

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC145512.4 + phase: 0 /pseudo
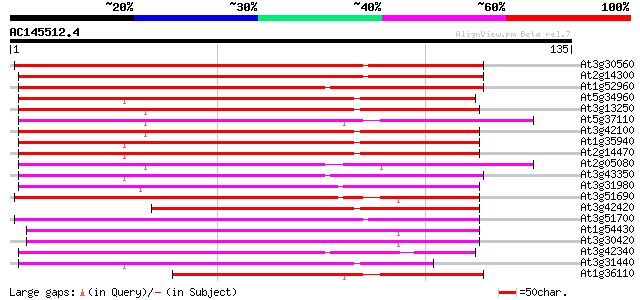
(135 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g30560 hypothetical protein 93 4e-20
At2g14300 pseudogene; similar to MURA transposase of maize Muta... 92 1e-19
At1g52960 hypothetical protein 91 2e-19
At5g34960 putative protein 87 2e-18
At3g13250 hypothetical protein 81 2e-16
At5g37110 putative helicase 80 2e-16
At3g42100 putative protein 80 3e-16
At1g35940 hypothetical protein 80 3e-16
At2g14470 pseudogene 80 4e-16
At2g05080 putative helicase 78 1e-15
At3g43350 putative protein 75 8e-15
At3g31980 hypothetical protein 75 8e-15
At3g51690 putative protein 70 4e-13
At3g42420 putative protein 69 6e-13
At3g51700 unknown protein 68 2e-12
At1g54430 hypothetical protein 67 2e-12
At3g30420 hypothetical protein 65 1e-11
At3g42340 putative protein 64 3e-11
At3g31440 hypothetical protein 60 3e-10
At1g36110 hypothetical protein 58 2e-09
>At3g30560 hypothetical protein
Length = 1473
Score = 93.2 bits (230), Expect = 4e-20
Identities = 52/113 (46%), Positives = 71/113 (62%), Gaps = 1/113 (0%)
Query: 2 KVEIPEDLLISTTTNLLMSLIDFVYPDLNDNLGDQLFFQERGILAPTLDSVEHVNEFMMS 61
+++I +D+LI N + S+I VY D D FFQ+R IL PT D V +N+ M+S
Sbjct: 1220 QIQIRDDILIPEGDNPIESIIKAVYGTSFDEERDPKFFQDRAILCPTNDDVNSINDHMLS 1279
Query: 62 LIPGEEKEYLTSNSIFRSGENSDV*SEWFTPEFLNGIKSSGIPNHRLKLKVGC 114
+ GEEK Y +S+SI S +D + +TP+FLN IK SG+PNH L LKVGC
Sbjct: 1280 KLTGEEKIYRSSDSIDPSDTRADK-NPVYTPDFLNKIKISGLPNHLLWLKVGC 1331
>At2g14300 pseudogene; similar to MURA transposase of maize Mutator
transposon
Length = 1230
Score = 91.7 bits (226), Expect = 1e-19
Identities = 50/112 (44%), Positives = 69/112 (60%), Gaps = 1/112 (0%)
Query: 3 VEIPEDLLISTTTNLLMSLIDFVYPDLNDNLGDQLFFQERGILAPTLDSVEHVNEFMMSL 62
++IP+D+LI N + S+I VY D FFQ + IL PT D V +N+ M+S
Sbjct: 1020 IQIPDDILIPEGDNPIESIIKAVYGTTFAQKRDPKFFQHKAILCPTNDDVNSINDHMLSK 1079
Query: 63 IPGEEKEYLTSNSIFRSGENSDV*SEWFTPEFLNGIKSSGIPNHRLKLKVGC 114
+ GEE+ Y +SNSI S +D + +TP+FLN IK SG+ NH L+LKVGC
Sbjct: 1080 LTGEERIYRSSNSIDPSDTRADK-NPIYTPDFLNKIKISGLANHLLRLKVGC 1130
>At1g52960 hypothetical protein
Length = 924
Score = 90.9 bits (224), Expect = 2e-19
Identities = 47/112 (41%), Positives = 73/112 (64%), Gaps = 1/112 (0%)
Query: 3 VEIPEDLLISTTTNLLMSLIDFVYPDLNDNLGDQLFFQERGILAPTLDSVEHVNEFMMSL 62
++IPE+ LI+ + + S+I+ VY + D FFQ R IL PT + V +NE MMS+
Sbjct: 696 IDIPEEFLINGDNDPVESIIEAVYGNTFMEEKDPKFFQGRAILCPTNEDVNSINEHMMSM 755
Query: 63 IPGEEKEYLTSNSIFRSGENSDV*SEWFTPEFLNGIKSSGIPNHRLKLKVGC 114
+ GEE+ YL+S+SI + S ++ ++ +FLN ++ SG+PNH L+LKVGC
Sbjct: 756 LDGEERIYLSSDSI-DPADTSSANNDAYSADFLNSVRVSGLPNHCLRLKVGC 806
>At5g34960 putative protein
Length = 1033
Score = 87.4 bits (215), Expect = 2e-18
Identities = 47/112 (41%), Positives = 73/112 (64%), Gaps = 3/112 (2%)
Query: 3 VEIPEDLLISTTTNLLMSLIDFVY--PDLNDNLGDQLFFQERGILAPTLDSVEHVNEFMM 60
++IPEDLLI+ T + S+ + +Y P + + D FFQ R ILAPT + V +NE+++
Sbjct: 778 IDIPEDLLITNTDKPIESITNEIYGDPKILHEITDPKFFQGRAILAPTNEDVNTINEYLL 837
Query: 61 SLIPGEEKEYLTSNSIFRSGENSDV*SEWFTPEFLNGIKSSGIPNHRLKLKV 112
+ EE+ YL+++SI + NS + + TP+FLN IK +G+PNH L+LKV
Sbjct: 838 EQLHAEERIYLSADSIDPTDSNS-LNNPVITPDFLNSIKLAGLPNHSLRLKV 888
>At3g13250 hypothetical protein
Length = 1419
Score = 80.9 bits (198), Expect = 2e-16
Identities = 45/113 (39%), Positives = 70/113 (61%), Gaps = 3/113 (2%)
Query: 3 VEIPEDLLISTTTNLLMSLIDFVYPDLND--NLGDQLFFQERGILAPTLDSVEHVNEFMM 60
++IPE+LLI N + ++ +Y D + D FFQ R ILAP + V +N++M+
Sbjct: 1165 IDIPEELLIQEADNPIEAISREIYGDPTKLHEISDPKFFQRRAILAPKNEDVNTINQYML 1224
Query: 61 SLIPGEEKEYLTSNSIFRSGENSDV*SEWFTPEFLNGIKSSGIPNHRLKLKVG 113
+ EE+ YL+++SI S +S + + TP+FLN IK SG+P+H L+LKVG
Sbjct: 1225 EHLDSEERIYLSADSIDPSDSDS-LKNPVITPDFLNSIKVSGMPHHSLRLKVG 1276
>At5g37110 putative helicase
Length = 1307
Score = 80.5 bits (197), Expect = 2e-16
Identities = 45/129 (34%), Positives = 75/129 (57%), Gaps = 9/129 (6%)
Query: 3 VEIPEDLLISTTTNLLMSLIDFVYPDLND--NLGDQLFFQERGILAPTLDSVEHVNEFMM 60
+ IP + LI+ + + ++ +Y D+ D +FFQE+ IL PT + V +NE M+
Sbjct: 1063 IVIPSEFLITKAKDPIEAICTEIYGDITKIHEQNDPIFFQEKAILCPTNEDVNQINETML 1122
Query: 61 SLIPGEEKEYLTSNSIFRS---GENSDV*SEWFTPEFLNGIKSSGIPNHRLKLKVGCL*G 117
+ GEE +L+S+S+ + G+N+ TP+FLN +K S +PNH+L+LK+GC
Sbjct: 1123 DNLQGEEFTFLSSDSLDPADIGGKNNPA----LTPDFLNSVKVSRLPNHKLRLKIGCPVM 1178
Query: 118 WQLDPDPMG 126
+ DP+G
Sbjct: 1179 LLRNIDPIG 1187
>At3g42100 putative protein
Length = 1752
Score = 80.1 bits (196), Expect = 3e-16
Identities = 43/113 (38%), Positives = 73/113 (64%), Gaps = 3/113 (2%)
Query: 3 VEIPEDLLISTTTNLLMSLIDFVYPDLND--NLGDQLFFQERGILAPTLDSVEHVNEFMM 60
++IPE+LLI N + ++ +Y D ++ + D FFQ R ILAPT + V +N++M+
Sbjct: 1497 IDIPEELLIKEAGNPIEAISKEIYGDPSELHMINDPKFFQRRAILAPTNEDVNTINQYML 1556
Query: 61 SLIPGEEKEYLTSNSIFRSGENSDV*SEWFTPEFLNGIKSSGIPNHRLKLKVG 113
+ EE+ YL+++SI + +S + + TP+FLN I+ +G+P+H L+LKVG
Sbjct: 1557 EHLKSEERIYLSADSIDPTDSDS-LANPVITPDFLNSIQLTGMPHHALRLKVG 1608
>At1g35940 hypothetical protein
Length = 1678
Score = 80.1 bits (196), Expect = 3e-16
Identities = 44/113 (38%), Positives = 70/113 (61%), Gaps = 3/113 (2%)
Query: 3 VEIPEDLLISTTTNLLMSLIDFVY--PDLNDNLGDQLFFQERGILAPTLDSVEHVNEFMM 60
++IPEDLLI+ + ++ + +Y P + + D FFQ R ILAP + V +NE+++
Sbjct: 1423 IDIPEDLLITNADKPIETITNEIYGDPKILHEITDPKFFQGRAILAPKNEDVNTINEYLL 1482
Query: 61 SLIPGEEKEYLTSNSIFRSGENSDV*SEWFTPEFLNGIKSSGIPNHRLKLKVG 113
+ EE+ YL+++SI + +S + + TP+FLN IK G+PNH L LKVG
Sbjct: 1483 EQLDAEERIYLSADSIDPTDSDS-LNNPVITPDFLNSIKLPGLPNHSLCLKVG 1534
>At2g14470 pseudogene
Length = 1265
Score = 79.7 bits (195), Expect = 4e-16
Identities = 44/113 (38%), Positives = 70/113 (61%), Gaps = 3/113 (2%)
Query: 3 VEIPEDLLISTTTNLLMSLIDFVY--PDLNDNLGDQLFFQERGILAPTLDSVEHVNEFMM 60
++IPEDLLI+ + S+ + +Y P + + D FFQ R ILA + V +NE+++
Sbjct: 1027 IDIPEDLLITNADKPIESITNEIYGDPKILHEITDPKFFQGRAILASKNEDVNTINEYLL 1086
Query: 61 SLIPGEEKEYLTSNSIFRSGENSDV*SEWFTPEFLNGIKSSGIPNHRLKLKVG 113
+ EE+ YL+++SI + +S + + TP+FLN IK G+PNH L+LKVG
Sbjct: 1087 DQLHAEERIYLSADSIDPTDSDS-LSNPVITPDFLNSIKLPGLPNHSLRLKVG 1138
>At2g05080 putative helicase
Length = 1219
Score = 78.2 bits (191), Expect = 1e-15
Identities = 45/129 (34%), Positives = 74/129 (56%), Gaps = 9/129 (6%)
Query: 3 VEIPEDLLISTTTNLLMSLIDFVYPDLND--NLGDQLFFQERGILAPTLDSVEHVNEFMM 60
++IP + LI+ + + ++ +Y D+ D +FFQER IL PT + V +NE M+
Sbjct: 958 IDIPSEFLITKAKDPIQAICTEIYGDITKIHEQKDPVFFQERAILCPTNEDVNQINETML 1017
Query: 61 SLIPGEEKEYLTSNSIFRSGENSDV*SE---WFTPEFLNGIKSSGIPNHRLKLKVGCL*G 117
+ GEE +L+S+S+ + +D+ S TPEFLN +K G+ NH+L+LK+G
Sbjct: 1018 DNLQGEELTFLSSDSL----DTADIGSRNNPVLTPEFLNNVKVLGLSNHKLRLKIGSPVM 1073
Query: 118 WQLDPDPMG 126
+ DP+G
Sbjct: 1074 LLRNIDPIG 1082
>At3g43350 putative protein
Length = 830
Score = 75.5 bits (184), Expect = 8e-15
Identities = 43/117 (36%), Positives = 69/117 (58%), Gaps = 6/117 (5%)
Query: 3 VEIPEDLLISTTTNLLMSLIDFVY-----PDLNDNLGDQLFFQERGILAPTLDSVEHVNE 57
++IPE+ LI+ + + S+I+ VY + + D +Q R IL PT + V +NE
Sbjct: 400 IDIPEEFLINGDNDPVESIIEAVYGNTFMEEKDPKKTDYPQYQGRAILCPTNEDVNSINE 459
Query: 58 FMMSLIPGEEKEYLTSNSIFRSGENSDV*SEWFTPEFLNGIKSSGIPNHRLKLKVGC 114
MM ++ GEE+ YL+S+SI + S + + +FLN ++ G+PNH L+LKVGC
Sbjct: 460 HMMRMLDGEERIYLSSDSI-DPADISSANNAAYLADFLNNVRVYGLPNHCLRLKVGC 515
>At3g31980 hypothetical protein
Length = 1099
Score = 75.5 bits (184), Expect = 8e-15
Identities = 43/113 (38%), Positives = 66/113 (58%), Gaps = 3/113 (2%)
Query: 3 VEIPEDLLISTTTNLLMSLIDFVYPDLN--DNLGDQLFFQERGILAPTLDSVEHVNEFMM 60
++IPE+LLI + + ++ VY DL+ D FFQ+R IL P V +N+ M+
Sbjct: 843 IDIPEELLIKDANDPIEAITKAVYGDLDLLQPNNDPKFFQQRAILCPRNTDVNTINDIML 902
Query: 61 SLIPGEEKEYLTSNSIFRSGENSDV*SEWFTPEFLNGIKSSGIPNHRLKLKVG 113
+ GE YL+++SI + + + + TP+FLN IK SG+PNH L LK+G
Sbjct: 903 DKLNGELVTYLSADSIDPQ-DAASLNNPVLTPDFLNSIKLSGLPNHNLTLKIG 954
>At3g51690 putative protein
Length = 374
Score = 69.7 bits (169), Expect = 4e-13
Identities = 43/113 (38%), Positives = 70/113 (61%), Gaps = 6/113 (5%)
Query: 2 KVEIPEDLLISTTTNLLMSLIDFVYPDLNDNLGDQLFFQERGILAPTLDSVEHVNEFMMS 61
+++I +DLLI+ + + + +L+ VY + + F + IL D V+ +N++M+S
Sbjct: 80 EIDISKDLLITESKDPIKTLLKEVYGEYFAKSYNPDFCHDSAILCHRDDDVDQINDYMLS 139
Query: 62 LIPGEEKEYLTSNSIFRSGENSDV*SEWFTP-EFLNGIKSSGIPNHRLKLKVG 113
L+PGEEKE L+++SI S N D+ F P E LN IK G+P+ +L+LKVG
Sbjct: 140 LLPGEEKECLSTDSISPS-PNDDM----FVPLEVLNSIKVPGLPDFKLRLKVG 187
>At3g42420 putative protein
Length = 1018
Score = 69.3 bits (168), Expect = 6e-13
Identities = 35/79 (44%), Positives = 56/79 (70%), Gaps = 1/79 (1%)
Query: 35 DQLFFQERGILAPTLDSVEHVNEFMMSLIPGEEKEYLTSNSIFRSGENSDV*SEWFTPEF 94
DQ FF +R IL+PT D V +N++M++ + GEE+ +L+++SI S +S + + TP+
Sbjct: 859 DQKFFLKRVILSPTNDDVHTINQYMLNNMEGEERIFLSADSIDPSDSDS-LKNPVITPDL 917
Query: 95 LNGIKSSGIPNHRLKLKVG 113
LN IK SG+P+H L+LK+G
Sbjct: 918 LNSIKLSGLPHHELRLKIG 936
>At3g51700 unknown protein
Length = 344
Score = 67.8 bits (164), Expect = 2e-12
Identities = 38/112 (33%), Positives = 67/112 (58%), Gaps = 1/112 (0%)
Query: 2 KVEIPEDLLISTTTNLLMSLIDFVYPDLNDNLGDQLFFQERGILAPTLDSVEHVNEFMMS 61
K++I EDLLI+ + + +++D VY + + F+QER IL T D + +N++M+S
Sbjct: 81 KIDIHEDLLITECKDPIKTIVDEVYGESFTESYNPDFYQERAILCHTNDVADEINDYMLS 140
Query: 62 LIPGEEKEYLTSNSIFRSGENSDV*SEWFTPEFLNGIKSSGIPNHRLKLKVG 113
+ GEE + +++I+ + + + + EFLN IK G P+ +L+LKVG
Sbjct: 141 QLQGEETKCYGADTIYPTHASPND-KMLYPLEFLNSIKIPGFPDFKLRLKVG 191
>At1g54430 hypothetical protein
Length = 1639
Score = 67.4 bits (163), Expect = 2e-12
Identities = 38/110 (34%), Positives = 63/110 (56%), Gaps = 1/110 (0%)
Query: 5 IPEDLLISTTTNLLMSLIDFVYPDLNDNLGDQLFFQERGILAPTLDSVEHVNEFMMSLIP 64
I ++LL+ T N L L V+PD + D + +L P ++V+ +N++++S +P
Sbjct: 1379 IDKNLLLPETENPLEVLCRSVFPDFTNTFQDLENLKGTAVLTPRNETVDEINDYLLSKVP 1438
Query: 65 GEEKEYLTSNSIFRSGENSDV*SEWFTP-EFLNGIKSSGIPNHRLKLKVG 113
G KEY +++SI R ++ E P E+LN ++ G+P HRL LKVG
Sbjct: 1439 GLAKEYFSADSIDRDEALTEEGFEMSYPMEYLNSLEFPGLPAHRLCLKVG 1488
>At3g30420 hypothetical protein
Length = 837
Score = 64.7 bits (156), Expect = 1e-11
Identities = 37/110 (33%), Positives = 62/110 (55%), Gaps = 1/110 (0%)
Query: 5 IPEDLLISTTTNLLMSLIDFVYPDLNDNLGDQLFFQERGILAPTLDSVEHVNEFMMSLIP 64
I ++LL+ T N L L V PD + D + +L P ++V+ +N++++S +P
Sbjct: 577 IDKNLLLPETENPLEVLCQSVSPDFTNTFQDLENLKGTAVLTPRNETVDEINDYLLSKVP 636
Query: 65 GEEKEYLTSNSIFRSGENSDV*SEWFTP-EFLNGIKSSGIPNHRLKLKVG 113
G KEY +++SI + ++ E P E+LN ++ G+P HRL LKVG
Sbjct: 637 GLAKEYFSADSIDQDEALTEEGFEMSYPMEYLNSLEFPGLPAHRLCLKVG 686
>At3g42340 putative protein
Length = 244
Score = 63.5 bits (153), Expect = 3e-11
Identities = 39/110 (35%), Positives = 64/110 (57%), Gaps = 4/110 (3%)
Query: 3 VEIPEDLLISTTTNLLMSLIDFVYPDLNDNLGDQLFFQERGILAPTLDSVEHVNEFMMSL 62
++I E+ I+ + + S+I+ VY + D+ FFQ R IL PT + V +NE MMS+
Sbjct: 114 IDIREEFFINGDNDPVESIIEAVYGNTFMEEKDRKFFQGRVILCPTNEDVNSINEHMMSM 173
Query: 63 IPGEEKEYLTSNSIFRSGENSDV*SEWFTPEFLNGIKSSGIPNHRLKLKV 112
+ GEE+ YL+SNSI + S ++ ++ +F K + P+H L+ KV
Sbjct: 174 LDGEERIYLSSNSI-DPADTSSANNDAYSADF---FKQNMDPHHTLRYKV 219
>At3g31440 hypothetical protein
Length = 536
Score = 60.5 bits (145), Expect = 3e-10
Identities = 35/102 (34%), Positives = 60/102 (58%), Gaps = 3/102 (2%)
Query: 3 VEIPEDLLISTTTNLLMSLIDFVY--PDLNDNLGDQLFFQERGILAPTLDSVEHVNEFMM 60
++IP+DLLI+ + + + +Y P + + D FFQ R ILAP + V +NE+++
Sbjct: 305 IDIPKDLLITNADKPIEWITNEIYGDPKILHEITDPKFFQGRAILAPKNEDVNTINEYLL 364
Query: 61 SLIPGEEKEYLTSNSIFRSGENSDV*SEWFTPEFLNGIKSSG 102
+ EE+ YL+++SI + +S + + TP+FLN IK G
Sbjct: 365 EQLHAEERIYLSADSIDPTDSDS-LNNPVITPDFLNSIKLPG 405
>At1g36110 hypothetical protein
Length = 745
Score = 57.8 bits (138), Expect = 2e-09
Identities = 33/78 (42%), Positives = 48/78 (61%), Gaps = 7/78 (8%)
Query: 40 QERGILAPTLDSVEHVNEFMMSLIPGEEKEYLTSNSIFRS---GENSDV*SEWFTPEFLN 96
+E + P + + + M+S + GEE+ Y +S+SI S G+N+ V +TP+FLN
Sbjct: 659 KELSLCVPPMTMLTQLTHHMLSKLTGEERIYRSSDSIDPSDTRGDNNLV----YTPDFLN 714
Query: 97 GIKSSGIPNHRLKLKVGC 114
IK SG+PNH L LKVGC
Sbjct: 715 KIKFSGVPNHILWLKVGC 732
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.325 0.144 0.451
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,135,422
Number of Sequences: 26719
Number of extensions: 125169
Number of successful extensions: 293
Number of sequences better than 10.0: 34
Number of HSP's better than 10.0 without gapping: 30
Number of HSP's successfully gapped in prelim test: 4
Number of HSP's that attempted gapping in prelim test: 233
Number of HSP's gapped (non-prelim): 36
length of query: 135
length of database: 11,318,596
effective HSP length: 88
effective length of query: 47
effective length of database: 8,967,324
effective search space: 421464228
effective search space used: 421464228
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 54 (25.4 bits)
Medicago: description of AC145512.4


