

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC145512.8 - phase: 0 /pseudo
(164 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
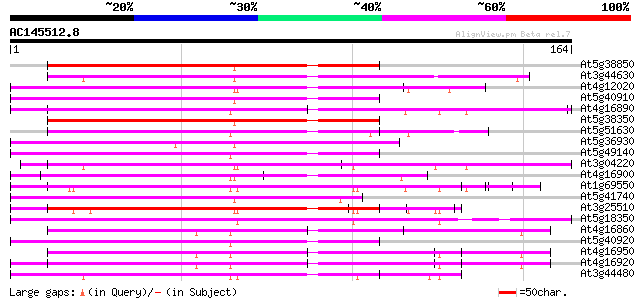
Score E
Sequences producing significant alignments: (bits) Value
At5g38850 disease resistance protein - like 79 8e-16
At3g44630 putative disease resistance protein 75 2e-14
At4g12020 putative disease resistance protein 74 3e-14
At5g40910 disease resistance protein-like 74 5e-14
At4g16890 disease resistance RPP5 like protein 74 5e-14
At5g38350 disease resistance protein-like 73 6e-14
At5g51630 putative protein 72 1e-13
At5g36930 disease resistance like protein 72 2e-13
At5g49140 disease resistance protein-like 71 3e-13
At3g04220 putative disease resistance protein 71 3e-13
At4g16900 disease resistance RPP5 like protein 70 5e-13
At1g69550 putative disease resistance protein 70 5e-13
At5g41740 disease resistance protein-like 70 7e-13
At3g25510 unknown protein 70 7e-13
At5g18350 disease resistance protein -like 69 9e-13
At4g16860 disease resistance RPP5 like protein 69 1e-12
At5g40920 disease resistance protein-like 67 3e-12
At4g16950 disease resistance RPP5 like protein 67 4e-12
At4g16920 disease resistance RPP5 like protein 67 4e-12
At3g44480 disease resistance protein -like 67 6e-12
>At5g38850 disease resistance protein - like
Length = 986
Score = 79.3 bits (194), Expect = 8e-16
Identities = 40/98 (40%), Positives = 64/98 (64%), Gaps = 4/98 (4%)
Query: 12 LTQIPDVSGLVNLEEFSFQGCVNLITVHDSIGLLGRLKTLRVMCCIKLRSIPP-LNLASL 70
L ++PD+S NLEE + C NL+ + S L +LK L +M C +L+ +PP +NL SL
Sbjct: 627 LKKLPDLSNATNLEELDLRACQNLVELPSSFSYLHKLKYLNMMGCRRLKEVPPHINLKSL 686
Query: 71 EELDLSECSCLESFPEILGEMRNVTNLNLEYTPIKELP 108
E +++ CS L+SFP+I N+++L++ YT ++ELP
Sbjct: 687 ELVNMYGCSRLKSFPDI---STNISSLDISYTDVEELP 721
>At3g44630 putative disease resistance protein
Length = 1214
Score = 74.7 bits (182), Expect = 2e-14
Identities = 52/145 (35%), Positives = 82/145 (55%), Gaps = 8/145 (5%)
Query: 12 LTQIPDVSG-LVNLEEFSFQGCVNLITVHDSIGLLGRLKTLRVMCCIKLRSIPP-LNLAS 69
L ++P G + NL+EF C NL+ + SIG L +L LR+ C KL ++P +NL S
Sbjct: 846 LVKLPSSIGDMTNLKEFDLSNCSNLVELPSSIGNLQKLFMLRMRGCSKLETLPTNINLIS 905
Query: 70 LEELDLSECSCLESFPEILGEMRNVTNLNLEYTPIKELPFPFQNLTLPKIYPCNCGFVSL 129
L LDL++CS L+SFPEI +++ L L+ T IKE+P + + +Y + F SL
Sbjct: 906 LRILDLTDCSQLKSFPEI---STHISELRLKGTAIKEVPLSITSWSRLAVYEMSY-FESL 961
Query: 130 PTSNYAMSKLVECTIQAE--QKVIP 152
+A+ + + + +E Q+V P
Sbjct: 962 KEFPHALDIITDLLLVSEDIQEVPP 986
Score = 26.9 bits (58), Expect = 4.9
Identities = 33/129 (25%), Positives = 58/129 (44%), Gaps = 7/129 (5%)
Query: 32 CVNLITVHDSIGLLGRLKTLRVMCCIKLRSIPPL--NLASLEELDLSECSCLESFPEILG 89
C L + + L LK + + L+ +P L SL+ LDL +CS L P +
Sbjct: 725 CSKLRKLWEGTKQLRNLKWMDLSDSRDLKELPSSIEKLTSLQILDLRDCSSLVKLPPSI- 783
Query: 90 EMRNVTNLNL-EYTPIKELPFPFQNLT-LPKIYPCNC-GFVSLPTSNYAMSKLVECTIQA 146
N+ L+L + + +LP +N+T L ++ NC + LP S + L + I+
Sbjct: 784 NANNLQGLSLTNCSRVVKLP-AIENVTNLHQLKLQNCSSLIELPLSIGTANNLWKLDIRG 842
Query: 147 EQKVIPIQS 155
++ + S
Sbjct: 843 CSSLVKLPS 851
>At4g12020 putative disease resistance protein
Length = 1895
Score = 74.3 bits (181), Expect = 3e-14
Identities = 53/142 (37%), Positives = 79/142 (55%), Gaps = 6/142 (4%)
Query: 1 MRVLNFDHHGLLTQIPDVSGLVNLEEFSFQGCVNLITVHDSIGLLGRLKTLRVMCCIKLR 60
++ + + LT+IP +S NLE +GC +L+++ SI L +L L + C KL
Sbjct: 1260 LKKMRLSYSDQLTKIPRLSSATNLEHIDLEGCNSLLSLSQSISYLKKLVFLNLKGCSKLE 1319
Query: 61 SIPPL-NLASLEELDLSECSCLESFPEILGEMRNVTNLNLEYTPIKELPFPFQNLT-LPK 118
+IP + +L SLE L+LS CS L +FPEI NV L + T I+E+P +NL L K
Sbjct: 1320 NIPSMVDLESLEVLNLSGCSKLGNFPEI---SPNVKELYMGGTMIQEIPSSIKNLVLLEK 1376
Query: 119 IYPCNCGFV-SLPTSNYAMSKL 139
+ N + +LPTS Y + L
Sbjct: 1377 LDLENSRHLKNLPTSIYKLKHL 1398
Score = 43.5 bits (101), Expect = 5e-05
Identities = 38/117 (32%), Positives = 56/117 (47%), Gaps = 5/117 (4%)
Query: 1 MRVLNFDHHGLLTQIPDVSGLVNLEEFSFQGCVNLITVHDSIGLLGRLKTLRVMCCIKLR 60
+ VLN L P++S N++E + G + + SI L L+ L + L+
Sbjct: 1330 LEVLNLSGCSKLGNFPEISP--NVKEL-YMGGTMIQEIPSSIKNLVLLEKLDLENSRHLK 1386
Query: 61 SIPP--LNLASLEELDLSECSCLESFPEILGEMRNVTNLNLEYTPIKELPFPFQNLT 115
++P L LE L+LS C LE FP+ M+ + L+L T IKELP LT
Sbjct: 1387 NLPTSIYKLKHLETLNLSGCISLERFPDSSRRMKCLRFLDLSRTDIKELPSSISYLT 1443
>At5g40910 disease resistance protein-like
Length = 1104
Score = 73.6 bits (179), Expect = 5e-14
Identities = 43/109 (39%), Positives = 65/109 (59%), Gaps = 4/109 (3%)
Query: 1 MRVLNFDHHGLLTQIPDVSGLVNLEEFSFQGCVNLITVHDSIGLLGRLKTLRVMCCIKLR 60
++ +N + L +IP++S NL+ + GC +L+ + SI L +L+ L CIKL+
Sbjct: 611 LKKINLGYSSNLKEIPNLSKATNLKTLTLTGCESLVEIPSSIWNLQKLEMLYASGCIKLQ 670
Query: 61 SIPP-LNLASLEELDLSECSCLESFPEILGEMRNVTNLNLEYTPIKELP 108
IP +NLASLEE+++S CS L SFP+I N+ L + T IKE P
Sbjct: 671 VIPTNINLASLEEVNMSNCSRLRSFPDI---SSNIKRLYVAGTMIKEFP 716
>At4g16890 disease resistance RPP5 like protein
Length = 1301
Score = 73.6 bits (179), Expect = 5e-14
Identities = 56/156 (35%), Positives = 83/156 (52%), Gaps = 7/156 (4%)
Query: 12 LTQIPDVSGLVNLEEFSFQGCVNLITVHDSIGLLGRLKTLRVMCCIKLRSIP-PLNLASL 70
LT+IPD+S LE C +L+T+ +IG L RL L + C L +P +NL+SL
Sbjct: 786 LTEIPDLSKATKLESLILNNCKSLVTLPSTIGNLHRLVRLEMKECTGLEVLPTDVNLSSL 845
Query: 71 EELDLSECSCLESFPEILGEMRNVTNLNLEYTPIKELPFPFQNL-TLPKIYPCNC-GFVS 128
E LDLS CS L SFP I N+ L LE T I+E+P NL L ++ C G
Sbjct: 846 ETLDLSGCSSLRSFPLI---STNIVWLYLENTAIEEIPSTIGNLHRLVRLEMKKCTGLEV 902
Query: 129 LPTS-NYAMSKLVECTIQAEQKVIPIQSLHVEYICL 163
LPT N + + ++ + + + P+ S ++++ L
Sbjct: 903 LPTDVNLSSLETLDLSGCSSLRSFPLISESIKWLYL 938
Score = 68.9 bits (167), Expect = 1e-12
Identities = 52/157 (33%), Positives = 82/157 (52%), Gaps = 7/157 (4%)
Query: 12 LTQIPDVSGLVNLEEFSFQGCVNLITVHDSIGLLGRLKTLRVMCCIKLRSIP-PLNLASL 70
+ +IPD+S NL+ C +L+T+ +IG L +L + + C L +P +NL+SL
Sbjct: 943 IEEIPDLSKATNLKNLKLNNCKSLVTLPTTIGNLQKLVSFEMKECTGLEVLPIDVNLSSL 1002
Query: 71 EELDLSECSCLESFPEILGEMRNVTNLNLEYTPIKELPFPFQNL-TLPKIYPCNC-GFVS 128
LDLS CS L +FP I N+ L LE T I+E+P NL L K+ C G
Sbjct: 1003 MILDLSGCSSLRTFPLI---STNIVWLYLENTAIEEIPSTIGNLHRLVKLEMKECTGLEV 1059
Query: 129 LPTS-NYAMSKLVECTIQAEQKVIPIQSLHVEYICLR 164
LPT N + +++ + + + P+ S +E + L+
Sbjct: 1060 LPTDVNLSSLMILDLSGCSSLRTFPLISTRIECLYLQ 1096
Score = 60.5 bits (145), Expect = 4e-10
Identities = 36/88 (40%), Positives = 51/88 (57%), Gaps = 1/88 (1%)
Query: 1 MRVLNFDHHGLLTQIPDVSGLVNLEEFSFQGCVNLITVHDSIGLLGRLKTLRVMCCIKLR 60
++ +N + L +IPD+S +NLEE GC +L+T+ SI +L L + C KL
Sbjct: 615 LKEMNLRYSNNLKEIPDLSLAINLEELDLVGCKSLVTLPSSIQNATKLIYLDMSDCKKLE 674
Query: 61 SIP-PLNLASLEELDLSECSCLESFPEI 87
S P LNL SLE L+L+ C L +FP I
Sbjct: 675 SFPTDLNLESLEYLNLTGCPNLRNFPAI 702
Score = 33.9 bits (76), Expect = 0.040
Identities = 22/54 (40%), Positives = 30/54 (54%), Gaps = 4/54 (7%)
Query: 45 LGRLKTLRVMCCIKLRSIPPLNLA-SLEELDLSECSCLESFPEILGEMRNVTNL 97
LG LK + + L+ IP L+LA +LEELDL C L + P ++N T L
Sbjct: 612 LGSLKEMNLRYSNNLKEIPDLSLAINLEELDLVGCKSLVTLP---SSIQNATKL 662
>At5g38350 disease resistance protein-like
Length = 833
Score = 73.2 bits (178), Expect = 6e-14
Identities = 40/98 (40%), Positives = 60/98 (60%), Gaps = 4/98 (4%)
Query: 12 LTQIPDVSGLVNLEEFSFQGCVNLITVHDSIGLLGRLKTLRVMCCIKLRSIPP-LNLASL 70
L ++PD+S NLE GC++L+ + SIG L +L L + C KL ++P +NL SL
Sbjct: 491 LKELPDLSTATNLEYLIMSGCISLVELPSSIGKLRKLLMLSLRGCSKLEALPTNINLESL 550
Query: 71 EELDLSECSCLESFPEILGEMRNVTNLNLEYTPIKELP 108
+ LDL++C ++ FPEI N+ +L L T IKE+P
Sbjct: 551 DYLDLTDCLLIKKFPEI---STNIKDLKLTKTAIKEVP 585
>At5g51630 putative protein
Length = 1239
Score = 72.0 bits (175), Expect = 1e-13
Identities = 52/132 (39%), Positives = 75/132 (56%), Gaps = 7/132 (5%)
Query: 12 LTQIPDVSGLVNLEEFSFQGCVNLITVHDSIGLLGRLKTLRVMCCIKLRSIP-PLNLASL 70
L +IPD+S VNLEE C +L+T+ S+ L +L+ LR+ C + +P LNL SL
Sbjct: 637 LKEIPDLSYAVNLEEMDLCSCKSLVTLPSSVRNLDKLRVLRMSSCSNVEVLPTDLNLESL 696
Query: 71 EELDLSECSCLESFPEILGEMRNVTNLNLEYTPI-KELPFPFQNLT-LPKIYPCNCGFVS 128
+ L+L +CS L SFP+I RN++ LNL T I +E +N++ L + C S
Sbjct: 697 DLLNLEDCSQLRSFPQI---SRNISILNLSGTAIDEESSLWIENMSRLTHLRWDFCPLKS 753
Query: 129 LPTSNYAMSKLV 140
LP SN+ LV
Sbjct: 754 LP-SNFRQEHLV 764
Score = 67.8 bits (164), Expect = 3e-12
Identities = 40/98 (40%), Positives = 58/98 (58%), Gaps = 4/98 (4%)
Query: 12 LTQIPDVSGLVNLEEFSFQGCVNLITVHDSIGLLGRLKTLRVMCCIKLRSIP-PLNLASL 70
L + P++S + NL+ GC +L+TV SI L +L L + C L ++P +NL SL
Sbjct: 797 LKEFPNLSKVTNLDTLDLYGCKSLVTVPSSIQSLSKLTELNMRRCTGLEALPTDVNLESL 856
Query: 71 EELDLSECSCLESFPEILGEMRNVTNLNLEYTPIKELP 108
LDLS CS L +FP+I RN+ L L+ T I+E+P
Sbjct: 857 HTLDLSGCSKLTTFPKI---SRNIERLLLDDTAIEEVP 891
>At5g36930 disease resistance like protein
Length = 1164
Score = 71.6 bits (174), Expect = 2e-13
Identities = 42/117 (35%), Positives = 64/117 (53%), Gaps = 3/117 (2%)
Query: 1 MRVLNFDHHGLLTQIPDVSGLVNLEEFSFQGCVNLITVHDSIGLLGR-LKTLRVMCCIKL 59
++ L+ H L + PD S N+E+ C +L+ VH SIG+L + L L + CI+L
Sbjct: 622 VKYLDLSHSVYLRETPDFSYFPNVEKLILINCKSLVLVHKSIGILDKKLVLLNLSSCIEL 681
Query: 60 RSIPP--LNLASLEELDLSECSCLESFPEILGEMRNVTNLNLEYTPIKELPFPFQNL 114
+P L SLE L LS CS LE + LGE+ ++T L ++T ++E+P L
Sbjct: 682 DVLPEEIYKLKSLESLFLSNCSKLERLDDALGELESLTTLLADFTALREIPSTINQL 738
>At5g49140 disease resistance protein-like
Length = 980
Score = 70.9 bits (172), Expect = 3e-13
Identities = 40/109 (36%), Positives = 65/109 (58%), Gaps = 4/109 (3%)
Query: 1 MRVLNFDHHGLLTQIPDVSGLVNLEEFSFQGCVNLITVHDSIGLLGRLKTLRVMCCIKLR 60
++ ++ L ++PD+S ++LE +GC +L + S+ L RLK LR+ C KL
Sbjct: 633 LKTIDLSFSNNLVEVPDLSKAISLETLCLEGCQSLAELPSSVLNLHRLKWLRLTMCEKLE 692
Query: 61 SIP-PLNLASLEELDLSECSCLESFPEILGEMRNVTNLNLEYTPIKELP 108
IP +NLASLE LD+ C L+SFP+I +N+ + ++ T I+E+P
Sbjct: 693 VIPLHINLASLEVLDMEGCLKLKSFPDI---SKNIERIFMKNTGIEEIP 738
>At3g04220 putative disease resistance protein
Length = 896
Score = 70.9 bits (172), Expect = 3e-13
Identities = 51/163 (31%), Positives = 80/163 (48%), Gaps = 10/163 (6%)
Query: 12 LTQIPDVSGLVNLEEFSFQGCVNLITVHDSIGLLGRLKTLRVMCCIKLRSIPPL--NLAS 69
L ++PD+S NL+ S + C +L+ + SIG LK + + C+ L +P NL +
Sbjct: 710 LKELPDLSTATNLQRLSIERCSSLVKLPSSIGEATNLKKINLRECLSLVELPSSFGNLTN 769
Query: 70 LEELDLSECSCLESFPEILGEMRNVTNLNL-EYTPIKELPFPFQNLTLPKIYPCN--CGF 126
L+ELDL ECS L P G + NV +L E + + +LP F NLT ++
Sbjct: 770 LQELDLRECSSLVELPTSFGNLANVESLEFYECSSLVKLPSTFGNLTNLRVLGLRECSSM 829
Query: 127 VSLPTS-----NYAMSKLVECTIQAEQKVIPIQSLHVEYICLR 164
V LP+S N + L +C+ E + ++E + LR
Sbjct: 830 VELPSSFGNLTNLQVLNLRKCSTLVELPSSFVNLTNLENLDLR 872
Score = 43.1 bits (100), Expect = 7e-05
Identities = 33/97 (34%), Positives = 46/97 (47%), Gaps = 8/97 (8%)
Query: 4 LNFDHHGLLTQIPDVSG-LVNLEEFSFQGCVNLITVHDSIGLLGRLKTLRVMCCIKLRSI 62
L F L ++P G L NL + C +++ + S G L L+ L + C L +
Sbjct: 797 LEFYECSSLVKLPSTFGNLTNLRVLGLRECSSMVELPSSFGNLTNLQVLNLRKCSTLVEL 856
Query: 63 PP--LNLASLEELDLSECSCLESFPEILGEMRNVTNL 97
P +NL +LE LDL +CS L P G NVT L
Sbjct: 857 PSSFVNLTNLENLDLRDCSSL--LPSSFG---NVTYL 888
>At4g16900 disease resistance RPP5 like protein
Length = 1072
Score = 70.1 bits (170), Expect = 5e-13
Identities = 47/117 (40%), Positives = 67/117 (57%), Gaps = 7/117 (5%)
Query: 10 GLLTQIPDVSGLVNLEEFSFQGCVNLITVHDSIGLLGRLKTLRVMCCIKLRSIP-PLNLA 68
G LT+IPD+S NL C +L+TV +IG L +L L + C L +P +NL+
Sbjct: 720 GNLTEIPDLSKATNLVNLYLSNCKSLVTVPSTIGNLQKLVRLEMKECTGLEVLPTDVNLS 779
Query: 69 SLEELDLSECSCLESFPEILGEMRNVTNLNLEYTPIKELPFPFQN---LTLPKIYPC 122
SL+ LDLS CS L +FP I +++ L LE T I+E+P +N LT+ +Y C
Sbjct: 780 SLKMLDLSGCSSLRTFPLI---SKSIKWLYLENTAIEEVPCCIENFSWLTVLMMYCC 833
Score = 42.4 bits (98), Expect = 1e-04
Identities = 28/75 (37%), Positives = 40/75 (53%), Gaps = 1/75 (1%)
Query: 1 MRVLNFDHHGLLTQIPDVSGLVNLEEFSFQGCVNLITVHDSIGLLGRLKTLRVMCCIKLR 60
++ +N L +I D+S NLEE + C +L+T+ SI +L L + C KL
Sbjct: 583 LKRMNMHGSRYLREISDLSNARNLEELNLSECRSLVTLSSSIQNAIKLIYLDMRGCTKLE 642
Query: 61 SIPP-LNLASLEELD 74
S P LNL SLE L+
Sbjct: 643 SFPTHLNLESLEYLE 657
Score = 33.9 bits (76), Expect = 0.040
Identities = 30/102 (29%), Positives = 52/102 (50%), Gaps = 5/102 (4%)
Query: 66 NLASLEELDLSECSCLESFPEILGEMRNVTNLNL-EYTPIKELPFPFQNL-TLPKIYPCN 123
+LASL E+D+SEC L P+ L + N+ NL L + +P NL L ++
Sbjct: 707 SLASLVEMDMSECGNLTEIPD-LSKATNLVNLYLSNCKSLVTVPSTIGNLQKLVRLEMKE 765
Query: 124 C-GFVSLPTS-NYAMSKLVECTIQAEQKVIPIQSLHVEYICL 163
C G LPT N + K+++ + + + P+ S ++++ L
Sbjct: 766 CTGLEVLPTDVNLSSLKMLDLSGCSSLRTFPLISKSIKWLYL 807
Score = 27.7 bits (60), Expect = 2.9
Identities = 18/38 (47%), Positives = 22/38 (57%), Gaps = 1/38 (2%)
Query: 45 LGRLKTLRVMCCIKLRSIPPL-NLASLEELDLSECSCL 81
LG LK + + LR I L N +LEEL+LSEC L
Sbjct: 580 LGSLKRMNMHGSRYLREISDLSNARNLEELNLSECRSL 617
>At1g69550 putative disease resistance protein
Length = 1398
Score = 70.1 bits (170), Expect = 5e-13
Identities = 50/138 (36%), Positives = 73/138 (52%), Gaps = 6/138 (4%)
Query: 1 MRVLNFDHHGLLTQIPD-VSGLVNLEEFSFQGCVNLITVHDSIGLLGRLKTLRVMCCIKL 59
+++L L +IP + L+NL+ + GC +L+ + SIG L LK L + C L
Sbjct: 836 LKILYLKRISSLVEIPSSIGNLINLKLLNLSGCSSLVELPSSIGNLINLKKLDLSGCSSL 895
Query: 60 RSIPPL--NLASLEELDLSECSCLESFPEILGEMRNVTNLNL-EYTPIKELPFPFQNL-T 115
+P NL +L+EL LSECS L P +G + N+ LNL E + + ELP NL
Sbjct: 896 VELPLSIGNLINLQELYLSECSSLVELPSSIGNLINLKTLNLSECSSLVELPSSIGNLIN 955
Query: 116 LPKIYPCNC-GFVSLPTS 132
L ++Y C V LP+S
Sbjct: 956 LQELYLSECSSLVELPSS 973
Score = 67.4 bits (163), Expect = 3e-12
Identities = 52/145 (35%), Positives = 74/145 (50%), Gaps = 5/145 (3%)
Query: 1 MRVLNFDHHGLLTQIPDVSGLVNLEEFSFQGCVNLITVHDSIGLLGRLKTLRVMCCIKLR 60
++ LN L ++P G +NL++ GC +L+ + SIG L LK L + C L
Sbjct: 1076 LKTLNLSGCSSLVELPSSIGNLNLKKLDLSGCSSLVELPSSIGNLINLKKLDLSGCSSLV 1135
Query: 61 SIPPL--NLASLEELDLSECSCLESFPEILGEMRNVTNLNL-EYTPIKELPFPFQNL-TL 116
+P NL +L+EL LSECS L P +G + N+ L L E + + ELP NL L
Sbjct: 1136 ELPLSIGNLINLQELYLSECSSLVELPSSIGNLINLQELYLSECSSLVELPSSIGNLINL 1195
Query: 117 PKIYPCNC-GFVSLPTSNYAMSKLV 140
K+ C VSLP ++S LV
Sbjct: 1196 KKLDLNKCTKLVSLPQLPDSLSVLV 1220
Score = 64.7 bits (156), Expect = 2e-11
Identities = 47/149 (31%), Positives = 76/149 (50%), Gaps = 5/149 (3%)
Query: 12 LTQIPD-VSGLVNLEEFSFQGCVNLITVHDSIGLLGRLKTLRVMCCIKLRSIP-PLNLAS 69
L ++P + L+NL++ GC +L+ + SIG L LKTL + C L +P + +
Sbjct: 1039 LVELPSSIGNLINLKKLDLSGCSSLVELPLSIGNLINLKTLNLSGCSSLVELPSSIGNLN 1098
Query: 70 LEELDLSECSCLESFPEILGEMRNVTNLNLE-YTPIKELPFPFQNL-TLPKIYPCNC-GF 126
L++LDLS CS L P +G + N+ L+L + + ELP NL L ++Y C
Sbjct: 1099 LKKLDLSGCSSLVELPSSIGNLINLKKLDLSGCSSLVELPLSIGNLINLQELYLSECSSL 1158
Query: 127 VSLPTSNYAMSKLVECTIQAEQKVIPIQS 155
V LP+S + L E + ++ + S
Sbjct: 1159 VELPSSIGNLINLQELYLSECSSLVELPS 1187
Score = 64.3 bits (155), Expect = 3e-11
Identities = 49/136 (36%), Positives = 71/136 (52%), Gaps = 8/136 (5%)
Query: 12 LTQIPD-VSGLVNLEEFSFQGCVNLITVHDSIGLLGRLKTLRVMCCIKLRSIPPL--NLA 68
L ++P + L+NL++ GC +L+ + SIG L LKTL + C L +P NL
Sbjct: 967 LVELPSSIGNLINLKKLDLSGCSSLVELPLSIGNLINLKTLNLSECSSLVELPSSIGNLI 1026
Query: 69 SLEELDLSECSCLESFPEILGEMRNVTNLNLE-YTPIKELPFPFQNL-TLPKIYPCNC-G 125
+L+EL LSECS L P +G + N+ L+L + + ELP NL L + C
Sbjct: 1027 NLQELYLSECSSLVELPSSIGNLINLKKLDLSGCSSLVELPLSIGNLINLKTLNLSGCSS 1086
Query: 126 FVSLPTS--NYAMSKL 139
V LP+S N + KL
Sbjct: 1087 LVELPSSIGNLNLKKL 1102
Score = 64.3 bits (155), Expect = 3e-11
Identities = 52/147 (35%), Positives = 73/147 (49%), Gaps = 11/147 (7%)
Query: 12 LTQIPD-VSGLVNLEEFSFQGCVNLITVHDSIGLLGRLKTLRVMCCIKLRSIPPL--NLA 68
L ++P + L+NLE F F GC +L+ + SIG L LK L + L IP NL
Sbjct: 799 LVELPSSIGNLINLEAFYFHGCSSLLELPSSIGNLISLKILYLKRISSLVEIPSSIGNLI 858
Query: 69 SLEELDLSECSCLESFPEILGEMRNVTNLNLE-YTPIKELPFPFQNL-TLPKIYPCNC-G 125
+L+ L+LS CS L P +G + N+ L+L + + ELP NL L ++Y C
Sbjct: 859 NLKLLNLSGCSSLVELPSSIGNLINLKKLDLSGCSSLVELPLSIGNLINLQELYLSECSS 918
Query: 126 FVSLPTS-----NYAMSKLVECTIQAE 147
V LP+S N L EC+ E
Sbjct: 919 LVELPSSIGNLINLKTLNLSECSSLVE 945
Score = 63.9 bits (154), Expect = 4e-11
Identities = 49/150 (32%), Positives = 74/150 (48%), Gaps = 6/150 (4%)
Query: 12 LTQIP-DVSGLVNLEEFSFQGCVNLITVHDSIGLLGRLKTLRVMCCIKLRSIPPL--NLA 68
L ++P + L+NL+E C +L+ + SIG L LKTL + C L +P NL
Sbjct: 895 LVELPLSIGNLINLQELYLSECSSLVELPSSIGNLINLKTLNLSECSSLVELPSSIGNLI 954
Query: 69 SLEELDLSECSCLESFPEILGEMRNVTNLNLE-YTPIKELPFPFQNL-TLPKIYPCNC-G 125
+L+EL LSECS L P +G + N+ L+L + + ELP NL L + C
Sbjct: 955 NLQELYLSECSSLVELPSSIGNLINLKKLDLSGCSSLVELPLSIGNLINLKTLNLSECSS 1014
Query: 126 FVSLPTSNYAMSKLVECTIQAEQKVIPIQS 155
V LP+S + L E + ++ + S
Sbjct: 1015 LVELPSSIGNLINLQELYLSECSSLVELPS 1044
Score = 60.1 bits (144), Expect = 5e-10
Identities = 43/137 (31%), Positives = 68/137 (49%), Gaps = 5/137 (3%)
Query: 1 MRVLNFDHHGLLTQIPDVSGLVNLEEFSFQGCVNLITVHDSIGLLGRLKTLRVMCCIKLR 60
++V++ + L ++P++S +NL E C +LI + SIG +K+L + C L
Sbjct: 693 LKVMDLRYSSHLKELPNLSTAINLLEMVLSDCSSLIELPSSIGNATNIKSLDIQGCSSLL 752
Query: 61 SIPPL--NLASLEELDLSECSCLESFPEILGEMRNVTNLNL-EYTPIKELPFPFQNL-TL 116
+P NL +L LDL CS L P +G + N+ L+L + + ELP NL L
Sbjct: 753 KLPSSIGNLITLPRLDLMGCSSLVELPSSIGNLINLPRLDLMGCSSLVELPSSIGNLINL 812
Query: 117 PKIYPCNC-GFVSLPTS 132
Y C + LP+S
Sbjct: 813 EAFYFHGCSSLLELPSS 829
>At5g41740 disease resistance protein-like
Length = 1018
Score = 69.7 bits (169), Expect = 7e-13
Identities = 38/106 (35%), Positives = 64/106 (59%), Gaps = 3/106 (2%)
Query: 1 MRVLNFDHHGLLTQIPDVSGLVNLEEFSFQGCVNLITVHDSIGLLGRLKTLRVMCCIKLR 60
++++N + L +IP++S NLE + + C++L+ + SI L +L+ L V C L+
Sbjct: 586 LKIINLNRSYRLKEIPNLSKATNLERLTLESCLSLVELPSSISNLHKLEILDVKFCSMLQ 645
Query: 61 SIPP-LNLASLEELDLSECSCLESFPEILGEMRNVT--NLNLEYTP 103
IP +NLASLE LD+S CS L +FP+I ++ + N+ +E P
Sbjct: 646 VIPTNINLASLERLDVSGCSRLRTFPDISSNIKTLIFGNIKIEDVP 691
>At3g25510 unknown protein
Length = 1981
Score = 69.7 bits (169), Expect = 7e-13
Identities = 42/123 (34%), Positives = 63/123 (51%), Gaps = 4/123 (3%)
Query: 12 LTQIPDVSGLVNLEEFSFQGCVNLITVHDSIGLLGRLKTLRVMCCIKLRSIPPL--NLAS 69
L ++PD+S NLEE + CV+L+ V +G LG+L+ L + C + +P N+
Sbjct: 677 LKELPDLSTATNLEELILKYCVSLVKVPSCVGKLGKLQVLCLHGCTSILELPSFTKNVTG 736
Query: 70 LEELDLSECSCLESFPEILGEMRNVTNLNLEYTPIKELPFPFQNLTLPKIYPCN--CGFV 127
L+ LDL+ECS L P +G N+ NL+L + +LP T K + N V
Sbjct: 737 LQSLDLNECSSLVELPSSIGNAINLQNLDLGCLRLLKLPLSIVKFTNLKKFILNGCSSLV 796
Query: 128 SLP 130
LP
Sbjct: 797 ELP 799
Score = 67.4 bits (163), Expect = 3e-12
Identities = 40/99 (40%), Positives = 60/99 (60%), Gaps = 5/99 (5%)
Query: 12 LTQIPD-VSGLVNLEEFSFQGCVNLITVHDSIGLLGRLKTLRVMCCIKLRSIPP-LNLAS 69
L ++P + + NL+E + C NL+ + SIG L L TL + C KL ++P +NL S
Sbjct: 938 LVELPSSIGNITNLQELNLCNCSNLVKLPSSIGNLHLLFTLSLARCQKLEALPSNINLKS 997
Query: 70 LEELDLSECSCLESFPEILGEMRNVTNLNLEYTPIKELP 108
LE LDL++CS +SFPEI N+ L L+ T ++E+P
Sbjct: 998 LERLDLTDCSQFKSFPEI---STNIECLYLDGTAVEEVP 1033
Score = 52.8 bits (125), Expect = 8e-08
Identities = 39/126 (30%), Positives = 61/126 (47%), Gaps = 5/126 (3%)
Query: 12 LTQIPDVSGLVNLEEFSFQGCVNLITVHDSIGLLGRLKTLRVMCCIKLRSIPPL--NLAS 69
L ++P + NL+ C +L+ + SIG L+ L + C L +P N +
Sbjct: 795 LVELPFMGNATNLQNLDLGNCSSLVELPSSIGNAINLQNLDLSNCSSLVKLPSFIGNATN 854
Query: 70 LEELDLSECSCLESFPEILGEMRNVTNLNLE-YTPIKELPFPFQNLT-LPKIYPCNC-GF 126
LE LDL +CS L P +G + N+ L+L + + ELP N++ L + NC
Sbjct: 855 LEILDLRKCSSLVEIPTSIGHVTNLWRLDLSGCSSLVELPSSVGNISELQVLNLHNCSNL 914
Query: 127 VSLPTS 132
V LP+S
Sbjct: 915 VKLPSS 920
Score = 51.6 bits (122), Expect = 2e-07
Identities = 37/120 (30%), Positives = 57/120 (46%), Gaps = 4/120 (3%)
Query: 1 MRVLNFDHHGLLTQIPDVSGLV-NLEEFSFQGCVNLITVHDSIGLLGRLKTLRVMCCIKL 59
+ +L+ L +IP G V NL GC +L+ + S+G + L+ L + C L
Sbjct: 855 LEILDLRKCSSLVEIPTSIGHVTNLWRLDLSGCSSLVELPSSVGNISELQVLNLHNCSNL 914
Query: 60 RSIPPL--NLASLEELDLSECSCLESFPEILGEMRNVTNLNL-EYTPIKELPFPFQNLTL 116
+P + +L LDLS CS L P +G + N+ LNL + + +LP NL L
Sbjct: 915 VKLPSSFGHATNLWRLDLSGCSSLVELPSSIGNITNLQELNLCNCSNLVKLPSSIGNLHL 974
Score = 47.0 bits (110), Expect = 5e-06
Identities = 32/101 (31%), Positives = 47/101 (45%), Gaps = 11/101 (10%)
Query: 1 MRVLNFDHHGLLTQIPDVSGLVNLEEFSFQGCVNLITVHDSIGLLGRLKTLRVMCCIKLR 60
++ +N H L ++PD S NL+ GC +L+ + SIG L+ L + C L
Sbjct: 1869 LKWMNLFHSKNLKELPDFSTATNLQTLILCGCSSLVELPYSIGSANNLQKLHLCRCTSLV 1928
Query: 61 SIPPL--NLASLEELDLSECSCLESFPEILGEMRNVTNLNL 99
+P NL L+ + L CS LE P TN+NL
Sbjct: 1929 ELPASIGNLHKLQNVTLKGCSKLEVVP---------TNINL 1960
Score = 45.1 bits (105), Expect = 2e-05
Identities = 38/137 (27%), Positives = 63/137 (45%), Gaps = 6/137 (4%)
Query: 1 MRVLNFDHHGLLTQIPD-VSGLVNLEEFSFQGCVNLITVHDSIGLLGRLKTLRVMCCIKL 59
++ L+ + L ++P + +NL+ GC+ L+ + SI LK + C L
Sbjct: 737 LQSLDLNECSSLVELPSSIGNAINLQNLDL-GCLRLLKLPLSIVKFTNLKKFILNGCSSL 795
Query: 60 RSIPPL-NLASLEELDLSECSCLESFPEILGEMRNVTNLNL-EYTPIKELPFPFQNLTLP 117
+P + N +L+ LDL CS L P +G N+ NL+L + + +LP N T
Sbjct: 796 VELPFMGNATNLQNLDLGNCSSLVELPSSIGNAINLQNLDLSNCSSLVKLPSFIGNATNL 855
Query: 118 KIYPCN--CGFVSLPTS 132
+I V +PTS
Sbjct: 856 EILDLRKCSSLVEIPTS 872
>At5g18350 disease resistance protein -like
Length = 1193
Score = 69.3 bits (168), Expect = 9e-13
Identities = 50/168 (29%), Positives = 80/168 (46%), Gaps = 11/168 (6%)
Query: 1 MRVLNFDHHGLLTQIPDVSGLVNLEEFSFQGCVNLITVHDSIGLLGRLKTLRVMCCIKLR 60
++ ++ H L +IPD+S NLEE C L+ + DSIG LK L++ CC L+
Sbjct: 647 LKRMDLSHSKDLKEIPDLSNATNLEELDLSSCSGLLELTDSIGKATNLKRLKLACCSLLK 706
Query: 61 SIPPL--NLASLEELDLSECSCLESFPEILGEMRNVTNLNL-EYTPIKELPFPFQNLTLP 117
+P + +L+ LDL C E P+ +G++ N+ L L + LP + LP
Sbjct: 707 KLPSSIGDATNLQVLDLFHCESFEELPKSIGKLTNLKVLELMRCYKLVTLPNSIKTPKLP 766
Query: 118 KIYPCNC-GFVSLPTSNYAMSKLVECTIQAEQKVIPIQSLHVEYICLR 164
+ C + PT L +CT + K+ P S +V+ + LR
Sbjct: 767 VLSMSECEDLQAFPT----YINLEDCT---QLKMFPEISTNVKELDLR 807
Score = 34.7 bits (78), Expect = 0.024
Identities = 38/154 (24%), Positives = 62/154 (39%), Gaps = 42/154 (27%)
Query: 1 MRVLNFDHHGLLTQIPDVSG-LVNLEEFSFQGCVNLITVHDSIGLLGRLKTLRVMCCIKL 59
++VL+ H ++P G L NL+ C L+T+ +SI +L L + C L
Sbjct: 718 LQVLDLFHCESFEELPKSIGKLTNLKVLELMRCYKLVTLPNSIKT-PKLPVLSMSECEDL 776
Query: 60 RSIP----------------------------------PLNLAS---LEELDLSECSCLE 82
++ P P ++ S L LD+SEC L+
Sbjct: 777 QAFPTYINLEDCTQLKMFPEISTNVKELDLRNTAIENVPSSICSWSCLYRLDMSECRNLK 836
Query: 83 SFPEILGEMRNVTNLNLEYTPIKELPFPFQNLTL 116
FP + ++ L+L T I+E+P +NL L
Sbjct: 837 EFPNV---PVSIVELDLSKTEIEEVPSWIENLLL 867
Score = 28.1 bits (61), Expect = 2.2
Identities = 25/86 (29%), Positives = 40/86 (46%), Gaps = 9/86 (10%)
Query: 67 LASLEELDLSECSCLESFPEILGEMRNVTNLNLEYTPIKELPFPFQNLTLPKIYPCNCGF 126
L L ELD+S C L S P++ G + ++ N E ++ + FQN P+I F
Sbjct: 961 LPGLSELDVSGCRNLVSLPQLPGSLLSLDANNCE--SLERINGSFQN---PEIC---LNF 1012
Query: 127 VSLPTSNYAMSKLVECTIQAEQKVIP 152
+ N KL++ T E ++P
Sbjct: 1013 ANCINLNQEARKLIQ-TSACEYAILP 1037
>At4g16860 disease resistance RPP5 like protein
Length = 1256
Score = 68.6 bits (166), Expect = 1e-12
Identities = 44/105 (41%), Positives = 61/105 (57%), Gaps = 4/105 (3%)
Query: 12 LTQIPDVSGLVNLEEFSFQGCVNLITVHDSIGLLGRLKTLRVMCCIKLRSIP-PLNLASL 70
LT+IPD+S NL+ GC +L+T+ +IG L RL L + C L +P +NL+SL
Sbjct: 928 LTEIPDLSKATNLKRLYLNGCKSLVTLPSTIGNLHRLVRLEMKECTGLELLPTDVNLSSL 987
Query: 71 EELDLSECSCLESFPEILGEMRNVTNLNLEYTPIKELPFPFQNLT 115
LDLS CS L +FP I + L LE T I+E+P ++LT
Sbjct: 988 IILDLSGCSSLRTFPLI---STRIECLYLENTAIEEVPCCIEDLT 1029
Score = 55.8 bits (133), Expect = 1e-08
Identities = 34/77 (44%), Positives = 45/77 (58%), Gaps = 1/77 (1%)
Query: 12 LTQIPDVSGLVNLEEFSFQGCVNLITVHDSIGLLGRLKTLRVMCCIKLRSIP-PLNLASL 70
L +IPD+S +NLE GC +L+T+ SI +L L + C KL S P LNL SL
Sbjct: 768 LKEIPDLSLAINLERLYLFGCESLVTLPSSIQNATKLINLDMRDCKKLESFPTDLNLESL 827
Query: 71 EELDLSECSCLESFPEI 87
E L+L+ C L +FP I
Sbjct: 828 EYLNLTGCPNLRNFPAI 844
Score = 46.6 bits (109), Expect = 6e-06
Identities = 43/151 (28%), Positives = 72/151 (47%), Gaps = 7/151 (4%)
Query: 12 LTQIPDVSGLVNLEEFSFQGCVNLITVHDSIGLLGRLKTLRV--MCCIKLRSIPPLNLAS 69
L +IPD+S +NLEE + C +L+T+ SI +L+TL + I L+S+ + +
Sbjct: 631 LKEIPDLSLAINLEELNLSKCESLVTLPSSIQNAIKLRTLYCSGVLLIDLKSLE--GMCN 688
Query: 70 LEELDLSECSCLESFPEILGEMRNVTNLNLEYTPIKELPFPFQNLTLPKIYPCNCGFVSL 129
LE L + + S +E ++ R + L +Y P+K LP F+ L ++ N L
Sbjct: 689 LEYLSV-DWSSMEGTQGLIYLPRKLKRLWWDYCPVKRLPSNFKAEYLVELRMENSDLEKL 747
Query: 130 PTSNYAMSKLVECTIQAEQ--KVIPIQSLHV 158
+ L E + + K IP SL +
Sbjct: 748 WDGTQPLGSLKEMYLHGSKYLKEIPDLSLAI 778
>At5g40920 disease resistance protein-like
Length = 876
Score = 67.4 bits (163), Expect = 3e-12
Identities = 39/109 (35%), Positives = 62/109 (56%), Gaps = 4/109 (3%)
Query: 1 MRVLNFDHHGLLTQIPDVSGLVNLEEFSFQGCVNLITVHDSIGLLGRLKTLRVMCCIKLR 60
++ +N ++ L +IP++S NLE GC +L+ + SI L +L+ L C KL
Sbjct: 407 LKKINLEYSSNLKEIPNLSKATNLETLRLTGCESLMEIPSSISNLHKLEVLDASGCSKLH 466
Query: 61 SIP-PLNLASLEELDLSECSCLESFPEILGEMRNVTNLNLEYTPIKELP 108
IP +NL+SL+ + + +CS L SFP+I N+ L++ T IKE P
Sbjct: 467 VIPTKINLSSLKMVGMDDCSRLRSFPDI---STNIKILSIRGTKIKEFP 512
>At4g16950 disease resistance RPP5 like protein
Length = 1317
Score = 67.0 bits (162), Expect = 4e-12
Identities = 46/123 (37%), Positives = 69/123 (55%), Gaps = 5/123 (4%)
Query: 12 LTQIPDVSGLVNLEEFSFQGCVNLITVHDSIGLLGRLKTLRVMCCIKLRSIP-PLNLASL 70
LT+IPD+S NL+ C +L+T+ +IG L +L L + C L +P +NL+SL
Sbjct: 929 LTEIPDLSKATNLKHLYLNNCKSLVTLPSTIGNLQKLVRLEMKECTGLEVLPTDVNLSSL 988
Query: 71 EELDLSECSCLESFPEILGEMRNVTNLNLEYTPIKELPFPFQNLTLPKIYPCNC-GFVSL 129
E LDLS CS L +FP I +++ L LE T I+E+ + L + NC V+L
Sbjct: 989 ETLDLSGCSSLRTFPLI---SKSIKWLYLENTAIEEILDLSKATKLESLILNNCKSLVTL 1045
Query: 130 PTS 132
P++
Sbjct: 1046 PST 1048
Score = 58.5 bits (140), Expect = 2e-09
Identities = 40/114 (35%), Positives = 59/114 (51%), Gaps = 4/114 (3%)
Query: 12 LTQIPDVSGLVNLEEFSFQGCVNLITVHDSIGLLGRLKTLRVMCCIKLRSIP-PLNLASL 70
+ +I D+S LE C +L+T+ +IG L L+ L + C L +P +NL+SL
Sbjct: 1019 IEEILDLSKATKLESLILNNCKSLVTLPSTIGNLQNLRRLYMKRCTGLEVLPTDVNLSSL 1078
Query: 71 EELDLSECSCLESFPEILGEMRNVTNLNLEYTPIKELPFPFQNLTLPKIYPCNC 124
LDLS CS L +FP I N+ L LE T I E+P ++ T ++ C
Sbjct: 1079 GILDLSGCSSLRTFPLI---STNIVWLYLENTAIGEVPCCIEDFTRLRVLLMYC 1129
Score = 52.4 bits (124), Expect = 1e-07
Identities = 33/77 (42%), Positives = 44/77 (56%), Gaps = 1/77 (1%)
Query: 12 LTQIPDVSGLVNLEEFSFQGCVNLITVHDSIGLLGRLKTLRVMCCIKLRSIP-PLNLASL 70
L +IPD+S +NLEE C +L+T S+ +L L + C KL S P LNL SL
Sbjct: 769 LKEIPDLSLAINLEEVDICKCESLVTFPSSMQNAIKLIYLDISDCKKLESFPTDLNLESL 828
Query: 71 EELDLSECSCLESFPEI 87
E L+L+ C L +FP I
Sbjct: 829 EYLNLTGCPNLRNFPAI 845
Score = 41.2 bits (95), Expect = 3e-04
Identities = 42/151 (27%), Positives = 70/151 (45%), Gaps = 7/151 (4%)
Query: 12 LTQIPDVSGLVNLEEFSFQGCVNLITVHDSIGLLGRLKTLRV--MCCIKLRSIPPLNLAS 69
L +IPD+S NLEE +GC +L+T+ SI +L+ L + I L+S+ + +
Sbjct: 632 LKEIPDLSNARNLEELDLEGCESLVTLPSSIQNAIKLRKLHCSGVILIDLKSLE--GMCN 689
Query: 70 LEELDLSECSCLESFPEILGEMRNVTNLNLEYTPIKELPFPFQNLTLPKIYPCNCGFVSL 129
LE L + +CS +E I+ + L P+K L F+ L K+ N L
Sbjct: 690 LEYLSV-DCSRVEGTQGIVYFPSKLRLLLWNNCPLKRLHSNFKVEYLVKLRMENSDLEKL 748
Query: 130 PTSNYAMSKLVECTIQAEQ--KVIPIQSLHV 158
+ +L + ++ + K IP SL +
Sbjct: 749 WDGTQPLGRLKQMFLRGSKYLKEIPDLSLAI 779
Score = 37.0 bits (84), Expect = 0.005
Identities = 19/42 (45%), Positives = 25/42 (59%), Gaps = 1/42 (2%)
Query: 45 LGRLKTLRVMCCIKLRSIPPL-NLASLEELDLSECSCLESFP 85
LG LK + ++C L+ IP L N +LEELDL C L + P
Sbjct: 618 LGSLKKMNLLCSKNLKEIPDLSNARNLEELDLEGCESLVTLP 659
Score = 28.1 bits (61), Expect = 2.2
Identities = 29/102 (28%), Positives = 50/102 (48%), Gaps = 5/102 (4%)
Query: 66 NLASLEELDLSECSCLESFPEILGEMRNVTNLNL-EYTPIKELPFPFQNL-TLPKIYPCN 123
+L SLEE+DLSE L P+ L + N+ +L L + LP NL L ++
Sbjct: 914 SLGSLEEMDLSESENLTEIPD-LSKATNLKHLYLNNCKSLVTLPSTIGNLQKLVRLEMKE 972
Query: 124 C-GFVSLPTS-NYAMSKLVECTIQAEQKVIPIQSLHVEYICL 163
C G LPT N + + ++ + + + P+ S ++++ L
Sbjct: 973 CTGLEVLPTDVNLSSLETLDLSGCSSLRTFPLISKSIKWLYL 1014
Score = 26.6 bits (57), Expect = 6.4
Identities = 13/34 (38%), Positives = 20/34 (58%), Gaps = 2/34 (5%)
Query: 47 RLKTLRVMCCIKLRSIPP--LNLASLEELDLSEC 78
RL+ L + CC +L++I P L SL D ++C
Sbjct: 1121 RLRVLLMYCCQRLKNISPNIFRLRSLMFADFTDC 1154
>At4g16920 disease resistance RPP5 like protein
Length = 1304
Score = 67.0 bits (162), Expect = 4e-12
Identities = 46/123 (37%), Positives = 69/123 (55%), Gaps = 5/123 (4%)
Query: 12 LTQIPDVSGLVNLEEFSFQGCVNLITVHDSIGLLGRLKTLRVMCCIKLRSIP-PLNLASL 70
LT+IPD+S NL+ C +L+T+ +IG L +L L + C L +P +NL+SL
Sbjct: 923 LTEIPDLSKATNLKHLYLNNCKSLVTLPSTIGNLQKLVRLEMKECTGLEVLPTDVNLSSL 982
Query: 71 EELDLSECSCLESFPEILGEMRNVTNLNLEYTPIKELPFPFQNLTLPKIYPCNC-GFVSL 129
E LDLS CS L +FP I +++ L LE T I+E+ + L + NC V+L
Sbjct: 983 ETLDLSGCSSLRTFPLI---SKSIKWLYLENTAIEEILDLSKATKLESLILNNCKSLVTL 1039
Query: 130 PTS 132
P++
Sbjct: 1040 PST 1042
Score = 58.5 bits (140), Expect = 2e-09
Identities = 40/114 (35%), Positives = 59/114 (51%), Gaps = 4/114 (3%)
Query: 12 LTQIPDVSGLVNLEEFSFQGCVNLITVHDSIGLLGRLKTLRVMCCIKLRSIP-PLNLASL 70
+ +I D+S LE C +L+T+ +IG L L+ L + C L +P +NL+SL
Sbjct: 1013 IEEILDLSKATKLESLILNNCKSLVTLPSTIGNLQNLRRLYMKRCTGLEVLPTDVNLSSL 1072
Query: 71 EELDLSECSCLESFPEILGEMRNVTNLNLEYTPIKELPFPFQNLTLPKIYPCNC 124
LDLS CS L +FP I N+ L LE T I E+P ++ T ++ C
Sbjct: 1073 GILDLSGCSSLRTFPLI---STNIVWLYLENTAIGEVPCCIEDFTRLRVLLMYC 1123
Score = 52.4 bits (124), Expect = 1e-07
Identities = 33/77 (42%), Positives = 44/77 (56%), Gaps = 1/77 (1%)
Query: 12 LTQIPDVSGLVNLEEFSFQGCVNLITVHDSIGLLGRLKTLRVMCCIKLRSIP-PLNLASL 70
L +IPD+S +NLEE C +L+T S+ +L L + C KL S P LNL SL
Sbjct: 763 LKEIPDLSLAINLEEVDICKCESLVTFPSSMQNAIKLIYLDISDCKKLESFPTDLNLESL 822
Query: 71 EELDLSECSCLESFPEI 87
E L+L+ C L +FP I
Sbjct: 823 EYLNLTGCPNLRNFPAI 839
Score = 40.4 bits (93), Expect = 4e-04
Identities = 42/162 (25%), Positives = 75/162 (45%), Gaps = 7/162 (4%)
Query: 1 MRVLNFDHHGLLTQIPDVSGLVNLEEFSFQGCVNLITVHDSIGLLGRLKTLRV--MCCIK 58
++ +N + +IPD+S +NLEE + C +L+T+ SI +L+TL + I
Sbjct: 615 LKKMNLWYSKYFKEIPDLSLAINLEELNLSECESLVTLPSSIQNAIKLRTLYCSGVLLID 674
Query: 59 LRSIPPLNLASLEELDLSECSCLESFPEILGEMRNVTNLNLEYTPIKELPFPFQNLTLPK 118
L+S+ + +LE L + +CS +E I+ + L P+K L F+ L K
Sbjct: 675 LKSLE--GMCNLEYLSV-DCSRMEGTQGIVYFPSKLRLLLWNNCPLKRLHSNFKVEYLVK 731
Query: 119 IYPCNCGFVSLPTSNYAMSKLVECTIQAEQ--KVIPIQSLHV 158
+ N L + +L + ++ + K IP SL +
Sbjct: 732 LRMENSDLEKLWDGTQPLGRLKQMFLRGSKYLKEIPDLSLAI 773
Score = 28.1 bits (61), Expect = 2.2
Identities = 29/102 (28%), Positives = 50/102 (48%), Gaps = 5/102 (4%)
Query: 66 NLASLEELDLSECSCLESFPEILGEMRNVTNLNL-EYTPIKELPFPFQNL-TLPKIYPCN 123
+L SLEE+DLSE L P+ L + N+ +L L + LP NL L ++
Sbjct: 908 SLGSLEEMDLSESENLTEIPD-LSKATNLKHLYLNNCKSLVTLPSTIGNLQKLVRLEMKE 966
Query: 124 C-GFVSLPTS-NYAMSKLVECTIQAEQKVIPIQSLHVEYICL 163
C G LPT N + + ++ + + + P+ S ++++ L
Sbjct: 967 CTGLEVLPTDVNLSSLETLDLSGCSSLRTFPLISKSIKWLYL 1008
Score = 26.6 bits (57), Expect = 6.4
Identities = 13/34 (38%), Positives = 20/34 (58%), Gaps = 2/34 (5%)
Query: 47 RLKTLRVMCCIKLRSIPP--LNLASLEELDLSEC 78
RL+ L + CC +L++I P L SL D ++C
Sbjct: 1115 RLRVLLMYCCQRLKNISPNIFRLRSLMFADFTDC 1148
>At3g44480 disease resistance protein -like
Length = 1220
Score = 66.6 bits (161), Expect = 6e-12
Identities = 42/110 (38%), Positives = 65/110 (58%), Gaps = 5/110 (4%)
Query: 1 MRVLNFDHHGLLTQIPDVSG-LVNLEEFSFQGCVNLITVHDSIGLLGRLKTLRVMCCIKL 59
++ LN L ++P G + +LE F C +L+T+ SIG L L L + C KL
Sbjct: 810 LKQLNISGCSSLVKLPSSIGDITDLEVFDLSNCSSLVTLPSSIGNLQNLCKLIMRGCSKL 869
Query: 60 RSIP-PLNLASLEELDLSECSCLESFPEILGEMRNVTNLNLEYTPIKELP 108
++P +NL SL+ L+L++CS L+SFPEI +++ L L+ T IKE+P
Sbjct: 870 EALPININLKSLDTLNLTDCSQLKSFPEI---STHISELRLKGTAIKEVP 916
Score = 58.9 bits (141), Expect = 1e-09
Identities = 39/136 (28%), Positives = 69/136 (50%), Gaps = 4/136 (2%)
Query: 1 MRVLNFDHHGLLTQIPDVSGLVNLEEFSFQGCVNLITVHDSIGLLGRLKTLRVMCCIKLR 60
++ ++ + L ++P++S NLEE + C +L+ + SI L L+ L + C L
Sbjct: 716 LKWMDLSYSSYLKELPNLSTATNLEELKLRNCSSLVELPSSIEKLTSLQILDLENCSSLE 775
Query: 61 SIPPL-NLASLEELDLSECSCLESFPEILGEMRNVTNLNLE-YTPIKELPFPFQNLTLPK 118
+P + N L EL L CS L P +G N+ LN+ + + +LP ++T +
Sbjct: 776 KLPAIENATKLRELKLQNCSSLIELPLSIGTATNLKQLNISGCSSLVKLPSSIGDITDLE 835
Query: 119 IYP-CNC-GFVSLPTS 132
++ NC V+LP+S
Sbjct: 836 VFDLSNCSSLVTLPSS 851
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.324 0.142 0.430
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,724,500
Number of Sequences: 26719
Number of extensions: 150449
Number of successful extensions: 1437
Number of sequences better than 10.0: 286
Number of HSP's better than 10.0 without gapping: 203
Number of HSP's successfully gapped in prelim test: 83
Number of HSP's that attempted gapping in prelim test: 557
Number of HSP's gapped (non-prelim): 663
length of query: 164
length of database: 11,318,596
effective HSP length: 92
effective length of query: 72
effective length of database: 8,860,448
effective search space: 637952256
effective search space used: 637952256
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 56 (26.2 bits)
Medicago: description of AC145512.8


