

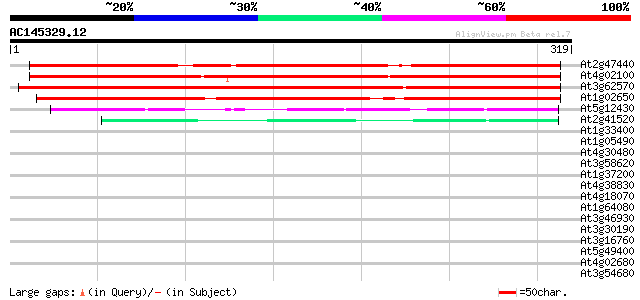
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC145329.12 - phase: 0
(319 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g47440 unknown protein 411 e-115
At4g02100 unknown protein 409 e-114
At3g62570 unknown protein 378 e-105
At1g02650 hypothetical protein 331 3e-91
At5g12430 putative protein 54 1e-07
At2g41520 Unknown protein 48 8e-06
At1g33400 unknown protein 37 0.014
At1g05490 hypothetical protein 35 0.054
At4g30480 unknown protein 32 0.35
At3g58620 putative protein 32 0.35
At1g37200 hypothetical protein 32 0.35
At4g38830 receptor-like protein kinase - like protein 32 0.46
At4g18070 unknown protein 32 0.46
At1g64080 unknown protein 32 0.46
At3g46930 protein kinase 6-like protein 32 0.60
At3g30190 hypothetical protein 32 0.60
At3g16760 unknown protein 32 0.60
At5g49400 putative protein 31 0.78
At4g02680 unknown protein 31 0.78
At3g54680 unknown protein 31 0.78
>At2g47440 unknown protein
Length = 526
Score = 411 bits (1057), Expect = e-115
Identities = 209/302 (69%), Positives = 252/302 (83%), Gaps = 21/302 (6%)
Query: 12 EKKHWWLTNRKIVEKYIKDARSLIATQEQSEILSALNLLDAALAISPRLDQALELRARSL 71
+KKHWW T+RK+V+KYIKDA +L+A++E +++ SAL+LLDAAL+ISPRL+ ALEL+ARSL
Sbjct: 10 DKKHWWFTHRKLVDKYIKDATTLMASEEANDVASALHLLDAALSISPRLETALELKARSL 69
Query: 72 LYLRRFKDVADMLQDYIPSLRMTNEDPSSGSSSSSSSSDSSSSREGVKLLSSDSPVRDQS 131
L+LRRFKDVADMLQDYIPSL++ +E S+SS SSS +G+ LLS S S
Sbjct: 70 LFLRRFKDVADMLQDYIPSLKLDDE--------GSASSQGSSSSDGINLLSDASS--PGS 119
Query: 132 FKCFSVSDLKKKVMAGLCKSCEKEGQWRYLVLGEACCHLGLMEDAMVLLQTGKRIASAAF 191
FKCFSVSDLKKKVMAG+CK C+KEGQWRY+VLG+ACCHLGLMEDAMVLLQTGKR+ASA F
Sbjct: 120 FKCFSVSDLKKKVMAGICKKCDKEGQWRYVVLGQACCHLGLMEDAMVLLQTGKRLASAEF 179
Query: 192 RRESVCWSDDSFPLLTIPLAGDTPNQQPTTPPRAPLNETESVTHLLSHIKFLLRRRAAAL 251
RR S+CWSDDSF LL+ + +P PP E+E+ THLL+HIK LLRRRAAA+
Sbjct: 180 RRRSICWSDDSFLLLSESSSASSP------PP-----ESENFTHLLAHIKLLLRRRAAAI 228
Query: 252 AALDAGLYSEAIRHFSKIVDGRRAAPQGFLAECYMHRASAHRSDGRIAESIADCNRTLSL 311
AALDAGL+SE+IRHFSKIVDGRR APQGFLAECYMHRA+A+RS GRIAE+IADCN+TL+L
Sbjct: 229 AALDAGLFSESIRHFSKIVDGRRPAPQGFLAECYMHRAAAYRSAGRIAEAIADCNKTLAL 288
Query: 312 DP 313
+P
Sbjct: 289 EP 290
>At4g02100 unknown protein
Length = 546
Score = 409 bits (1050), Expect = e-114
Identities = 208/305 (68%), Positives = 251/305 (82%), Gaps = 5/305 (1%)
Query: 12 EKKHWWLTNRKIVEKYIKDARSLIATQEQSEILSALNLLDAALAISPRLDQALELRARSL 71
EKKHWWL N+KIV+KY+K+A+SLIA+Q+ +++ SALNLL++AL++SPR + ALEL+ARSL
Sbjct: 6 EKKHWWLRNKKIVDKYMKEAKSLIASQDPNDVKSALNLLESALSVSPRYELALELKARSL 65
Query: 72 LYLRRFKDVADMLQDYIPSLRMTNEDPSSGSSSSSSSSDSSSSREGVKLLS---SDSPVR 128
LYLRR+KDVADMLQDYIPSL++ SG SS S + SSRE VKLL+ S
Sbjct: 66 LYLRRYKDVADMLQDYIPSLKLAGGGEDSGIGSSELSF-THSSRESVKLLNDLPSHHHHH 124
Query: 129 DQSFKCFSVSDLKKKVMAGLCKSCEKEGQWRYLVLGEACCHLGLMEDAMVLLQTGKRIAS 188
D SFKCFSVSDLKKKVMAGL K+C+++GQWRYLVLG+ACCHLGLMEDAMVLLQTGKR+A+
Sbjct: 125 DSSFKCFSVSDLKKKVMAGLTKNCDEQGQWRYLVLGQACCHLGLMEDAMVLLQTGKRLAT 184
Query: 189 AAFRRESVCWSDDSFPLLTIPLAGDTPNQQPTTPPRAPLNETESVTHLLSHIKFLLRRRA 248
AAFRR+S+CWSDDSF L + G +P A +ES+ H+LSHIK LLRRRA
Sbjct: 185 AAFRRQSICWSDDSFILFSSEDGGSSP-PSSVVVTSASQPRSESIAHVLSHIKLLLRRRA 243
Query: 249 AALAALDAGLYSEAIRHFSKIVDGRRAAPQGFLAECYMHRASAHRSDGRIAESIADCNRT 308
AALAALDAGLY+E+IRHFSKI+D RR APQGFLA+C+MHRASA+RS GRIAESIADCN+T
Sbjct: 244 AALAALDAGLYTESIRHFSKILDSRRGAPQGFLAQCFMHRASAYRSAGRIAESIADCNKT 303
Query: 309 LSLDP 313
L+LDP
Sbjct: 304 LALDP 308
>At3g62570 unknown protein
Length = 552
Score = 378 bits (971), Expect = e-105
Identities = 191/309 (61%), Positives = 247/309 (79%), Gaps = 2/309 (0%)
Query: 6 HSLTATEKKHWWLTNRKIVEKYIKDARSLIATQEQSEILSALNLLDAALAISPRLDQALE 65
+S+ +KKHWW T++KIV+KYIKDARSL+ ++EQ+++ SA++LLDAAL+ISPR + ALE
Sbjct: 4 YSVHNGDKKHWWFTHKKIVDKYIKDARSLMESEEQNDVASAIHLLDAALSISPRSETALE 63
Query: 66 LRARSLLYLRRFKDVADMLQDYIPSLRMTNEDPSSGSSSSSSSSDSSSSREGVKLLSSDS 125
L+ARSLL+LRRFKDV DMLQDYIPSL++ + S SS SSSS+ KLLS S
Sbjct: 64 LKARSLLFLRRFKDVVDMLQDYIPSLKLAVNEEDGSYSYEGSSYSSSSSQLSRKLLSDSS 123
Query: 126 PVRDQSFKCFSVSDLKKKVMAGLCKSCEKEGQWRYLVLGEACCHLGLMEDAMVLLQTGKR 185
P RD SFKCFSVS LKKK+MAG+CK+ +++ QWRY+VLG+ACCHLGLMEDA+VLLQTGKR
Sbjct: 124 PRRDSSFKCFSVSYLKKKIMAGICKNRDQDKQWRYVVLGQACCHLGLMEDALVLLQTGKR 183
Query: 186 IASAAFRRESVCWSDDSFPLLTIPLAGDTPNQQPTTPPRAPLNETESVTHLLSHIKFLLR 245
+A+ FRR SV SDDS LL + + + PPR ++E E+VT+LL+H K LLR
Sbjct: 184 LATVEFRRLSVSLSDDSVSLLLSESSSSSSSSSYAFPPR-KVSECETVTNLLAHTKNLLR 242
Query: 246 RRAAALAALDAGLYSEAIRHFSKIVDG-RRAAPQGFLAECYMHRASAHRSDGRIAESIAD 304
RR+A AA DAGL++++IRHFSKI+DG RR APQGFLA+CYMHRA+A++S G+IAE+IAD
Sbjct: 243 RRSAGFAAFDAGLFADSIRHFSKILDGRRRPAPQGFLADCYMHRAAAYKSAGKIAEAIAD 302
Query: 305 CNRTLSLDP 313
CN+TL+L+P
Sbjct: 303 CNKTLALEP 311
>At1g02650 hypothetical protein
Length = 513
Score = 331 bits (849), Expect = 3e-91
Identities = 176/298 (59%), Positives = 220/298 (73%), Gaps = 18/298 (6%)
Query: 16 WWLTNRKIVEKYIKDARSLIATQEQSEILSALNLLDAALAISPRLDQALELRARSLLYLR 75
WW NRK V+KY+K+A+ LI +Q+ ++I+SAL+LL++ L+ISP + ALEL+ARSLLYLR
Sbjct: 6 WWKRNRKKVDKYMKNAKDLITSQDPNDIVSALSLLNSTLSISPHHELALELKARSLLYLR 65
Query: 76 RFKDVADMLQDYIPSLRMTNEDPSSGSSSSSSSSDSSSSREGVKLLSSDSPVRDQSFKCF 135
RFKDVA +L +YIPSLR+ NED SS ++SS S + LL S SP D SFKCF
Sbjct: 66 RFKDVAVLLHNYIPSLRIDNEDVSSVFAASSELSSL------MLLLPSGSPSHDSSFKCF 119
Query: 136 SVSDLKKKVMAGLCKSCEKEGQWRYLVLGEACCHLGLMEDAMVLLQTGKRIASAAFRRES 195
S S LKKKVMAGL + + +GQWRYLVLG+AC HLGLM+DA++LLQTGKR+A+A RRES
Sbjct: 120 SYSYLKKKVMAGLSNNSQVQGQWRYLVLGQACYHLGLMDDAIILLQTGKRLATAELRRES 179
Query: 196 VCWSDDSFPLLTIPLAGDTPNQQPTTPPRAPLNETESVTHLLSHIKFLLRRRAAALAALD 255
+CWS+DSF L T QP P+ E+E V+ +LS K LRRR AALAALD
Sbjct: 180 ICWSEDSFNL-------STSESQP-----QPITESEIVSQMLSQTKLFLRRRTAALAALD 227
Query: 256 AGLYSEAIRHFSKIVDGRRAAPQGFLAECYMHRASAHRSDGRIAESIADCNRTLSLDP 313
AGLYSE+IRHFSKI+D RR APQ FL C + RA A++S GRIA+SIADCN L+L+P
Sbjct: 228 AGLYSESIRHFSKIIDSRRGAPQSFLVYCLIRRAFAYKSAGRIADSIADCNLILALEP 285
>At5g12430 putative protein
Length = 1187
Score = 53.9 bits (128), Expect = 1e-07
Identities = 60/289 (20%), Positives = 124/289 (42%), Gaps = 58/289 (20%)
Query: 24 VEKYIKDARSLIATQEQSEILSALNLLDAALAISPRLDQALELRARSLLYLRRFKDVADM 83
V + + +A + + ++ AL +L+ +L IS ++ L ++ +LL L ++ D A
Sbjct: 739 VSECMHEAGRRLQLRTLTDAEKALEILEDSLLISTYSEKLLTMKGEALLMLEKY-DAAIK 797
Query: 84 LQDYIPSLRMTNEDPSSGSSSSSSSSDSSSSREGVKLLSSDSPVRDQSFKCFSVSDLKKK 143
L + L N P S D+P +D +F+
Sbjct: 798 LCEQTVDLAGKNSPPDS----------------------HDTP-KDINFRI--------- 825
Query: 144 VMAGLCKSCEKEGQWRYLVLGEACCHLGLMEDAMVLLQTGKRIASAAFRRESVCWSDDSF 203
W+ ++ ++ ++G +E+A+ L+ +++ SA +RE + S
Sbjct: 826 --------------WQCHLMLKSSFYMGKLEEAIASLEKQEQLLSAT-KREGNKTLESSI 870
Query: 204 PLLTIPLAGDTPNQQPTTPPRAPLNETESVTHLLSHIKFLLRRRAAALAALDAGLYSEAI 263
PL P++ LN H+ F ++ AA A +G ++EA+
Sbjct: 871 PLAATIRELLRLKVLPSSSMSIALN---------LHLLFRIQLPAAGNEAFQSGRHTEAV 921
Query: 264 RHFSKIVDGRRAAPQGFLAECYMHRASAHRSDGRIAESIADCNRTLSLD 312
H++ + + F A C+ +RA+A+++ G+ +++IADC+ ++LD
Sbjct: 922 EHYTAAL-ACNVESRPFTAVCFCNRAAAYKALGQFSDAIADCSLAIALD 969
Score = 32.0 bits (71), Expect = 0.46
Identities = 13/29 (44%), Positives = 19/29 (64%)
Query: 284 CYMHRASAHRSDGRIAESIADCNRTLSLD 312
CY +RA+ + GR+ E+IADC S+D
Sbjct: 654 CYSNRAATRMALGRMREAIADCTMASSID 682
>At2g41520 Unknown protein
Length = 1108
Score = 47.8 bits (112), Expect = 8e-06
Identities = 50/260 (19%), Positives = 104/260 (39%), Gaps = 72/260 (27%)
Query: 53 ALAISPRLDQALELRARSLLYLRRFKDVADMLQDYIPSLRMTNEDPSSGSSSSSSSSDSS 112
AL+IS D+ L+++A +L +RR+K+V ++ ++ + + G +++ +
Sbjct: 713 ALSISSCSDKLLQMKAEALFMIRRYKEVIELCENTLQTAERNFVSAGIGGTTNVN----- 767
Query: 113 SSREGVKLLSSDSPVRDQSFKCFSVSDLKKKVMAGLCKSCEKEGQWRYLVLGEACCHLGL 172
GL + WR+ + ++ +LG
Sbjct: 768 ----------------------------------GLGSTYHSLIVWRWNKISKSHFYLGN 793
Query: 173 MEDAMVLLQTGKRIASAAFRRESVCWSDDSFPLLTIPLAGDTPNQQPTTPPRAPLNETES 232
+E A+ +L+ +++ + C ES
Sbjct: 794 LEKALDILEKLQQVEYTCNENQEEC--------------------------------RES 821
Query: 233 VTHLLSHIKFLLRRRAAALAALDAGLYSEAIRHFSKIVDGRRAAPQGFLAECYMHRASAH 292
L++ I LLR + A A+ Y EA+ ++ + R + F A C+ +RA+A+
Sbjct: 822 PASLVATISELLRYKNAGNEAVRDRKYMEAVEQYTAALS-RNVDSRPFAAICFCNRAAAN 880
Query: 293 RSDGRIAESIADCNRTLSLD 312
++ +IA++IADC+ ++LD
Sbjct: 881 QALVQIADAIADCSLAMALD 900
Score = 38.1 bits (87), Expect = 0.006
Identities = 18/33 (54%), Positives = 23/33 (69%)
Query: 281 LAECYMHRASAHRSDGRIAESIADCNRTLSLDP 313
LA CY +RA+A S GR+ E+I+DC SLDP
Sbjct: 596 LALCYGNRAAARISLGRLREAISDCEMAASLDP 628
>At1g33400 unknown protein
Length = 798
Score = 37.0 bits (84), Expect = 0.014
Identities = 21/62 (33%), Positives = 35/62 (55%), Gaps = 10/62 (16%)
Query: 259 YSEAIRHFSKIVDGRRAAP-------QGFLAECYMHRASAHRSDGRIAESIADCNRTLSL 311
+ EA+R +SK + R AP + LA +++RA+ + G + ES+ DC+R L +
Sbjct: 79 FDEALRLYSKAL---RVAPLDAIDGDKSLLASLFLNRANVLHNLGLLKESLRDCHRALRI 135
Query: 312 DP 313
DP
Sbjct: 136 DP 137
>At1g05490 hypothetical protein
Length = 731
Score = 35.0 bits (79), Expect = 0.054
Identities = 18/30 (60%), Positives = 23/30 (76%)
Query: 85 QDYIPSLRMTNEDPSSGSSSSSSSSDSSSS 114
+D + +++EDPSS SSSSSSSS SSSS
Sbjct: 270 KDNVTVESLSSEDPSSSSSSSSSSSSSSSS 299
Score = 30.8 bits (68), Expect = 1.0
Identities = 21/57 (36%), Positives = 29/57 (50%)
Query: 89 PSLRMTNEDPSSGSSSSSSSSDSSSSREGVKLLSSDSPVRDQSFKCFSVSDLKKKVM 145
PS ++ SS SSSSSSS D S +E V D +R S VS +++K +
Sbjct: 283 PSSSSSSSSSSSSSSSSSSSDDESYVKEVVGDNRDDDDLRKASSPIKRVSLVERKAL 339
>At4g30480 unknown protein
Length = 277
Score = 32.3 bits (72), Expect = 0.35
Identities = 16/59 (27%), Positives = 30/59 (50%), Gaps = 2/59 (3%)
Query: 257 GLYSEAIRHFSKIVDGRRAAPQGFLAE--CYMHRASAHRSDGRIAESIADCNRTLSLDP 313
GLY EA+ ++ ++ + P+ CY++R G+ E+I +C + L L+P
Sbjct: 119 GLYEEALSKYAFALELVQELPESIELRSICYLNRGVCFLKLGKCEETIKECTKALELNP 177
>At3g58620 putative protein
Length = 677
Score = 32.3 bits (72), Expect = 0.35
Identities = 21/86 (24%), Positives = 38/86 (43%), Gaps = 5/86 (5%)
Query: 228 NETESVTHLLSHIKFLLRRRAAALAALDAGLYSEAIRHFSKIVDGRRAAPQGFLAECYMH 287
+ + V +L+++K + + R +G YSEA + G F + Y +
Sbjct: 434 SNSPEVVSVLNNVKNVAKARTRGNELFSSGRYSEA-----SVAYGDGLKLDAFNSVLYCN 488
Query: 288 RASAHRSDGRIAESIADCNRTLSLDP 313
RA+ G +S+ DCN+ L + P
Sbjct: 489 RAACWFKLGMWEKSVDDCNQALRIQP 514
>At1g37200 hypothetical protein
Length = 1564
Score = 32.3 bits (72), Expect = 0.35
Identities = 22/75 (29%), Positives = 37/75 (49%), Gaps = 4/75 (5%)
Query: 63 ALELRARSLLYLRRFKDVADMLQDYIPSLRMTNEDPSSGSSSSSSSSDSSSSR----EGV 118
A+EL + Y R A +L D++ L + + D ++ + D SS+R EG+
Sbjct: 999 AIELSEYDIEYRTRTSFKAQVLADFVIELPLADLDGTNSNKKWLLHVDGSSNRQGSGEGI 1058
Query: 119 KLLSSDSPVRDQSFK 133
+L S V +QSF+
Sbjct: 1059 QLTSPTGEVIEQSFR 1073
>At4g38830 receptor-like protein kinase - like protein
Length = 665
Score = 32.0 bits (71), Expect = 0.46
Identities = 14/26 (53%), Positives = 16/26 (60%)
Query: 209 PLAGDTPNQQPTTPPRAPLNETESVT 234
P+ D P+ P TP R P NET SVT
Sbjct: 247 PVDTDDPSSVPATPSRPPKNETRSVT 272
>At4g18070 unknown protein
Length = 262
Score = 32.0 bits (71), Expect = 0.46
Identities = 21/50 (42%), Positives = 27/50 (54%), Gaps = 4/50 (8%)
Query: 95 NEDPSSGSSSSSSSSDSSSSREG----VKLLSSDSPVRDQSFKCFSVSDL 140
+ D SS SSSSSSSS SSSS + VK + D D + +VS +
Sbjct: 61 DHDKSSSSSSSSSSSSSSSSSDDESREVKKIDDDKKTGDDVSEVITVSSV 110
>At1g64080 unknown protein
Length = 411
Score = 32.0 bits (71), Expect = 0.46
Identities = 21/55 (38%), Positives = 30/55 (54%)
Query: 88 IPSLRMTNEDPSSGSSSSSSSSDSSSSREGVKLLSSDSPVRDQSFKCFSVSDLKK 142
+P + + + + SS +SSSSSSS SSSS K ++S V D+ LKK
Sbjct: 219 VPIISLFSRENSSKNSSSSSSSSSSSSSSLKKQNGNESVVSDEKRFVMMQKYLKK 273
>At3g46930 protein kinase 6-like protein
Length = 475
Score = 31.6 bits (70), Expect = 0.60
Identities = 26/100 (26%), Positives = 43/100 (43%), Gaps = 14/100 (14%)
Query: 97 DPSSGSSSSSSSSDSSSSREGVKL--LSSDSPVRDQSFKCFSVSDLKKKVMAGLCKSCEK 154
+P G + SSS SS ++ V L LS P+RD S +K++ + + K K
Sbjct: 82 EPEKGMKAKSSSRKDSSEKKSVNLRSLSHSGPIRD-----LSTQKVKERGKSKIDKKSSK 136
Query: 155 EGQWRYLVLGEACCHLGLMEDAMV---LLQTGKRIASAAF 191
+R G G++E+ ++ L G R A +
Sbjct: 137 SVDYR----GSKVSSAGVLEECLIDVSKLSYGDRFAHGKY 172
>At3g30190 hypothetical protein
Length = 263
Score = 31.6 bits (70), Expect = 0.60
Identities = 20/47 (42%), Positives = 24/47 (50%)
Query: 94 TNEDPSSGSSSSSSSSDSSSSREGVKLLSSDSPVRDQSFKCFSVSDL 140
++ SS SSSSSSSS SSSS S+ S SF + V L
Sbjct: 28 SSSSSSSSSSSSSSSSSSSSSSSSSSSSSTSSSSSSSSFSSYFVVPL 74
Score = 30.4 bits (67), Expect = 1.3
Identities = 19/40 (47%), Positives = 20/40 (49%)
Query: 92 RMTNEDPSSGSSSSSSSSDSSSSREGVKLLSSDSPVRDQS 131
+ N SS SSSSSSSS SSSS SS S S
Sbjct: 22 KQCNGSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSTSSSS 61
Score = 30.0 bits (66), Expect = 1.7
Identities = 18/34 (52%), Positives = 20/34 (57%)
Query: 92 RMTNEDPSSGSSSSSSSSDSSSSREGVKLLSSDS 125
+MT + S SSSSSSSS SSSS SS S
Sbjct: 19 KMTKQCNGSSSSSSSSSSSSSSSSSSSSSSSSSS 52
Score = 28.1 bits (61), Expect = 6.6
Identities = 19/39 (48%), Positives = 20/39 (50%)
Query: 100 SGSSSSSSSSDSSSSREGVKLLSSDSPVRDQSFKCFSVS 138
+GSSSSSSSS SSSS SS S S S S
Sbjct: 25 NGSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSTSSSSSS 63
>At3g16760 unknown protein
Length = 475
Score = 31.6 bits (70), Expect = 0.60
Identities = 18/57 (31%), Positives = 30/57 (52%), Gaps = 1/57 (1%)
Query: 257 GLYSEAIRHFS-KIVDGRRAAPQGFLAECYMHRASAHRSDGRIAESIADCNRTLSLD 312
G Y++AI+ S ++ RA + AE RAS ++ G +++ADC + L D
Sbjct: 367 GQYADAIKWLSWAVILMDRAGDEAGSAEVLSTRASCYKEVGEYKKAVADCTKVLDHD 423
>At5g49400 putative protein
Length = 280
Score = 31.2 bits (69), Expect = 0.78
Identities = 22/53 (41%), Positives = 28/53 (52%), Gaps = 2/53 (3%)
Query: 65 ELRARSLLYLRRFKDVADMLQDYIPSLRMTNEDPSSGSSSS--SSSSDSSSSR 115
E+ RS R+ + +D D S+ T+ D SSG SSS SSSSDS R
Sbjct: 168 EVEKRSKKSKRKHRSKSDSESDSEASVFETDSDGSSGESSSEYSSSSDSEDER 220
>At4g02680 unknown protein
Length = 888
Score = 31.2 bits (69), Expect = 0.78
Identities = 26/88 (29%), Positives = 41/88 (46%), Gaps = 12/88 (13%)
Query: 228 NETESVTHLLSHIKFLLRRRAAALAALDAGLYSEAIRHFSKIVDGRRAAPQGFLAECYMH 287
++ E VT L + R RAA L +D+ EAI S+ + F A+ ++
Sbjct: 794 SDLEMVTRLDPLRVYPYRYRAAVL--MDSRKEREAITELSRAI--------AFKADLHLL 843
Query: 288 --RASAHRSDGRIAESIADCNRTLSLDP 313
RA+ H G + ++ DC LS+DP
Sbjct: 844 HLRAAFHEHIGDVTSALRDCRAALSVDP 871
>At3g54680 unknown protein
Length = 211
Score = 31.2 bits (69), Expect = 0.78
Identities = 19/41 (46%), Positives = 26/41 (63%), Gaps = 4/41 (9%)
Query: 95 NEDPSSGSSSSSSSSDSSSSREGVKLL----SSDSPVRDQS 131
++DP S SSSSSS+S SS +R ++L SS P+R S
Sbjct: 19 HKDPPSSSSSSSSASYSSVARSNGRMLVLTKSSPKPLRSPS 59
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.130 0.376
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,823,162
Number of Sequences: 26719
Number of extensions: 281194
Number of successful extensions: 2868
Number of sequences better than 10.0: 84
Number of HSP's better than 10.0 without gapping: 43
Number of HSP's successfully gapped in prelim test: 41
Number of HSP's that attempted gapping in prelim test: 2524
Number of HSP's gapped (non-prelim): 180
length of query: 319
length of database: 11,318,596
effective HSP length: 99
effective length of query: 220
effective length of database: 8,673,415
effective search space: 1908151300
effective search space used: 1908151300
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC145329.12


