

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC145330.4 + phase: 0
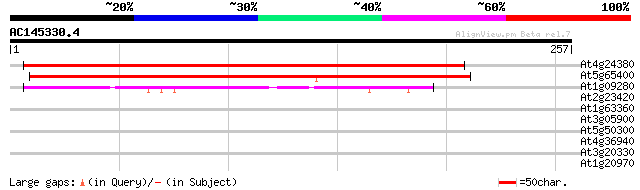
(257 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g24380 unknown protein 216 1e-56
At5g65400 putative protein 207 6e-54
At1g09280 unknown protein 67 9e-12
At2g23420 unknown protein 29 2.8
At1g63360 disease resistance protein, putative 29 2.8
At3g05900 unknown protein 28 3.7
At5g50300 transmembrane transport protein-like 28 4.8
At4g36940 unknown protein 28 6.3
At3g20330 aspartate carbamoyltransferase precursor (aspartate tr... 28 6.3
At1g20970 hypothetical protein 27 8.2
>At4g24380 unknown protein
Length = 234
Score = 216 bits (549), Expect = 1e-56
Identities = 98/202 (48%), Positives = 137/202 (67%)
Query: 7 RKLKILCLHGFRTSGSFIKKQISKWDPSIFSQFHLEFPDGKFPAGGKSDIEGIFPPPYFE 66
RK + LCLHGFRTSG +K Q+ KW S+ + L F D FP GKSD+EGIF PPY+E
Sbjct: 10 RKPRFLCLHGFRTSGEIMKIQLHKWPKSVIDRLDLVFLDAPFPCQGKSDVEGIFDPPYYE 69
Query: 67 WFQFDKDFTVYTNLDECISYLTEYIIANGPFDGFLGFSQGATLSALLIGYQAQGKLLKEH 126
WFQF+K+FT YTN ++C+ YL + +I GPFDG +GFSQGA LS L G QA+G ++
Sbjct: 70 WFQFNKEFTEYTNFEKCLEYLEDRMIKLGPFDGLIGFSQGAILSGGLPGLQAKGIAFQKV 129
Query: 127 PPIKFLVSISGSKFRDPSICDVAYKDPIKAKSVHFIGEKDWLKIPSEELASAFDKPLIIR 186
P IKF++ I G+K + + + AY ++ S+HF+GE D+LK +L ++ P+++
Sbjct: 130 PKIKFVIIIGGAKLKSAKLAENAYSSSLETLSLHFLGETDFLKPYGTQLIESYKNPVVVH 189
Query: 187 HPQGHTVPRLDEVSTGQLQNFV 208
HP+GHTVPRLDE S ++ F+
Sbjct: 190 HPKGHTVPRLDEKSLEKVTAFI 211
>At5g65400 putative protein
Length = 234
Score = 207 bits (526), Expect = 6e-54
Identities = 98/207 (47%), Positives = 134/207 (64%), Gaps = 5/207 (2%)
Query: 10 KILCLHGFRTSGSFIKKQISKWDPSIFSQFHLEFPDGKFPAGGKSDIEGIFPPPYFEWFQ 69
+ILCLHGFRTSG ++ I KW +I L+F D FPA GKSD+E F PPY+EW+Q
Sbjct: 13 RILCLHGFRTSGRILQAGIGKWPDTILRDLDLDFLDAPFPATGKSDVERFFDPPYYEWYQ 72
Query: 70 FDKDFTVYTNLDECISYLTEYIIANGPFDGFLGFSQGATLSALLIGYQAQGKLLKEHPPI 129
+K F Y N +EC++Y+ +Y+I NGPFDG LGFSQGA L+A + G Q QG L + P +
Sbjct: 73 ANKGFKEYRNFEECLAYIEDYMIKNGPFDGLLGFSQGAFLTAAIPGMQEQGSALTKVPKV 132
Query: 130 KFLVSISGSK-----FRDPSICDVAYKDPIKAKSVHFIGEKDWLKIPSEELASAFDKPLI 184
KFLV ISG+K F +P A+ P++ S+HFIGE+D+LKI E L +F +P++
Sbjct: 133 KFLVIISGAKIPGLMFGEPKAAVNAFSSPVRCPSLHFIGERDFLKIEGEVLVESFVEPVV 192
Query: 185 IRHPQGHTVPRLDEVSTGQLQNFVAEI 211
I H GH +P+LD + + +F I
Sbjct: 193 IHHSGGHIIPKLDTKAEETMLSFFQSI 219
>At1g09280 unknown protein
Length = 581
Score = 67.0 bits (162), Expect = 9e-12
Identities = 65/220 (29%), Positives = 90/220 (40%), Gaps = 39/220 (17%)
Query: 7 RKLKILCLHGFRTSGSFIKKQISKWDPSIFSQFHLEFPDGKFPAGGKSDIEGIFPP---- 62
RKL+ILCLHGFR + S K + + + L F D P + + PP
Sbjct: 352 RKLRILCLHGFRQNASSFKGRTGSLAKKLKNIAELVFIDA--PHELQFIYQTATPPSGVC 409
Query: 63 -PYFEWF---QFDKDF-----------------TVYTNLDECISYLTEYIIANGPFDGFL 101
F W FDK T D+ ++YL GPFDG L
Sbjct: 410 NKKFAWLVSSDFDKPSETGWTVAQCQFDPLQYQTQTEGFDKSLTYLKTAFEEKGPFDGIL 469
Query: 102 GFSQGATLSALLIGYQAQGKLLKEHPPIKFLVSISGSKFRDPSICDVAYKDPIKAKSVHF 161
GFSQGA ++A + G Q Q L +F V SG F + ++ K IK S+H
Sbjct: 470 GFSQGAAMAAAVCGKQEQ---LVGEIDFRFCVLCSG--FTPWPLLEMKEKRSIKCPSLHI 524
Query: 162 IG-----EKDWLKIPSEELASAFDK--PLIIRHPQGHTVP 194
G ++ + S +LA F+ I+ H GH +P
Sbjct: 525 FGSQPGKDRQIVTQASSDLAGLFEDGCATIVEHDFGHIIP 564
>At2g23420 unknown protein
Length = 557
Score = 28.9 bits (63), Expect = 2.8
Identities = 13/47 (27%), Positives = 24/47 (50%), Gaps = 1/47 (2%)
Query: 52 GKSDIEGIFPPPYFEWFQFDKDFTVYTNLDECISYLTEYIIANGPFD 98
GK + +F YF F ++TV+ L+EC+ +L + + + D
Sbjct: 43 GKHNERSVFDL-YFRKNPFGGEYTVFAGLEECVKFLANFKLTDEEID 88
>At1g63360 disease resistance protein, putative
Length = 884
Score = 28.9 bits (63), Expect = 2.8
Identities = 26/107 (24%), Positives = 48/107 (44%), Gaps = 10/107 (9%)
Query: 155 KAKSVHFIGEKDWLKIPSEELASAFDKPLIIRHPQG-----HTVPRLDEVSTGQLQNFVA 209
K +S+ KD I +EE A ++ I+ P+ H +P+L ++ L
Sbjct: 763 KIRSLSVWHAKDLEDIINEEKACEGEESGILPFPELNFLTLHDLPKLKKIYWRPLPFLCL 822
Query: 210 EILSQPKVGVSICEHESKVEVDGTNGDKGVNGVEINQGNAETVEVVQ 256
E ++ + C + K+ +D T+G +G NG I ++ E V+
Sbjct: 823 E-----EINIRECPNLRKLPLDSTSGKQGENGCIIRNKDSRWFEGVK 864
>At3g05900 unknown protein
Length = 660
Score = 28.5 bits (62), Expect = 3.7
Identities = 25/104 (24%), Positives = 44/104 (42%), Gaps = 14/104 (13%)
Query: 164 EKDWLKIPSEELASAFDKPLIIRHPQGHTVPRLDEVSTGQLQNFVAEILSQPKVGVSICE 223
EK+ + S ++ + DKP +P +D+ G+ + ++ + VG +
Sbjct: 16 EKEIISSDSADVVN--DKPASDSNPVVTKEEEIDQTPAGEPEKESPAVVEE--VGAVVKA 71
Query: 224 HESKVEVDGTNGDKGVNGVEINQG----------NAETVEVVQA 257
ES V NG+KG VE+ + N ETV+ +A
Sbjct: 72 EESTETVKHENGEKGAEQVELKEPILVKETVAEVNVETVDTEKA 115
>At5g50300 transmembrane transport protein-like
Length = 530
Score = 28.1 bits (61), Expect = 4.8
Identities = 14/51 (27%), Positives = 25/51 (48%)
Query: 72 KDFTVYTNLDECISYLTEYIIANGPFDGFLGFSQGATLSALLIGYQAQGKL 122
KD V T+L + L ++AN PF G A ++ ++G++ G +
Sbjct: 107 KDLVVATSLSAMVGSLAMGLLANLPFGLAPGMGANAYIAYNVVGFRGSGSI 157
>At4g36940 unknown protein
Length = 559
Score = 27.7 bits (60), Expect = 6.3
Identities = 12/39 (30%), Positives = 20/39 (50%), Gaps = 1/39 (2%)
Query: 52 GKSDIEGIFPPPYFEWFQFDKDFTVYTNLDECISYLTEY 90
GK +F YF F ++T++ L+ECI +L +
Sbjct: 45 GKQSERSVFDL-YFRKNPFGGEYTIFAGLEECIKFLANF 82
>At3g20330 aspartate carbamoyltransferase precursor (aspartate
transcarbamylase)
Length = 390
Score = 27.7 bits (60), Expect = 6.3
Identities = 32/120 (26%), Positives = 51/120 (41%), Gaps = 21/120 (17%)
Query: 116 YQAQGKLLKEHPPIKFLVSISGSKFRDPSI-------CDVAYKDPIKAKSVHFIGEKDWL 168
Y +++K IK ++ SG ++ + S CDV Y+ I+ + GE+ L
Sbjct: 266 YFVSPEIVKMKDDIKDYLTSSGVEWEESSDLMEVASKCDVVYQTRIQRER---FGERLDL 322
Query: 169 K-------IPSEELASAFDKPLIIRHPQGHTVPRLDEVSTGQLQNFVAEILSQPKVGVSI 221
I ++L K II HP +PRLDE++ + A Q K G+ I
Sbjct: 323 YEAARGKYIVDKDLLGVMQKKAIIMHP----LPRLDEITADVDADPRAAYFRQAKNGLFI 378
>At1g20970 hypothetical protein
Length = 1498
Score = 27.3 bits (59), Expect = 8.2
Identities = 21/89 (23%), Positives = 42/89 (46%), Gaps = 18/89 (20%)
Query: 163 GEKDWLKIPSEELASAFDKPLIIRHP-QGH-------------TVPRLDEVSTGQLQNFV 208
G ++ ++IP E+ + +K + +HP GH V +L + G ++
Sbjct: 128 GPEEVVEIPKSEVEDSLEKSVDQQHPGNGHLESGLEGKVESKEEVEQLHDSEVGS-KDLT 186
Query: 209 AEILSQPKVGVSICEHESKVEVDGTNGDK 237
+ +P+V + E +S+ +V+G GDK
Sbjct: 187 KNNVEEPEVEI---ESDSETDVEGHQGDK 212
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.140 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,607,271
Number of Sequences: 26719
Number of extensions: 296342
Number of successful extensions: 552
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 6
Number of HSP's successfully gapped in prelim test: 4
Number of HSP's that attempted gapping in prelim test: 544
Number of HSP's gapped (non-prelim): 11
length of query: 257
length of database: 11,318,596
effective HSP length: 97
effective length of query: 160
effective length of database: 8,726,853
effective search space: 1396296480
effective search space used: 1396296480
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 59 (27.3 bits)
Medicago: description of AC145330.4


