFRHKAVNNND 704
L+V IH K+A + D + I F+ A N
Sbjct: 671 DLNVTMIHFRHKSA-------------------LQDEDYSDMSEYNLISDFKSYAANKG- 710
Query: 705 SIVYLEKEVHSDTGEEIPTLLNEIDKPGYDLYIVGQGSGKNKTIFLKLLEWCDHPELGVI 764
I Y+E+ V G E +++ + YD+ +VG+ ++ L +W + PELGVI
Sbjct: 711 KIHYVEEIVRD--GVETTQVISSLG-DAYDMVLVGRDHDLESSVLYGLTDWSECPELGVI 767
Query: 765 GDILASTSFGTHSSVLIVQQ 784
GD+L S F H SVL+V Q
Sbjct: 768 GDMLTSPDF--HFSVLVVHQ 785
>At5g37060 putative transporter protein
Length = 859
Score = 256 bits (654), Expect = 4e-68
Identities = 212/838 (25%), Positives = 380/838 (45%), Gaps = 96/838 (11%)
Query: 17 MTDDVMIKRVPLLCLQI-------------AYNLLVSRFL-------FFI------LKPL 50
+++ V ++P++C ++ A N S FL FFI L+P
Sbjct: 30 VSNRVFSPKIPVVCRKLHSKQPFGMFKGENAMNYAFSTFLIEAIIIIFFIKVVSIALRPF 89
Query: 51 HVPLIVAQMLTGFSVSRSLLGYFEGVITLFYNPRGILAVETISNLGIMYYVFLSGLEMNS 110
P IV++++ G + S+ G + P I +G Y++FL+ + +
Sbjct: 90 RQPRIVSEIIGGMMIGPSMFGGIRNFNYYLFPPIANYICANIGLMGFFYFLFLTAAKTDV 149
Query: 111 DTILRSRKKGTSMAIAGIVTSMLF--GVGFLTLQQKLLDKKEKTHIKAYLFWCLTLSVTG 168
I ++ +K +A G++ ++ VG Q + ++ + I +F LS T
Sbjct: 150 GAIGKAPRKHKYIAAIGVIVPIICVGSVGMAMRDQMDENLQKPSSIGGVVF---ALSFTS 206
Query: 169 FPVLARILAKLKLLYTKLGKDTLTAAMLTD---AYGWVLFTLLIPAANNWGEGYYLSVIT 225
FPV+ +L + LL +++GK ++ A+L D Y V+F + A + +++
Sbjct: 207 FPVIYTVLRDMNLLNSEVGKFAMSVALLGDMAGVYVIVIFEAMTHADVGGAYSVFWFLVS 266
Query: 226 TFLFIAFCFTVVRPILTPIIENRTNKNMWRKSHMLDVFIGLFICSYITDFLGTHPIVGAF 285
+F AF VVR I+ + +++++ + +G+ ++TD G VG
Sbjct: 267 VVIFAAFMLLVVRRAFDWIVSQTPEGTLVNQNYIVMILMGVLASCFLTDMFGLSIAVGPI 326
Query: 286 VFGLILPHGK-FADMVMEMSDDFVTGILCPVYFSGFGFKLDLPVL----WNTPNSVLMML 340
GL++PHG + S+ F+ L P ++ G ++ L W S L +
Sbjct: 327 WLGLLVPHGPPLGSTLAVRSETFIYEFLMPFTYALVGQGTNIHFLRDETWRNQLSPLFYM 386
Query: 341 IMVLLCIPKVLSSLIVTFFFGMPARDGLSIGLLLNTKGVMAVILQNVAWDKRFLDPCSFM 400
+V I K LS+ FF +PAR+ +++GL++N +G M +++ DKR + +
Sbjct: 387 TVVGF-ITKFLSTAFAALFFKVPARESITLGLMMNLRGQMDLLVYLHWIDKRIVGFPGYT 445
Query: 401 VMTLAIILMTVIVSPLINAIYKPKFRFMQSQLRTVQKLRFDMELRVVACVHNAKHATNII 460
VM L +++T + +PLIN Y P + S+ RT+Q + E+ +V V + + + +I
Sbjct: 446 VMVLHTVVVTAVTTPLINFFYDPTRPYRSSKHRTIQHTPQNTEMGLVLAVSDHETLSGLI 505
Query: 461 HVLEATNATRISPVHVSAVHLLELTRHGTAILVSQMADLNNIAGAETTNYGPKAE----- 515
L+ T+ SP+ + AV L+EL T + + D E Y + E
Sbjct: 506 TFLDFAYPTKSSPLSIFAVQLVELAGRATPLFI----DHEQRKEEEEEEYEEEEEEPERK 561
Query: 516 ----FEIITTAFKEFVEQYN-AVRFDTSSVVSSYTTIHEDIYNVAEEKRASLILLPFHKE 570
+ + +AFK + E+ N V + + + +++DI +A K+ + ILLP+ KE
Sbjct: 562 QSGRIDQVQSAFKLYEEKRNECVTLRSYTAHAPKRLMYQDICELALGKKTAFILLPYQKE 621
Query: 571 YSTVGGSPEISHNEHCEINENVLQHAPCSVGIFVDRG--LGSLLKTKM------------ 616
E+ + +N +VL+H PCSV I+ D+G ++++ M
Sbjct: 622 RLEDAAPTELRDSGMLSVNADVLEHTPCSVCIYFDKGRLKNAVVRLSMDLQHSTNSIRMR 681
Query: 617 ----RIITLFIGGPDDREALSIAWRMAGHSGTQLHVVRIHLLGKAAEEKVLKKKISKSPH 672
R + LF+GG D+REAL +A RM+ + L V+R E++
Sbjct: 682 QETYRFVVLFLGGADNREALHLADRMSTNPDVTLTVIRFLSYNHEGEDE----------- 730
Query: 673 GMLSTVMDGVMQKELDEEYIFSFRHKAVNNNDSIVYLEKEVHSDTGEEIPTLLNEIDKPG 732
+K+LD+ + F K +N+ + Y KEV G E + ++
Sbjct: 731 ----------REKKLDDGVVTWFWVKN-ESNERVSY--KEVVVKNGAETLAAIQAMNVND 777
Query: 733 YDLYIVGQGSGKNKTIFLKLLEWCDHPELGVIGDILASTSFGTHSSVLIVQQYMVGRK 790
YDL+I G+ G N I L W + +LGVIGD +A++ F + SVL+VQQ + +K
Sbjct: 778 YDLWITGRREGINPKILEGLSTWSEDHQLGVIGDTVAASVFASEGSVLVVQQQVRNQK 835
>At5g22900 Na+/H+ antiporter-like protein
Length = 822
Score = 253 bits (646), Expect = 3e-67
Identities = 201/782 (25%), Positives = 367/782 (46%), Gaps = 72/782 (9%)
Query: 41 RFLFFILKPLHVPLIVAQMLTGFSVSRSLLGYFEGVITLFYNPRGI--LAVETISNLGIM 98
+FL F L+ L + + MLTG +S+S L F++ + + M
Sbjct: 70 QFLHFFLRRLGMIRFTSHMLTGVLLSKSFLKE-NSAARRFFSTEDYKEIVFSLTAACSYM 128
Query: 99 YYVFLSGLEMNSDTILRSRKKGTSMAIAGIVTSMLFGVGFLTLQQKLLDKKEKTHIKAYL 158
+ FL G++M++ I + +K ++ ++ ++ S L + + K H L
Sbjct: 129 MFWFLMGVKMDTGLIRTTGRKAITIGLSSVLLSTLVCSVIFFGNLRDVGTKNSDHTLNSL 188
Query: 159 FWCLTLSV---TGFPVLARILAKLKLLYTKLGKDTLTAAMLTDAYGWVLFTLLI------ 209
+ + S+ + FPV+ +L +L+L ++LG+ +++A+++D +L ++LI
Sbjct: 189 EYVVIYSIQCLSSFPVVGNLLFELRLQNSELGRLAISSAVISDFSTSILASVLIFMKELK 248
Query: 210 PAANNWGEGYYLSVIT----------TFLFIAFCFTVVRPILTPIIENRTNKNMWRKSHM 259
G + VI LF+ V RP++ II+ + + ++
Sbjct: 249 DEQTRLGSVFIGDVIAGNRPLMRAGIVVLFVCIAIYVFRPLMFYIIKQTPSGRPVKAIYL 308
Query: 260 LDVFIGLFICSYITDFLGTHPIVGAFVFGLILPHGK-FADMVMEMSDDFVTGILCPVYFS 318
+ + + + + ++ +G F+ GL +PHG +++ + + G P + +
Sbjct: 309 STIIVMVSGSAILANWCKQSIFMGPFILGLAVPHGPPLGSAIIQKYESAIFGTFLPFFIA 368
Query: 319 GFGFKLDLPVLWNTPNSVLMMLIMVLLCIPKVLSSLIVTFFFGMPARDGLSIGLLLNTKG 378
++D+ L+ ++LIMV + K + + + F+GMP D ++ L+++ KG
Sbjct: 369 SSSTEIDISALFGWEGLNGIILIMVTSFVVKFIFTTVPALFYGMPMEDCFALSLIMSFKG 428
Query: 379 VMAVILQNVAWDKRFLDPCSFMVMTLAIILMTVIVSPLINAIYKPKFRFMQSQLRTVQKL 438
+ + +A+ + + P +F V L I L + I+ P++ +Y P + + R +Q L
Sbjct: 429 IFELGAYALAYQRGSVRPETFTVACLYITLNSAIIPPILRYLYDPSRMYAGYEKRNMQHL 488
Query: 439 RFDMELRVVACVHNAKHATNIIHVLEATNATRISPVHVSAVHLLELTRHGTAILVSQMAD 498
+ + ELR+++C++ + +I++LEA +R SPV +HL+EL I +S
Sbjct: 489 KPNSELRILSCIYRTDDISPMINLLEAICPSRESPVATYVLHLMELVGQANPIFISHKLQ 548
Query: 499 LNNIAGAETTNYGPKAEFEIITTAFKEFVEQ-YNAVRFDTSSVVSSYTTIHEDIYNVAEE 557
E T+Y + +F++F + Y +V T + +S T+H DI +A
Sbjct: 549 TRR---TEETSYSNN-----VLVSFEKFRKDFYGSVFVSTYTALSMPDTMHGDICMLALN 600
Query: 558 KRASLILLPFHKEYSTVGGSPEISHNEHCEINENVLQHAPCSVGIFVDR------GLGSL 611
SLILLPFH+ +S G + ++N +N++VL APCSVG+FV R + S
Sbjct: 601 NTTSLILLPFHQTWSADGSALISNNNMIRNLNKSVLDVAPCSVGVFVYRSSSGRKNISSG 660
Query: 612 LKT---------KMRIITLFIGGPDDREALSIAWRMAGHSGTQLHVVRIHLLGKAAEEKV 662
KT I +F+GG DDREA+++A RMA + +VR+ + A E
Sbjct: 661 RKTINGTVPNLSSYNICMIFLGGKDDREAVTLATRMARDPRINITIVRLITTDEKARE-- 718
Query: 663 LKKKISKSPHGMLSTVMDGVMQKELDEEYIFSFRHKAVNNNDSIVYLEKEVHSDTGEEIP 722
+TV D K LD+E + R N I Y EK + + E
Sbjct: 719 -------------NTVWD----KMLDDELL---RDVKSNTLVDIFYSEKAI--EDAAETS 756
Query: 723 TLLNEIDKPGYDLYIVGQGSGKNKTIFLKLLEWCDHPELGVIGDILASTSFGTHSSVLIV 782
+LL + +D++IVG+G+G+ L EW + ELG+IGD+L S F +SVL++
Sbjct: 757 SLLRSM-VSDFDMFIVGRGNGRTSVFTEGLEEWSEFKELGIIGDLLTSQDFNCQASVLVI 815
Query: 783 QQ 784
QQ
Sbjct: 816 QQ 817
>At3g44900 putative protein
Length = 817
Score = 251 bits (641), Expect = 1e-66
Identities = 204/787 (25%), Positives = 374/787 (46%), Gaps = 86/787 (10%)
Query: 41 RFLFFILKPLHVPLIVAQMLTGFSVSRSLLGYFEGVITLFYNPRGILAVET--------I 92
+F F L+ L + + MLTG +S+S L R L+ E +
Sbjct: 69 QFFHFFLRRLGMIRFTSHMLTGILLSKSFLKENTPA-------RKFLSTEDYKETLFGLV 121
Query: 93 SNLGIMYYVFLSGLEMNSDTILRSRKKGTSMAIAGIVTSMLFG--VGFLTLQQKLLDKKE 150
M + FL G++M+ I + +K ++ ++ ++ S+ + FL L+ K E
Sbjct: 122 GACSYMMFWFLMGVKMDLSLIRSTGRKAVAIGLSSVLLSITVCALIFFLILRDVGTKKGE 181
Query: 151 KTH-----IKAYLFWCLTLSVTGFPVLARILAKLKLLYTKLGKDTLTAAMLTDAYGWVLF 205
I YL CL+ FPV+ +L +L+L ++LG+ +++A+++D +L
Sbjct: 182 PVMSFFEIIFIYLIQCLS----SFPVIGNLLFELRLQNSELGRLAMSSAVISDFSTSILS 237
Query: 206 TLLI------PAANNWGEGYYLSVI----------TTFLFIAFCFTVVRPILTPIIENRT 249
+L+ + G + VI T LF+ F + RP++ II+
Sbjct: 238 AVLVFLKELKDDKSRLGSVFIGDVIVGNRPMKRAGTVVLFVCFAIYIFRPLMFFIIKRTP 297
Query: 250 NKNMWRKSHMLDVFIGLFICSYITDFLGTHPIVGAFVFGLILPHGK-FADMVMEMSDDFV 308
+ +K ++ + I +F + + D+ +G F+ GL +PHG +++ + V
Sbjct: 298 SGRPVKKFYIYAIIILVFGSAILADWCKQSIFIGPFILGLAVPHGPPLGSAILQKFESVV 357
Query: 309 TGILCPVYFSGFGFKLDLPVLWNTPNSVLMMLIMVLLCIPKVLSSLIVTFFFGMPARDGL 368
G P + + ++D +L + + +++++ + I K + + F +GMPA+D +
Sbjct: 358 FGTFLPFFVATSAEEIDTSILQSWIDLKSIVILVSVSFIVKFALTTLPAFLYGMPAKDCI 417
Query: 369 SIGLLLNTKGVMAVILQNVAWDKRFLDPCSFMVMTLAIILMTVIVSPLINAIYKPKFRFM 428
++ L+++ KG+ A+ + + P +F V++L I+L + ++ PL+ IY P +
Sbjct: 418 ALSLIMSFKGIFEFGAYGYAYQRGTIRPVTFTVLSLYILLNSAVIPPLLKRIYDPSRMYA 477
Query: 429 QSQLRTVQKLRFDMELRVVACVHNAKHATNIIHVLEATNATRISPVHVSAVHLLELTRHG 488
+ R + ++ + ELR+++C++ +I++LEAT +R +PV +HL+EL
Sbjct: 478 GYEKRNMLHMKPNSELRILSCIYKTDDIRPMINLLEATCPSRENPVATYVLHLMELVGQA 537
Query: 489 TAILVSQMADLNNIAGAETTNYGPKAEFEIITTAFKEFVEQ-YNAVRFDTSSVVSSYTTI 547
+L+S +E +Y E + +F++F + +V T + +S +
Sbjct: 538 NPVLISHRLQTRK---SENMSYNS----ENVVVSFEQFHNDFFGSVFVSTYTALSVPKMM 590
Query: 548 HEDIYNVAEEKRASLILLPFHKEYSTVGGSPEISHNEHCEINENVLQHAPCSVGIFVDR- 606
H DI +A SLI+LPFH+ +S G + ++N++VL +PCSVGIFV R
Sbjct: 591 HGDICMLALNNTTSLIILPFHQTWSADGSAIVSDSLMIRQLNKSVLDLSPCSVGIFVYRS 650
Query: 607 --GLGSLLKT-----KMRIITLFIGGPDDREALSIAWRMAGHSGTQLHVVRIHLLGKAAE 659
G ++ +T ++ LF+GG DDREALS+A RMA S + VV +
Sbjct: 651 SNGRRTIKETAANFSSYQVCMLFLGGKDDREALSLAKRMARDSRITITVVSL-------- 702
Query: 660 EKVLKKKISKSPHGMLSTVMDGVMQKELDEEYIFSFRHKAVNNNDSIVYLEKEVH--SDT 717
IS +T D + LD E + + + D IV+ E+ V+ + T
Sbjct: 703 -------ISSEQRANQATDWD----RMLDLELLRDVKSNVLAGAD-IVFSEEVVNDANQT 750
Query: 718 GEEIPTLLNEIDKPGYDLYIVGQGSGKNKTIFLKLLEWCDHPELGVIGDILASTSFGTHS 777
+ + ++ NE YDL+IVG+ G+ L EW + ELG+IGD+L S +
Sbjct: 751 SQLLKSIANE-----YDLFIVGREKGRKSVFTEGLEEWSEFEELGIIGDLLTSQDLNCQA 805
Query: 778 SVLIVQQ 784
SVL++QQ
Sbjct: 806 SVLVIQQ 812
>At1g08150 hypothetical protein
Length = 815
Score = 249 bits (636), Expect = 4e-66
Identities = 205/783 (26%), Positives = 365/783 (46%), Gaps = 63/783 (8%)
Query: 26 VPLLCLQIAYNLLVSRFLFFILKP--LHVPLIVAQMLTGFSVSRSLLGYFEGVIT--LFY 81
+P L I L + +F + LK L VP I + M+ G ++S++ L + I LF
Sbjct: 65 LPHLESVIVLVLCLWQFFYLSLKKIGLPVPKITSMMIAGAALSQTNLLPNDWTIQHILFP 124
Query: 82 NPRGILAVETISNLGIMYYVFLSGLEMNSDTILRSRKKGTSMAIAGIVTSMLFGVGFLTL 141
+ ET+ ++Y F+ G++M+ + RK GT + + GI T +L + +
Sbjct: 125 DDTRPKVPETLGGFAFVFYWFIEGVKMDVGMV---RKTGTKVIVTGIATVILPIIAANMV 181
Query: 142 QQKLLDKKEK----THIKAYLFWCLTLSVTGFPVLARILAKLKLLYTKLGKDTLTAAMLT 197
KL + K + LF S++ F ++R+L L++ +++ G+ ++ AM+
Sbjct: 182 FGKLRETGGKYLTGMEYRTILFM---QSISAFTGISRLLRDLRINHSEFGRIVISTAMVA 238
Query: 198 DAYGWVLFTLLIPAANNWGEGYYLSVITTFLFIAFCFTVVRPILTPIIENRTNKNMWRKS 257
D G+ + + A +W L + ++ F VVRP + +I+ + ++
Sbjct: 239 DGTGFGVNLFALVAWMDWRVSA-LQGVGIIGYVIFMVWVVRPAMFWVIKRTPQERPVKEC 297
Query: 258 HMLDVFIGLFICSYITDFLGTHPIVGAFVFGLILPHGK-FADMVMEMSDDFVTGILCPVY 316
+ + I F Y + P VG F+ GL +PHG ++E + F TGIL P++
Sbjct: 298 FIYIILILAFGGYYFLKEIHMFPAVGPFLLGLCVPHGPPLGSQLVEKFESFNTGILLPLF 357
Query: 317 FSGFGFKLDLPVLWNTPNSVL--------MMLIMVLLCIPKVLSSLIVTFFFGMPARDGL 368
++D P L N + + I++++ + K++ S+I MP D
Sbjct: 358 LFFSMLQIDGPWLANQIGQLRHFDGQLYEALTIIIVVFVAKIIFSMIPALLAKMPLTDSF 417
Query: 369 SIGLLLNTKGVMAVILQNVAWDKRFLDPCSFMVMTLAIILMTVIVSPLINAIYKPKFRFM 428
+ L+L+ KG++ + + L SF +M I++ + I LI+ +Y RF+
Sbjct: 418 VMALILSNKGIVELCYFLYGVESNVLHVKSFTIMATMILVSSTISPVLIHYLYDSSKRFI 477
Query: 429 QSQLRTVQKLRFDMELRVVACVHNAKHATNIIHVLEATNATRISPVHVSAVHLLELTRHG 488
Q R + L+ EL+ + C+H A H + +I++L + S + +HL+EL
Sbjct: 478 SFQKRNLMSLKLGSELKFLVCIHKADHISGMINLLAQSFPLHESTISCYVIHLVELVGLD 537
Query: 489 TAILVSQMADLNNIAGAETTNYGPKAEFEIITTAFKEFVEQYNAVRFDTSSVVSSYTTIH 548
+ +S + G ++ + AF F + ++ + + +S+ +H
Sbjct: 538 NPVFISHQ--------MQKAEPGNRSYSNNVLIAFDNFKHYWKSISLELFTCISNPRYMH 589
Query: 549 EDIYNVAEEKRASLILLPFH----KEYSTVGGSPEISHNEHCEINENVLQHAPCSVGIFV 604
++IY++A +K+AS ++LPFH + +TV + N N NVL+ APCSVGIFV
Sbjct: 590 QEIYSLALDKQASFLMLPFHIIWSLDQTTVVSDDVMRRN----ANLNVLRQAPCSVGIFV 645
Query: 605 DR-GLGSLLKT--KMRIITLFIGGPDDREALSIAWRMAGHSGTQLHVVRIHLLGKAAEEK 661
R L S K+ + +F+GG DDREAL++ +M + L V+ K
Sbjct: 646 HRQKLLSAQKSSPSFEVCAIFVGGKDDREALALGRQMMRNPNVNLTVL-----------K 694
Query: 662 VLKKKISKSPHGMLSTVMDGVMQKELDEEYIFSFRHKAVNNNDSIVYLEKEVHSDTGEEI 721
++ K+ G ++D KE + + V + + Y+E+ V+ G +
Sbjct: 695 LIPAKMDGMTTGW-DQMLDSAEVKE-----VLRNNNNTVGQHSFVEYVEETVND--GSDT 746
Query: 722 PTLLNEIDKPGYDLYIVGQGSGKNKTIFLKLLEWCDHPELGVIGDILASTSFGTHSSVLI 781
TLL I +DL++VG+ +G + L EW + ELGVIGD+L S F SVL+
Sbjct: 747 STLLLSIAN-SFDLFVVGRSAGVGTDVVSALSEWTEFDELGVIGDLLVSQDFPRRGSVLV 805
Query: 782 VQQ 784
VQQ
Sbjct: 806 VQQ 808
>At2g28180 hypothetical protein
Length = 822
Score = 243 bits (621), Expect = 2e-64
Identities = 200/751 (26%), Positives = 352/751 (46%), Gaps = 72/751 (9%)
Query: 50 LHVPLIVAQMLTGF--SVSRSLLGYFEGVITLFYNPRGILAVETISNLGIMYYVFLSGLE 107
L +P + + ML G +V +L G + + I + + G + + FL G+
Sbjct: 120 LSIPKLSSMMLAGLLLNVLVTLSGENSIIADILVTKNRIDVAGCLGSFGFLIFWFLKGVR 179
Query: 108 MNSDTILRSRKKGTSMAIAGIVTSMLFGVGFLTLQQKLLDKKEKTHIKAYLFWCLTLSVT 167
M+ I ++ K +A + ++ VGFL K + T + Y L S+T
Sbjct: 180 MDVKRIFKAEAKARVTGVAAVTFPIV--VGFLLFNLKSAKNRPLT-FQEYDVMLLMESIT 236
Query: 168 GFPVLARILAKLKLLYTKLGKDTLTAAMLTDAYGWVLFTLLIPAAN-NWGEGY-YLSVIT 225
F +AR+L L + ++ +G+ L++A+++D G +L + ++ +G L+ IT
Sbjct: 237 SFSGIARLLRDLGMNHSSIGRVALSSALVSDIVGLLLLIANVSRSSATLADGLAILTEIT 296
Query: 226 TFLFIAFCFTVVRPILTPIIENRTNKNMWRKSHMLDVFIGLFICSYITDFLGTHPIVGAF 285
FL IAF VVRPI+ II+ + ++ V + + + + L P +GAF
Sbjct: 297 LFLVIAFA--VVRPIMFKIIKRKGEGRPIEDKYIHGVLVLVCLSCMYWEDLSQFPPLGAF 354
Query: 286 VFGLILPHGK-FADMVMEMSDDFVTGILCPVYFSGFGFKLD-------LPVLWNTPNSVL 337
GL +P+G ++E + F GI+ P++ + + D L
Sbjct: 355 FLGLAIPNGPPIGSALVERLESFNFGIILPLFLTAVMLRTDTTAWKGALTFFSGDDKKFA 414
Query: 338 MMLIMVLLCIPKVLSSLIVTFFFGMPARDGLSIGLLLNTKGVMAVILQNVAWDKRFLDPC 397
+ +++L+ + K+ S+IV + + MP RD + + L+++ K
Sbjct: 415 VASLVLLIFLLKLSVSVIVPYLYKMPLRDSIILALIMSHK-------------------- 454
Query: 398 SFMVMTLAIILMTVIVSPLINAIYKPKFRFMQSQLRTVQKLRFDMELRVVACVHNAKHAT 457
L+I+L ++++ I +Y P +F+ Q R + ++ EL+ + C+H H +
Sbjct: 455 -----VLSIVLNSLLIPMAIGFLYDPSKQFICYQKRNLASMKNMGELKTLVCIHRPDHIS 509
Query: 458 NIIHVLEATNATRISPVHVSAVHLLELTRHGTAILVSQMADLNNIAGAETTNYGPKAEFE 517
++I++LEA+ + SP+ +HL+EL L+S + + E
Sbjct: 510 SMINLLEASYQSEDSPLTCYVLHLVELRGQDVPTLISHKVQKLGVGAGNKYS-------E 562
Query: 518 IITTAFKEFVEQY-NAVRFDTSSVVSSYTTIHEDIYNVAEEKRASLILLPFHKEYSTVGG 576
+ +F+ F +++ DT + +++ + +DI +A +K +LI+LPFH+ +S
Sbjct: 563 NVILSFEHFHRSVCSSISIDTFTCIANANHMQDDICWLALDKAVTLIILPFHRTWSLDRT 622
Query: 577 SPEISHNEHCEINENVLQHAPCSVGIFVDRGLGSLLKT---KMRIITLFIGGPDDREALS 633
S +N NVL+ APCSVGI ++R L + + +++ +F+GG DDREAL+
Sbjct: 623 SIVSDVEAIRFLNVNVLKQAPCSVGILIERHLVNKKQEPHESLKVCVIFVGGKDDREALA 682
Query: 634 IAWRMAGHSGTQLHVVRIHLLGKAAEEKVLKKKISKSPHGMLSTVMDGVMQKELDEEYIF 693
A RMA L V+R+ GK SK G ++D V +EL I
Sbjct: 683 FAKRMARQENVTLTVLRLLASGK-----------SKDATGW-DQMLDTVELREL----IK 726
Query: 694 SFRHKAVNNNDSIVYLEKEVHSDTGEEIPTLLNEIDKPGYDLYIVGQGSGKNKTIFLKLL 753
S V S +YLE+E+ G + LL + YDL++VG+ G+N +
Sbjct: 727 SNNAGMVKEETSTIYLEQEILD--GADTSMLLRSMAFD-YDLFVVGRTCGENHEATKGIE 783
Query: 754 EWCDHPELGVIGDILASTSFGTHSSVLIVQQ 784
WC+ ELGVIGD LAS F + +SVL+VQQ
Sbjct: 784 NWCEFEELGVIGDFLASPDFPSKTSVLVVQQ 814
>At5g58460 putative protein
Length = 857
Score = 235 bits (600), Expect = 6e-62
Identities = 195/775 (25%), Positives = 352/775 (45%), Gaps = 58/775 (7%)
Query: 41 RFLFFILKPLHVPLIVAQMLTGFSVSRSLLGYFEGVITLFYNPRGILAVETISNLGIMYY 100
+ ++ +L+PL P IV +++ G + S+LG + P I +G Y+
Sbjct: 80 KIVYVLLRPLRQPRIVCEIIGGMMIGPSMLGRNRNFNYYLFPPIANYICANIGLMGFFYF 139
Query: 101 VFLSGLEMNSDTILRS-RKKGTSMAIAGIVTSMLFGVGFLTLQQKLLDKKEK-THIKAYL 158
FL+ + + I ++ RK A++ +V G L+ K+ + +K + I
Sbjct: 140 FFLTAAKTDVAEIFKAPRKHKYIAAVSVLVPIACVGSTGAALKHKMDIRLQKPSSIGGVT 199
Query: 159 FWCLTLSVTGFPVLARILAKLKLLYTKLGKDTLTAAMLTDAYGWVLFTLLIPAANNWGEG 218
F L T FPV+ +L + LL +++GK ++ +L D G + L A G G
Sbjct: 200 F---ALGFTSFPVIYTVLRDMNLLNSEIGKFAMSVTLLGDMVGVYVLVLFEAMAQADGGG 256
Query: 219 YYLSVI----TTFLFIAFCFTVVRPILTPIIENRTNKNMWRKSHMLDVFIGLFICSYITD 274
SVI + + A VV+ I+ + +++++++ +G+ + ++TD
Sbjct: 257 GAYSVIWFLISAAIMAACLLLVVKRSFEWIVAKTPEGGLVNQNYIVNILMGVLVSCFLTD 316
Query: 275 FLGTHPIVGAFVFGLILPHGKFADMVMEM-SDDFVTGILCPVYFSGFGFKLDLPVLWNT- 332
G VG GL++PHG + + S+ FV L P F+ G K ++ ++
Sbjct: 317 MFGMAIAVGPIWLGLVVPHGPPLGSTLAIRSETFVNEFLMPFSFALVGQKTNVNLISKET 376
Query: 333 -PNSVLMMLIMVLL-CIPKVLSSLIVTFFFGMPARDGLSIGLLLNTKGVMAVILQNVAWD 390
P + ++ M ++ + K +SS FF +P RD L++GL++N +G + ++L D
Sbjct: 377 WPKQISPLIYMSIVGFVTKFVSSTGAALFFKVPTRDSLTLGLMMNLRGQIDILLYLHWID 436
Query: 391 KRFLDPCSFMVMTLAIILMTVIVSPLINAIYKPKFRFMQSQLRTVQKLRFDMELRVVACV 450
K+ + + VM L I++T + +PLI+ +Y P + S+ RT+Q + E +V V
Sbjct: 437 KQMVGLPGYSVMVLYAIVVTGVTAPLISFLYDPTRPYRSSKRRTIQHTPQNTETGLVLAV 496
Query: 451 HNAKHATNIIHVLEATNATRISPVHVSAVHLLELTRHGTAILVS--QMADLNNIAGAETT 508
+ + +I L+ T+ SP V A+ L+EL + ++ + + E
Sbjct: 497 TDHDTFSGLITFLDFAYPTKTSPFSVFAIQLVELEGRAQPLFIAHDKKREEEYEEEEEPA 556
Query: 509 NYGPKAEFEIITTAFKEFVEQYN-AVRFDTSSVVSSYTTIHEDIYNVAEEKRASLILLPF 567
+ + +AFK + E+ + V + +S ++++I +A K+ + ILLP+
Sbjct: 557 ERMGSRRVDQVQSAFKLYQEKRSECVTMHAYTAHASKHNMYQNICELALTKKTAFILLPY 616
Query: 568 HKEYSTVGGSPEISHNEHCEINENVLQHAPCSVGIFVDRG--LGSLLKTKM--------- 616
KE E+ + +N +VL H PCSV I+ ++G +++++ M
Sbjct: 617 QKERLQDAALTELRDSGMLSVNADVLAHTPCSVCIYYEKGRLKNAMVRSSMDPQHTTNSS 676
Query: 617 -------RIITLFIGGPDDREALSIAWRMAGHSGTQLHVVRIHLLGKAAEEKVLKKKISK 669
R + LF+GG D+REAL +A RM + L V+R E++
Sbjct: 677 HMRQEMYRFVVLFLGGADNREALHLADRMTENPFINLTVIRFLAHNHEGEDE-------- 728
Query: 670 SPHGMLSTVMDGVMQKELDEEYIFSFRHKAVNNNDSIVYLEKEVHSDTGEEIPTLLNEID 729
+K+LD+ + F V N + KEV G E + ++
Sbjct: 729 -------------REKKLDDGVVTWF---WVKNESNARVSYKEVVVKNGAETLAAIQAMN 772
Query: 730 KPGYDLYIVGQGSGKNKTIFLKLLEWCDHPELGVIGDILASTSFGTHSSVLIVQQ 784
YDL+I G+ G N I L W + +LGVIGD +A + F + SVL+VQQ
Sbjct: 773 VNDYDLWITGRREGINPKILEGLSTWSEDHQLGVIGDTVAGSVFASEGSVLVVQQ 827
>At1g79400 hypothetical protein
Length = 783
Score = 228 bits (582), Expect = 8e-60
Identities = 194/776 (25%), Positives = 359/776 (46%), Gaps = 27/776 (3%)
Query: 20 DVMIKRVPLLCLQIAYNLLVSRFLFFILKPLHVPLIVAQMLTGFSVSRSLLGYFEGVITL 79
D + + + +Q+A L+ S+ + +LKP VAQ+L G +S LL V
Sbjct: 13 DELFNPLNTMFIQMACILVFSQLFYLLLKPCGQAGPVAQILAGIVLSPVLLSRIPKVKEF 72
Query: 80 FYNPRGILAVETISNLGIMYYVFLSGLEMNSDTILRSRKKGTSMAIAGIVTSMLFGVGFL 139
F S ++FL GLE++ + R+ KK + ++ V S L L
Sbjct: 73 FLQKNAADYYSFFSFALRTSFMFLIGLEVDLHFMRRNFKKAAVITLSSFVVSGLLSFASL 132
Query: 140 TLQQKLLDKKEKTHIKAYLFWCLTLSVTGFPVLARILAKLKLLYTKLGKDTLTAAMLTDA 199
L L KE + +L +TLS T PV+ R +A KL ++G+ T++ A+ +
Sbjct: 133 MLFIPLFGIKED-YFTFFLVLLVTLSNTASPVVVRSIADWKLNTCEIGRLTISCALFIEL 191
Query: 200 YGWVLFTLLIPAANNWGEGYYLSVITTFLFIAFCFTVVRPILTPIIENRTNKNMW-RKSH 258
VL+T+++ + G + + FL ++ +L P + R K + K+
Sbjct: 192 TNVVLYTIIMAFIS----GTIILELFLFLLATVALILINMVLAPWLPKRNPKEKYLSKAE 247
Query: 259 MLDVFIGLFICSYITDFLGTHPIVGAFVFGLILPH-GKFADMVMEMSDDFVTGILCPVYF 317
L FI L I + + V F G++ P GK +++ + + PVYF
Sbjct: 248 TLVFFIFLLIIGITIESYDVNSSVSVFAIGIMFPRQGKTHRTLIQRLSYPIHEFVLPVYF 307
Query: 318 SGFGFKLDLPVLWNTPNSVLMMLIMVLLCIP-KVLSSLIVTFFFGMPARDGLSIGLLLNT 376
GF+ + L T L ++I+V++ I K + + + +P + L + +L+
Sbjct: 308 GYIGFRFSIIAL--TKRFYLGIVIIVIVTIAGKFIGVISACMYLKIPKKYWLFLPTILSV 365
Query: 377 KGVMAVILQNVAW-DKRFLDPCSFMVMTLAIILMTVIVSPLINAIYKPKFRFMQSQLRTV 435
KG + ++L + + +K++ +M A+++ T++ L + + K + + + ++
Sbjct: 366 KGHVGLLLLDSNYSEKKWWTTTIHDMMVAALVITTLVSGVLASFLLKTREKDFAYEKTSL 425
Query: 436 QKLRFDMELRVVACVHNAKHATNIIHVLEATNATR--ISPVHVSAVHLLELTRHGTAILV 493
+ + ELR+++C + +HA I ++ A + +R P +HL+ L + + L+
Sbjct: 426 ESHNTNEELRILSCAYGVRHARGAISLVSALSGSRGASDPFTPLLMHLVPLPKKRKSELM 485
Query: 494 SQMADLNNIAGAETTNYGPKAEFEIITTAFKEFVEQYNAVRFDTSSVVSSYTTIHEDIYN 553
D + +G EI + F + + + +V+ +HE+I N
Sbjct: 486 YHEHDEDGGNANGDDEFGTNEGLEI-NDSIDSFAKD-SKILIQQVKLVTQMLNMHEEICN 543
Query: 554 VAEEKRASLILLPFHKEYSTVGGSPEISHNEHCEINENVLQHAPCSVGIFVDRGLGSL-- 611
E+ R S++ LPFHK + + G ++N NVL+H PCS+GIFVDR +
Sbjct: 544 ATEDLRVSIVFLPFHK-HQRIDGKTTNDGELFRQMNRNVLRHGPCSIGIFVDRNITGFQQ 602
Query: 612 ---LKTKMRIITLFIGGPDDREALSIAWRMAGHSGTQLHVVRIHLLGKAAEEKVLKKKIS 668
+ + TLF GGPDDREAL++ +A ++ L V++ AE V
Sbjct: 603 PHGFDSVQHVATLFFGGPDDREALALCRWLANNTLIHLTVIQFVSEESKAETPVGNAMTR 662
Query: 669 KSPHGMLSTVMDGVMQKELDEEYIFSFRHKAVNNNDSIVYLEKEVHSDTGEEIPTLLNEI 728
+ + + ++E D ++ F ++ V + ++EK V G T+L EI
Sbjct: 663 DNNEVFMEVLGRNQTEQETDRSFLEEFYNRFVTTG-QVGFIEKLV--SNGPHTLTILREI 719
Query: 729 DKPGYDLYIVGQGSGKNKTIFLKLLEWCDHPELGVIGDILASTSFGTHSSVLIVQQ 784
+ Y L++VG+ +G + + +++ +W + PELG +GD LAS S ++SVL+VQ+
Sbjct: 720 GEM-YSLFVVGKSTG-DCPMTVRMKDWEECPELGTVGDFLAS-SLDVNASVLVVQR 772
>At1g16380 putative Na/H antiporter
Length = 785
Score = 215 bits (547), Expect = 9e-56
Identities = 195/781 (24%), Positives = 359/781 (44%), Gaps = 33/781 (4%)
Query: 20 DVMIKRVPLLCLQIAYNLLVSRFLFFILKPLHVPLIVAQMLTGFSVSRSLLGYFEGVITL 79
D + + + +Q+A L+ S+F + LKP VAQ+L G +S LL V
Sbjct: 13 DALFNPLNTMFIQMACILVFSQFFYLFLKPCGQAGPVAQILAGIVLS--LLTIIRKVHEF 70
Query: 80 FYNPRGILAVETISNLGIMYYVFLSGLEMNSDTILRSRKKGTSMAIAGIVTSMLFGVGFL 139
F S L +VFL GLE++ D + R+ K + + +V S + + FL
Sbjct: 71 FLQKDSASYYIFFSFLLRTAFVFLIGLEIDLDFMKRNLKNSIVITLGSLVISGIIWLPFL 130
Query: 140 TLQQKLLDKKEKTHIKAYLFWCLTLSVTGFPVLARILAKLKLLYTKLGKDTLTAAMLTDA 199
+ + K + YL + +TLS T PV+ R + KL +++G+ ++ + +
Sbjct: 131 WFLIRFMQIKGD-FLTFYLAFLITLSNTAAPVVIRSIIDWKLHTSEIGRLAISCGLFIEI 189
Query: 200 YGWVLFTLLIPAANNWGEGYYLSVITTFLFIAFCFTVVRPILTPIIENRTNKNMW-RKSH 258
++T+++ ++ G + I + F + L + R K + K+
Sbjct: 190 TNIFIYTIVL----SFISGTMTADIFIYSFATGVIILTNRFLASWLPKRNPKEKYLSKAE 245
Query: 259 MLDVFIGLFICSYITDFLGTHPIVGAFVFGLILPH-GKFADMVMEMSDDFVTGILCPVYF 317
L I + I + + + + F+ GL+ P GK +++ + + PVYF
Sbjct: 246 TLAFIILILIIALTIESSNLNSTLFVFIIGLMFPREGKTYRTLIQRLSYPIHEFVLPVYF 305
Query: 318 SGFGFKLDLPVLWNTPNSVLMMLIMVLLCIPKVLSSLIVTFFFGMPARDGLSIGLLLNTK 377
GF+ + L VL M + + L + K+L L F +P + L + +L+ K
Sbjct: 306 GYIGFRFSVNSLTKRHYLVLGMTVALSL-LGKLLGVLFACSFLKIPKQYWLFLSTMLSVK 364
Query: 378 G-VMAVILQNVAWDKRFLDPCSFMVMTLAIILMTVIVSPLINAIYKPKFRFMQSQLRTVQ 436
G + V+L + K++ P + A+++MT++ + + + + + + +++
Sbjct: 365 GHIGLVLLDSNLMYKKWFTPVVHDMFVAALVIMTLLSGVITSLLLRSQEKSFAHIKTSLE 424
Query: 437 KLRFDMELRVVACVHNAKHATNIIHVLEATNA----TRISPVHVSAVHLLELTR-HGTAI 491
ELRV+ CV+ +HA I ++ A + T SP +HL+ L + T +
Sbjct: 425 LFDTTEELRVLTCVYGVRHARGSISLVSALSGFSPGTSSSPFTPYLMHLIPLPKKRKTEL 484
Query: 492 LVSQM-ADLNNIAGAETTNYGPKAEFEIITTAFKEFVEQYNAVRFDTSSVVSSYTTIHED 550
L ++ D N G + +G EI + F + +V+ +HE+
Sbjct: 485 LYHELDEDAGNSNGGDD-EFGTNEGLEI-NDSIDSFTRD-RKIMVRQVKLVAPMENMHEE 541
Query: 551 IYNVAEEKRASLILLPFHKEYSTVGGSPEISHNEHCEINENVLQHAPCSVGIFVDRGLGS 610
I N E+ R S++ LPFHK + + G +N VL+ A CS+GIFVDR +
Sbjct: 542 ICNATEDLRVSIVFLPFHK-HQRIDGKTTNDGEVFRHMNRKVLKQAQCSIGIFVDRNITG 600
Query: 611 LLK-----TKMRIITLFIGGPDDREALSIAWRMAGHSGTQLHVVRIHLLGKAAEEKVLKK 665
+ + + LF GGPDDREALS+ + +S L V++ + + EK++
Sbjct: 601 FHQLHGSDSVQHVAALFFGGPDDREALSLCKWLTNNSQIHLTVIQF-VADDSKTEKIVGD 659
Query: 666 KISKSPHGM-LSTVMDGVMQKELDEEYIFSFRHKAVNNNDSIVYLEKEVHSDTGEEIPTL 724
++K + + L V + + E D ++ F H+ V + ++EK V + G + T+
Sbjct: 660 AVTKENNEVFLEIVSEDQTENETDRIFLEEFYHRFVTTGQ-VGFIEKRVSN--GMQTLTI 716
Query: 725 LNEIDKPGYDLYIVGQGSGKNKTIFLKLLEWCDHPELGVIGDILASTSFGTHSSVLIVQQ 784
L EI + Y L++VG+ G + + + +W + PELG +GD LAS++ ++SVL+VQ+
Sbjct: 717 LREIGEM-YSLFVVGKNRG-DCPMTSGMNDWEECPELGTVGDFLASSNMDVNASVLVVQR 774
Query: 785 Y 785
+
Sbjct: 775 H 775
>At3g44910 putative protein
Length = 705
Score = 207 bits (527), Expect = 2e-53
Identities = 156/623 (25%), Positives = 298/623 (47%), Gaps = 51/623 (8%)
Query: 170 PVLARILAKLKLLYTKLGKDTLTAAMLTDAYGWV--LFTLLIPAANNWGE-GYYLSVITT 226
P + L++LK+L ++LG+ L+A+++ D + +F L+ N Y +I
Sbjct: 122 PTVVHFLSELKILNSELGRLVLSASLINDIFASTVSIFAYLVGTYKNISPMTAYRDLIAV 181
Query: 227 FLFIAFCFTVVRPILTPIIENRTNKNMWRKSHMLDVFIGLFICSYITDFLGTHPIVGAFV 286
+ I F V+RP++ I+E ++ V + + + + F ++G F+
Sbjct: 182 IILILVAFCVLRPVVEWIVERTPEGKPVADVYVHAVVLSVIASAAYSSFFNMKYLLGPFL 241
Query: 287 FGLILPHGKFADMVMEMSDDFVT-GILCPVYFSGFGFKLD-LPVLWNTPNSVLMMLIMVL 344
G+I+P G +E + +T +L P+ + + D + +++ + + +M
Sbjct: 242 LGIIIPEGPPIGSALEAKYEALTMNVLIPISITFSTMRCDVMKIVYQYDDIWYNIFLMTF 301
Query: 345 LCIPKVLSSLIVTFFFGMPARDGLSIGLLLNTKGVMAVILQNVAWDKRFLDPCSFMVMTL 404
K+ + ++ + +P ++ ++ LLL +K + L +D ++ ++ +
Sbjct: 302 TGFLKMATGMVPCLYCKIPFKEAIAASLLLCSKSFSEIFLYESTYDDSYISQATYTFLIT 361
Query: 405 AIILMTVIVSPLINAIYKPKFRFMQSQLRTVQKLRFDMELRVVACVHNAKHATNIIHVLE 464
++ + I+ + +Y PK +++ Q + + L+ D +LR++ C+H ++ + I L+
Sbjct: 362 CALINSGIIPTALAGLYDPKRKYVGYQKKNIMNLKPDSDLRILTCIHRPENISAAISFLQ 421
Query: 465 ATNATRISPVHVSAVHLLELTRHGTAILVSQMADLNNIAGAETTNYGPKAEFEIITTAFK 524
+T + V+ +HL++L +L+S +N + T +Y I TA
Sbjct: 422 FLPSTIV----VTVLHLVKLVGKTVPVLISHNKQINRVV---TNSY--------IHTANL 466
Query: 525 EFVEQYNAVRFDTSSVVSSYTTIHEDIYNVAEEKRASLILLPFHKEYSTVGGSPEISHNE 584
F Q +V + ++ +H++I VA E+ S+I++P +++ TV G+ E
Sbjct: 467 AF-SQLESVTMTMFTAITHENLMHDEICKVALEQATSIIIVPSGRKW-TVDGAFESEDEA 524
Query: 585 HCEINENVLQHAPCSVGIFVDRGLGSLLKTK---MRIITLFIGGPDDREALSIAWRMAGH 641
+NE++L+ A CS+GI VDRG SL T+ + + +FIGG DDREALS+ +M +
Sbjct: 525 IRRLNESLLKSASCSIGILVDRGQLSLKGTRKFNIDVGVIFIGGKDDREALSLVKKMKQN 584
Query: 642 SGTQLHVVRIHLLGKAAEEKVLKKKISKSPHGMLSTVMDGVMQKELDEEYIFSFRHKAVN 701
++ V+R+ S ST D ++ E+ E+ K
Sbjct: 585 PRVKITVIRLI-----------------SDRETESTNWDYILDHEVLEDL------KDTE 621
Query: 702 NNDSIVYLEKEVHSDTGEEIPTLLNEIDKPGYDLYIVGQGSGKNKTIFLKLLEWCDHPEL 761
+SI Y E+ V G E+ T + + + YDL +VG+ G F L+EW + PEL
Sbjct: 622 ATNSIAYTERIVTG--GPEVATTVRSLSED-YDLMVVGRDHGMASPDFDGLMEWVELPEL 678
Query: 762 GVIGDILASTSFGTHSSVLIVQQ 784
GVIGD+LAS + SVL+VQQ
Sbjct: 679 GVIGDLLASRELDSRVSVLVVQQ 701
>At1g08140 hypothetical protein
Length = 1536
Score = 205 bits (521), Expect = 9e-53
Identities = 172/642 (26%), Positives = 308/642 (47%), Gaps = 58/642 (9%)
Query: 165 SVTGFPVLARILAKLKLLYTKLGKDTLTAAMLTD--AYGWVLFTLLIPAANNWGEGYYLS 222
S++ F + +L L++ +++ G+ L+ AM+TD A+G V F I +G ++
Sbjct: 161 SISAFTSIDTLLKDLQIKHSEFGRIALSGAMVTDMLAFG-VTFFNAIYYEKLYG---FMQ 216
Query: 223 VITTFLFIAFCFTVVRPILTPIIENRTNKNMWRKSHMLDVFIGLFIC-SYITDFLGTHPI 281
+ LF+ VVRP + +I+ + ++ +F F C ++ +
Sbjct: 217 TVGFCLFVVVMICVVRPAMYWVIKQTPEGRPVKDFYLYSIFGIAFACFTFFNKVIHLFGP 276
Query: 282 VGAFVFGLILPHG-KFADMVMEMSDDFVTGILCPVYFSGFGFKLDLPVLWNTPNSVLMM- 339
G+FVFGL +P+G +++ + F G + P++ S ++DL L+ ++ M
Sbjct: 277 AGSFVFGLTVPNGYPLGTTLIQKFESFNLGSILPLFGSLTMMQVDLLRLFKESGDLIRME 336
Query: 340 -------LIMVLLCIPKVLSSLIVTFFFGMPARDGLSIGLLLNTKGVMAVILQNVAWDKR 392
++L+ K + + I + F MP RD ++ L+L+ KG+ + A + +
Sbjct: 337 GQIYEVISFILLVNTTKFVVTTITAYAFKMPLRDSFALALVLSNKGIFELAYYTYAVELK 396
Query: 393 FLDPCSFMVMTLAIILMTVIVSPLINAIYKPKFRFMQSQLRTVQKLRFDMELRVVACVHN 452
+ P F ++ +L ++ + L+ ++ P RF + R + L+ L+ + CV+
Sbjct: 397 LIRPEVFTILAAYTLLNSIFIPMLLELVHDPTKRFRCYRKRNLGILKDGAALQCLMCVYR 456
Query: 453 AKHATNIIHVLEATNATRISPVHVSAVHLLELTRHGTAILVSQMADLNNIAGAETTNYGP 512
H T++ +LE + ++ SP+ + +HL+EL + +S + G
Sbjct: 457 PDHITSMTDLLETFSPSQDSPMACNILHLVELVGQANPMFISHQ--------LQKPEPGS 508
Query: 513 KAEFEIITTAFKEFVEQ---YNAVRFDTSSVVSSYTTIHEDIYNVAEEKRASLILLPFHK 569
+ + + +F+ F Q Y ++ TS VS + +HEDI +A + SLI+LPFH+
Sbjct: 509 TSLSDNVIISFRGFQRQFFEYTSLDIFTSVSVSQH--MHEDICWLALSRSLSLIVLPFHR 566
Query: 570 EYSTVGGSPEISHNEHCE-INENVLQHAPCSVGIFVDRG---LGSLLKTKMRIITLFIGG 625
+S V S IS++++ +N NVL+ APCSVGIFV R + K+ +I +F GG
Sbjct: 567 TWS-VDRSTVISNDDNLRMLNVNVLRRAPCSVGIFVYRKPIVESHMAKSHSKICLIFNGG 625
Query: 626 PDDREALSIAWRM-AGHSGTQLHVVRIHLLGKAAEEKVLKKKISKSPHGMLSTVMDGVMQ 684
DDREAL+I RM T+L ++R + K++E M + +
Sbjct: 626 KDDREALAITNRMRLTEKRTRLTIIR--FIPKSSE--------------MDNDEWEQQQS 669
Query: 685 KELDEEYIFSFRHKAVNNNDSIVYLEKEVH--SDTGEEIPTLLNEIDKPGYDLYIVGQGS 742
L E N+ + Y++K V S+T + + N+ YDL+IVG GS
Sbjct: 670 INLKESVTSIVGSNIKENDAKVTYIDKAVSDGSETSRILRAMAND-----YDLFIVGSGS 724
Query: 743 GKNKTIFLKLLEWCDHPELGVIGDILASTSFGTHSSVLIVQQ 784
G + EW + ELG IGD+LAS + + +SVL+VQ+
Sbjct: 725 GIGTEATSGISEWTEFNELGPIGDLLASHEYPSSASVLVVQK 766
Score = 184 bits (466), Expect = 2e-46
Identities = 199/784 (25%), Positives = 348/784 (44%), Gaps = 107/784 (13%)
Query: 26 VPLLCLQIAYNLLVSRFLFFILKPLHVPL--IVAQMLTGFSVSRSLLGYFE----GVITL 79
+P L + I L+ R + K L VP+ + ML G +V + G + I +
Sbjct: 833 LPQLEIIILSIFLLWRLFDMLFKKLGVPIPKFTSMMLVG-AVLSEMFGSMQIPCLKHIFI 891
Query: 80 FYNPRGILAVETISNLGIMYYVFLSGLEMNSDTILRSRKKGTSMAIAGIVTSML-FGVGF 138
YN +TI + FL G+ + + +K GT + GI + ++ + +G
Sbjct: 892 HYNQYMTKVPDTIGAFAFVLDWFLRGVTTDVGIM---KKSGTKSVVIGITSMIIPWQIGK 948
Query: 139 LTLQQKLLDKKEKTHIKA-----YLFWCLTLSVTGFPVLARILAKLKLLYTKLGKDTLTA 193
L L +EK+ I Y T+S+T F + +L LK+++T G+ +A
Sbjct: 949 L-----LYSSREKSSILTMTEMEYTVMTFTMSMTPFTCVNMLLTDLKIVHTDFGQIAQSA 1003
Query: 194 AMLTDAYGWVL-FTLLIPAANNWGEGYYLSVITTFLFIAFCFTVVRPILTPIIENRTNKN 252
M+TD + L + + G L+ + F+F+ + ++ E KN
Sbjct: 1004 GMVTDLLAFFLTVSAYVSRDETQGVKMGLAFMAFFIFVYLVRQFMLWVIRHTPEGAPVKN 1063
Query: 253 MWRKSHMLDVFIGLFIC----SYITDFLGTHPIVGAFVFGLILPHGKFADMVMEMSDDFV 308
++ ++IGL + Y + FL P+ GAF GL +P+G + +F
Sbjct: 1064 VY-------LYIGLLLAYLSYLYWSRFLFFGPL-GAFALGLAVPNGP------PLGSEFG 1109
Query: 309 TG-ILCPVYFSGFGFKLDLPVLWNTPNSVLMMLIMVLLCIPKVLSSLIVTFFFGMPARDG 367
G L + F F LP+++ I K +S + +P RD
Sbjct: 1110 NGRHLHGHMYECFSF---LPIVY----------------IAKFATSFLAALATKIPLRDS 1150
Query: 368 LSIGLLLNTKGVMAVILQNVAWDKRFLDPCSFMVMTL--AIILMTVIVSPL-INAIYKPK 424
+ +G+++ TK + A++K D S V++L IL+ +++P+ I+ +Y
Sbjct: 1151 IILGVIMGTKSSFELGYVLTAFEK---DRISLEVLSLLGVYILVNSLLTPMAIHFLYDRS 1207
Query: 425 FRFMQSQLRTVQKLRFDMELRVVACVHNAKHATNIIHVLEATNATRISPVHVSAVHLLEL 484
RF+ R L+ E++ + C++ + T++I +L AT+ ++ SP+ +HL+EL
Sbjct: 1208 KRFVCYGRR---NLKEKPEMQTLVCINKPDNITSMISLLRATSPSKDSPMECCVLHLIEL 1264
Query: 485 TRHGTAILVSQMADLNNIAGAETTNYGPKAEFEIITTAFKEFVEQY-NAVRFDTSSVVSS 543
T +S + G ++ E + ++F+ F E Y ++ + + ++S
Sbjct: 1265 LGQATPTFISHQ--------LQKPKPGSRSYSENVISSFQLFQEVYWDSASINMFTSLTS 1316
Query: 544 YTTIHEDIYNVAEEKRASLILLPFHKEYSTVGGSPEISHNEHCEINENVLQHAPCSVGIF 603
+HE I A + ++LILL FH+ + G +N NVL+ APCSVGIF
Sbjct: 1317 AKEMHEQICWFALSQGSNLILLSFHRTWEPNGNVIISDDQTLRSLNLNVLKRAPCSVGIF 1376
Query: 604 VDR---GLGSLLKTKMRIITLFIGGPDDREALSIAWRMAGHSGTQLHVVRIHLLGKAAEE 660
V R L++ R+ +++GG DD+EAL++A M G+ L V+R+ A E
Sbjct: 1377 VYRKPIWQTKALESPCRVCLIYVGGNDDKEALALADHMRGNQQVILTVLRLIPTSYADES 1436
Query: 661 KVLKKKISKSPHGMLSTVMDGVMQKELDEEYIFSFRHKAVNNNDSIVYLEKEVHSDTGEE 720
+ + + + RH+ D ++ V G E
Sbjct: 1437 SL-----------------------RIHSQMVDMNRHEDQRPGDKSTIIDWTV--GDGTE 1471
Query: 721 IPTLLNEIDKPGYDLYIVGQGSGKNKTIFLKLLEWCDHPELGVIGDILASTSFGTHSSVL 780
+L+ + YDL+IVG+ SG T+ L +W + ELGVIGD+LAS F + +SVL
Sbjct: 1472 TSKILHSVSY-DYDLFIVGRRSGVGTTVTRGLGDWMEFEELGVIGDLLASEYFPSRASVL 1530
Query: 781 IVQQ 784
+VQQ
Sbjct: 1531 VVQQ 1534
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.325 0.140 0.416
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,141,630
Number of Sequences: 26719
Number of extensions: 729532
Number of successful extensions: 2416
Number of sequences better than 10.0: 41
Number of HSP's better than 10.0 without gapping: 29
Number of HSP's successfully gapped in prelim test: 12
Number of HSP's that attempted gapping in prelim test: 2180
Number of HSP's gapped (non-prelim): 56
length of query: 807
length of database: 11,318,596
effective HSP length: 107
effective length of query: 700
effective length of database: 8,459,663
effective search space: 5921764100
effective search space used: 5921764100
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 64 (29.3 bits)
Medicago: description of AC145767.3


