

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC145329.3 + phase: 0 /partial
(121 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
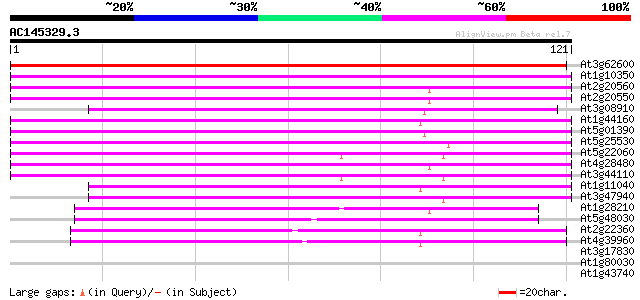
Score E
Sequences producing significant alignments: (bits) Value
At3g62600 unknown protein 192 3e-50
At1g10350 putative heat-shock protein 83 2e-17
At2g20560 putative heat shock protein 80 1e-16
At2g20550 putative heat shock protein 80 1e-16
At3g08910 putative heat shock protein 79 4e-16
At1g44160 putative protein 78 6e-16
At5g01390 heat shock protein 40-like 76 2e-15
At5g25530 heat-shock protein - like 76 3e-15
At5g22060 DNAJ PROTEIN HOMOLOG ATJ 76 3e-15
At4g28480 heat-shock protein 74 1e-14
At3g44110 dnaJ protein homolog atj3 70 1e-13
At1g11040 DnaJ isolog 64 1e-11
At3g47940 heat shock protein-like protein 62 3e-11
At1g28210 AtJ1 (atj) 54 1e-08
At5g48030 DnaJ protein-like 52 4e-08
At2g22360 DnaJ like protein 46 3e-06
At4g39960 DnaJ like protein 45 6e-06
At3g17830 DnaJ, putative 35 0.008
At1g80030 unknown protein 31 0.11
At1g43740 hypothetical protein 29 0.43
>At3g62600 unknown protein
Length = 346
Score = 192 bits (487), Expect = 3e-50
Identities = 89/120 (74%), Positives = 107/120 (89%)
Query: 1 EVLFYEDGEPIIDGESGDLRFRIRTAPHELFKREGNDLHTTVTITLVQALVGFEKTIKHL 60
EV FYEDGEPI+DG+ GDL+FRIRTAPH F+R+GNDLH V ITLV+ALVGFEK+ KHL
Sbjct: 225 EVSFYEDGEPILDGDPGDLKFRIRTAPHARFRRDGNDLHMNVNITLVEALVGFEKSFKHL 284
Query: 61 DEHLVDISSKGITNPKQVRKFKGEGMPLHTSTKKGDLYVTFEVLFPTTLSEEQKTKIKSI 120
D+H VDISSKGIT PK+V+KFKGEGMPLH STKKG+L+VTFEVLFP++L+++QK KIK +
Sbjct: 285 DDHEVDISSKGITKPKEVKKFKGEGMPLHYSTKKGNLFVTFEVLFPSSLTDDQKKKIKEV 344
>At1g10350 putative heat-shock protein
Length = 349
Score = 82.8 bits (203), Expect = 2e-17
Identities = 46/121 (38%), Positives = 63/121 (52%)
Query: 1 EVLFYEDGEPIIDGESGDLRFRIRTAPHELFKREGNDLHTTVTITLVQALVGFEKTIKHL 60
++ F E G DL F + PH +FKR+GNDL ++L+ AL G ++ L
Sbjct: 226 KITFPEKGNQEPGVTPADLIFVVDEKPHSVFKRDGNDLILEKKVSLIDALTGLTISVTTL 285
Query: 61 DEHLVDISSKGITNPKQVRKFKGEGMPLHTSTKKGDLYVTFEVLFPTTLSEEQKTKIKSI 120
D + I I P Q EGMP K+GDL VTFE+LFP+ L+ EQK +K +
Sbjct: 286 DGRSLTIPVLDIVKPGQEIVIPNEGMPTKDPLKRGDLRVTFEILFPSRLTSEQKNDLKRV 345
Query: 121 L 121
L
Sbjct: 346 L 346
>At2g20560 putative heat shock protein
Length = 337
Score = 80.5 bits (197), Expect = 1e-16
Identities = 47/122 (38%), Positives = 65/122 (52%), Gaps = 1/122 (0%)
Query: 1 EVLFYEDGEPIIDGESGDLRFRIRTAPHELFKREGNDLHTTVTITLVQALVGFEKTIKHL 60
++ F E G DL F I PH +F REGNDL T I+LV+AL G+ + L
Sbjct: 215 KITFPEKGNEQPGVIPADLVFIIDEKPHPVFTREGNDLIVTQKISLVEALTGYTVNLTTL 274
Query: 61 DEHLVDISSKGITNPKQVRKFKGEGMPLH-TSTKKGDLYVTFEVLFPTTLSEEQKTKIKS 119
D + I + +P+ EGMPL TK+G+L + F + FPT L+ EQKT +K
Sbjct: 275 DGRRLTIPVTNVVHPEYEEVVPKEGMPLQKDQTKRGNLRIKFNIKFPTRLTSEQKTGVKK 334
Query: 120 IL 121
+L
Sbjct: 335 LL 336
>At2g20550 putative heat shock protein
Length = 284
Score = 80.5 bits (197), Expect = 1e-16
Identities = 46/122 (37%), Positives = 65/122 (52%), Gaps = 1/122 (0%)
Query: 1 EVLFYEDGEPIIDGESGDLRFRIRTAPHELFKREGNDLHTTVTITLVQALVGFEKTIKHL 60
++ F E G DL F I PH +F REGNDL T I++++A G+ + L
Sbjct: 162 KITFSEKGNEQPGVIPADLVFIIDEKPHPVFTREGNDLVVTQKISVLEAFTGYTVNLTTL 221
Query: 61 DEHLVDISSKGITNPKQVRKFKGEGMPLH-TSTKKGDLYVTFEVLFPTTLSEEQKTKIKS 119
D + I + +P+ V EGMPL KKG+L + F + FPTTL+ EQKT +K
Sbjct: 222 DGRRLTIPVNTVIHPEYVEVVPNEGMPLQKDQAKKGNLRIKFNIKFPTTLTSEQKTGLKK 281
Query: 120 IL 121
+L
Sbjct: 282 LL 283
>At3g08910 putative heat shock protein
Length = 323
Score = 79.0 bits (193), Expect = 4e-16
Identities = 43/102 (42%), Positives = 59/102 (57%), Gaps = 1/102 (0%)
Query: 18 DLRFRIRTAPHELFKREGNDLHTTVTITLVQALVGFEKTIKHLDEHLVDISSKGITNPKQ 77
DL F + PH +FKR+GNDL T I LV+AL G+ + LD V + + +P
Sbjct: 216 DLVFIVDEKPHAVFKRDGNDLVMTQKIPLVEALTGYTAQVSTLDGRSVTVPINNVISPSY 275
Query: 78 VRKFKGEGMPL-HTSTKKGDLYVTFEVLFPTTLSEEQKTKIK 118
KGEGMP+ +KKG+L + F V FP+ L+ EQK+ IK
Sbjct: 276 EEVVKGEGMPIPKDPSKKGNLRIKFTVKFPSRLTTEQKSGIK 317
>At1g44160 putative protein
Length = 352
Score = 78.2 bits (191), Expect = 6e-16
Identities = 46/122 (37%), Positives = 68/122 (55%), Gaps = 1/122 (0%)
Query: 1 EVLFYEDGEPIIDGESGDLRFRIRTAPHELFKREGNDLHTTVTITLVQALVGFEKTIKHL 60
+V F G + DL F I HE+FKREG+DL V ++L++AL G E ++ L
Sbjct: 227 KVTFEGKGNEAMRSVPADLTFVIVEKEHEVFKREGDDLEMAVEVSLLEALTGCELSVALL 286
Query: 61 DEHLVDISSKGITNPKQVRKFKGEGMP-LHTSTKKGDLYVTFEVLFPTTLSEEQKTKIKS 119
D + + + + +P V +G+GMP L K+GDL V F FP L++EQ+ +I S
Sbjct: 287 DGDNMRLRIEDVIHPGYVTVVQGKGMPNLKEKGKRGDLRVRFRTKFPQHLTDEQRAEIHS 346
Query: 120 IL 121
IL
Sbjct: 347 IL 348
>At5g01390 heat shock protein 40-like
Length = 335
Score = 76.3 bits (186), Expect = 2e-15
Identities = 43/122 (35%), Positives = 65/122 (53%), Gaps = 1/122 (0%)
Query: 1 EVLFYEDGEPIIDGESGDLRFRIRTAPHELFKREGNDLHTTVTITLVQALVGFEKTIKHL 60
++ F E G DL F + PH +FKR+GNDL I+LV AL G+ + L
Sbjct: 212 KITFLEKGNEHRGVIPSDLVFIVDEKPHPVFKRDGNDLVVMQKISLVDALTGYTAQVTTL 271
Query: 61 DEHLVDISSKGITNPKQVRKFKGEGMPL-HTSTKKGDLYVTFEVLFPTTLSEEQKTKIKS 119
D + + + +P KGEGMP+ ++KG+L + F + FP+ L+ EQK+ IK
Sbjct: 272 DGRTLTVPVNNVISPSYEEVVKGEGMPIPKDPSRKGNLRIRFIIKFPSKLTTEQKSGIKR 331
Query: 120 IL 121
+L
Sbjct: 332 ML 333
>At5g25530 heat-shock protein - like
Length = 347
Score = 75.9 bits (185), Expect = 3e-15
Identities = 44/122 (36%), Positives = 66/122 (54%), Gaps = 1/122 (0%)
Query: 1 EVLFYEDGEPIIDGESGDLRFRIRTAPHELFKREGNDLHTTVTITLVQALVGFEKTIKHL 60
++ F + G ++ DL F I PH+LF R+GNDL T+ +TL +A+ G I L
Sbjct: 224 KIKFPDKGNEQVNQLPADLVFVIDEKPHDLFTRDGNDLITSRRVTLAEAIGGTTVNINTL 283
Query: 61 DEHLVDISSKGITNPKQVRKFKGEGMPLHTSTK-KGDLYVTFEVLFPTTLSEEQKTKIKS 119
D + + I +P GEGMP+ + KGDL + F+V FP L+ EQK+ +K
Sbjct: 284 DGRNLPVGVAEIVSPGYEFVVPGEGMPIAKEPRNKGDLKIKFDVQFPARLTTEQKSALKR 343
Query: 120 IL 121
+L
Sbjct: 344 VL 345
>At5g22060 DNAJ PROTEIN HOMOLOG ATJ
Length = 419
Score = 75.9 bits (185), Expect = 3e-15
Identities = 44/124 (35%), Positives = 67/124 (53%), Gaps = 3/124 (2%)
Query: 1 EVLFYEDGEPIIDGESGDLRFRIRTAPHELFKREGNDLHTTVTITLVQALVGFEKTIKHL 60
++ F + D +GD+ F I+ H FKR+G DL TI+L +AL GF+ + HL
Sbjct: 236 KITFSGQADEAPDTVTGDIVFVIQQKEHPKFKRKGEDLFVEHTISLTEALCGFQFVLTHL 295
Query: 61 DEHLVDISSK--GITNPKQVRKFKGEGMPLHTST-KKGDLYVTFEVLFPTTLSEEQKTKI 117
D+ + I SK + P + EGMP++ KG LY+ F V FP +LS +Q I
Sbjct: 296 DKRQLLIKSKPGEVVKPDSYKAISDEGMPIYQRPFMKGKLYIHFTVEFPESLSPDQTKAI 355
Query: 118 KSIL 121
+++L
Sbjct: 356 EAVL 359
>At4g28480 heat-shock protein
Length = 348
Score = 73.9 bits (180), Expect = 1e-14
Identities = 44/122 (36%), Positives = 60/122 (49%), Gaps = 1/122 (0%)
Query: 1 EVLFYEDGEPIIDGESGDLRFRIRTAPHELFKREGNDLHTTVTITLVQALVGFEKTIKHL 60
++ F E G DL F I PH +F REGNDL T ++L AL G+ I L
Sbjct: 226 KITFPEKGNEHPGVIPADLVFIIDEKPHPVFTREGNDLIVTQKVSLADALTGYTANIATL 285
Query: 61 DEHLVDISSKGITNPKQVRKFKGEGMPLH-TSTKKGDLYVTFEVLFPTTLSEEQKTKIKS 119
D + I + +P+ EGMPL TKKG+L + F + FP L+ EQK K
Sbjct: 286 DGRTLTIPITNVIHPEYEEVVPKEGMPLQKDQTKKGNLRIKFNIKFPARLTAEQKAGFKK 345
Query: 120 IL 121
++
Sbjct: 346 LI 347
>At3g44110 dnaJ protein homolog atj3
Length = 420
Score = 70.5 bits (171), Expect = 1e-13
Identities = 40/124 (32%), Positives = 65/124 (52%), Gaps = 3/124 (2%)
Query: 1 EVLFYEDGEPIIDGESGDLRFRIRTAPHELFKREGNDLHTTVTITLVQALVGFEKTIKHL 60
++ F + D +GD+ F ++ H FKR+G DL T++L +AL GF+ + HL
Sbjct: 235 KITFEGQADEAPDTVTGDIVFVLQQKEHPKFKRKGEDLFVEHTLSLTEALCGFQFVLTHL 294
Query: 61 DEHLVDISSK--GITNPKQVRKFKGEGMPLHTST-KKGDLYVTFEVLFPTTLSEEQKTKI 117
D + I S + P + EGMP++ KG LY+ F V FP +LS +Q +
Sbjct: 295 DGRSLLIKSNPGEVVKPDSYKAISDEGMPIYQRPFMKGKLYIHFTVEFPDSLSPDQTKAL 354
Query: 118 KSIL 121
+++L
Sbjct: 355 EAVL 358
>At1g11040 DnaJ isolog
Length = 438
Score = 63.9 bits (154), Expect = 1e-11
Identities = 38/105 (36%), Positives = 56/105 (53%), Gaps = 1/105 (0%)
Query: 18 DLRFRIRTAPHELFKREGNDLHTTVTITLVQALVGFEKTIKHLDEHLVDISSKGITNPKQ 77
D+ F + H LFKR G+DL V I L++AL G + ++ L + I+ +
Sbjct: 328 DITFVVEEKRHPLFKRRGDDLEIAVEIPLLKALTGCKLSVPLLSGESMSITVGDVIFHGF 387
Query: 78 VRKFKGEGMP-LHTSTKKGDLYVTFEVLFPTTLSEEQKTKIKSIL 121
+ KG+GMP K+GDL +TF V FP LSEEQ++ +L
Sbjct: 388 EKAIKGQGMPNAKEEGKRGDLRITFLVNFPEKLSEEQRSMAYEVL 432
>At3g47940 heat shock protein-like protein
Length = 350
Score = 62.4 bits (150), Expect = 3e-11
Identities = 36/105 (34%), Positives = 56/105 (53%), Gaps = 1/105 (0%)
Query: 18 DLRFRIRTAPHELFKREGNDLHTTVTITLVQALVGFEKTIKHLDEHLVDISSKGITNPKQ 77
D+ F + PH ++KR+GNDL + ITL++AL G + LD + I I P
Sbjct: 242 DIVFVVEEKPHPVYKRDGNDLLVSQEITLLEALTGKTVNLITLDGRTLMIPLTEIIKPDH 301
Query: 78 VRKFKGEGMPLHTST-KKGDLYVTFEVLFPTTLSEEQKTKIKSIL 121
EGMP+ KKG+L + V +P+ L+ +QK ++K +L
Sbjct: 302 EIVVPNEGMPISKEPGKKGNLKLKLSVKYPSRLTSDQKFELKRVL 346
>At1g28210 AtJ1 (atj)
Length = 427
Score = 53.9 bits (128), Expect = 1e-08
Identities = 31/101 (30%), Positives = 53/101 (51%), Gaps = 2/101 (1%)
Query: 15 ESGDLRFRIRTAPHELFKREGNDLHTTVTITLVQALVGFEKTIKHLDEHLVDISSKGITN 74
+ G+L +++ A F R+G+D++ I+ QA++G + + L + KG T
Sbjct: 278 QPGNLYIKLKVANDSTFTRDGSDIYVDANISFTQAILGGKVVVPTLSGKIQLDIPKG-TQ 336
Query: 75 PKQVRKFKGEGMPLH-TSTKKGDLYVTFEVLFPTTLSEEQK 114
P Q+ +G+G+P GD YV F V FPT ++E Q+
Sbjct: 337 PDQLLVLRGKGLPKQGFFVDHGDQYVRFRVNFPTEVNERQR 377
>At5g48030 DnaJ protein-like
Length = 456
Score = 52.4 bits (124), Expect = 4e-08
Identities = 28/100 (28%), Positives = 51/100 (51%), Gaps = 1/100 (1%)
Query: 15 ESGDLRFRIRTAPHELFKREGNDLHTTVTITLVQALVGFEKTIKHLDEHLVDISSKGITN 74
+ GDL ++ +F+REG+D+H +++ QA++G + L +V + + T
Sbjct: 334 QPGDLYVTLKVREDPVFRREGSDIHVDAVLSVTQAILGGTIQVPTLTGDVV-VKVRPGTQ 392
Query: 75 PKQVRKFKGEGMPLHTSTKKGDLYVTFEVLFPTTLSEEQK 114
P + +G+ STK GD YV F V P +++ Q+
Sbjct: 393 PGHKVVLRNKGIRARKSTKFGDQYVHFNVSIPANITQRQR 432
>At2g22360 DnaJ like protein
Length = 442
Score = 45.8 bits (107), Expect = 3e-06
Identities = 31/108 (28%), Positives = 51/108 (46%), Gaps = 2/108 (1%)
Query: 14 GESGDLRFRIRTAPHELFKREGNDLHTTVTITLVQALVGFEKTIKHLDEHLVDISSKGIT 73
G GDL I P + KR+ ++ T I+ + A++G + +D VD+ T
Sbjct: 322 GSPGDLFVVIEVIPDPILKRDDTNILYTCKISYIDAILGTTLKVPTVD-GTVDLKVPAGT 380
Query: 74 NPKQVRKFKGEGMP-LHTSTKKGDLYVTFEVLFPTTLSEEQKTKIKSI 120
P +G+P L+ S +GD V +V P LS+E+K I+ +
Sbjct: 381 QPSTTLVMAKKGVPVLNKSNMRGDQLVRVQVEIPKRLSKEEKKLIEEL 428
>At4g39960 DnaJ like protein
Length = 447
Score = 45.1 bits (105), Expect = 6e-06
Identities = 31/108 (28%), Positives = 51/108 (46%), Gaps = 2/108 (1%)
Query: 14 GESGDLRFRIRTAPHELFKREGNDLHTTVTITLVQALVGFEKTIKHLDEHLVDISSKGIT 73
G GDL I P + KR+ ++ T I+ V A++G + +D VD+ T
Sbjct: 328 GSPGDLFAVIEVIPDPVLKRDDTNILYTCKISYVDAILGTTLKVPTVDGE-VDLKVPAGT 386
Query: 74 NPKQVRKFKGEGMP-LHTSTKKGDLYVTFEVLFPTTLSEEQKTKIKSI 120
P +G+P L+ S +GD V +V P LS+E+K ++ +
Sbjct: 387 QPSTTLVMAKKGVPVLNKSKMRGDQLVRVQVEIPKRLSKEEKMLVEEL 434
>At3g17830 DnaJ, putative
Length = 517
Score = 34.7 bits (78), Expect = 0.008
Identities = 24/106 (22%), Positives = 49/106 (45%), Gaps = 2/106 (1%)
Query: 14 GESGDLRFRIRTAPHELFKREGNDLHTTVTITLVQALVGFEKTIKHLDEHLVDISSKGIT 73
G +GDL ++ +REG +L++ + I A++G ++ + E +D+ T
Sbjct: 311 GRAGDLFIVLQVDEKRGIRREGLNLYSNINIDFTDAILGATTKVETV-EGSMDLRIPPGT 369
Query: 74 NPKQVRKFKGEGMP-LHTSTKKGDLYVTFEVLFPTTLSEEQKTKIK 118
P K +G+P + +GD ++ P LSE ++ ++
Sbjct: 370 QPGDTVKLPRKGVPDTDRPSIRGDHCFVVKISIPKKLSERERKLVE 415
>At1g80030 unknown protein
Length = 500
Score = 30.8 bits (68), Expect = 0.11
Identities = 25/108 (23%), Positives = 52/108 (48%), Gaps = 2/108 (1%)
Query: 14 GESGDLRFRIRTAPHELFKREGNDLHTTVTITLVQALVGFEKTIKHLDEHLVDISSKGIT 73
G GDL + +R+G +L +T++I+ + A++G +K + E ++ T
Sbjct: 321 GPPGDLYVYLDVEDVRGIERDGINLLSTLSISYLDAILGAVVKVKTV-EGDTELQIPPGT 379
Query: 74 NPKQVRKFKGEGMP-LHTSTKKGDLYVTFEVLFPTTLSEEQKTKIKSI 120
P V +G+P L+ + +GD T +V P +S ++ ++ +
Sbjct: 380 QPGDVLVLAKKGVPKLNRPSIRGDHLFTVKVSVPNQISAGERELLEEL 427
>At1g43740 hypothetical protein
Length = 682
Score = 28.9 bits (63), Expect = 0.43
Identities = 19/73 (26%), Positives = 35/73 (47%), Gaps = 7/73 (9%)
Query: 34 EGNDLHTTVTITLVQALVGFEKTIK--HLDEHLVDISSKGITNPKQVRKFKGEGMPLHTS 91
EG+ T++++ L+ L+GF + H ++ + + G+ P + G+PLHT
Sbjct: 315 EGDRADTSLSVVLIDHLIGFREYAAGIHQSDYGGSLCAGGVITPILI----AVGVPLHTP 370
Query: 92 TKKGDLYVTFEVL 104
T Y+ E L
Sbjct: 371 THTA-TYIDMEYL 382
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.317 0.138 0.386
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,700,376
Number of Sequences: 26719
Number of extensions: 103036
Number of successful extensions: 273
Number of sequences better than 10.0: 39
Number of HSP's better than 10.0 without gapping: 31
Number of HSP's successfully gapped in prelim test: 8
Number of HSP's that attempted gapping in prelim test: 223
Number of HSP's gapped (non-prelim): 43
length of query: 121
length of database: 11,318,596
effective HSP length: 97
effective length of query: 24
effective length of database: 8,726,853
effective search space: 209444472
effective search space used: 209444472
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 52 (24.6 bits)
Medicago: description of AC145329.3


