

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC145329.11 - phase: 0
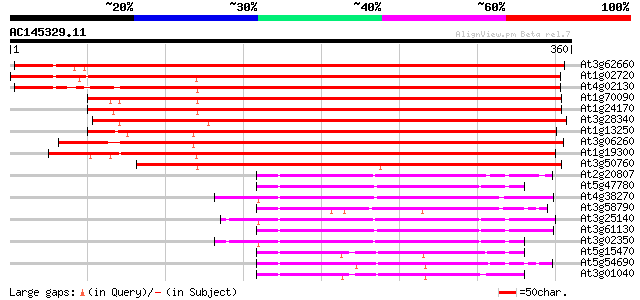
(360 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g62660 unknown protein 534 e-152
At1g02720 unknown protein 497 e-141
At4g02130 glycosyl transferase like protein 485 e-137
At1g70090 unknown protein 446 e-126
At1g24170 glycosyl transferase like protein 433 e-122
At3g28340 unknown protein 414 e-116
At1g13250 unknown protein 405 e-113
At3g06260 glycosyl transferase like protein 404 e-113
At1g19300 unknown protein 390 e-109
At3g50760 unknown protein 364 e-101
At2g20807 unknown protein 112 3e-25
At5g47780 unknown protein 106 2e-23
At4g38270 unknown protein 102 4e-22
At3g58790 unknown protein 96 4e-20
At3g25140 glycosyl transferase, putative 96 4e-20
At3g61130 unknown protein 93 2e-19
At3g02350 unknown protein 93 3e-19
At5g15470 unknown protein 91 1e-18
At5g54690 putative protein 90 2e-18
At3g01040 unknown protein 89 3e-18
>At3g62660 unknown protein
Length = 361
Score = 534 bits (1375), Expect = e-152
Identities = 260/359 (72%), Positives = 292/359 (80%), Gaps = 7/359 (1%)
Query: 4 IIKFSRFFSAVMTVIIISPSLHSSHYPALAIRSSIIH---HIPHHD---YRFYFHTSPFF 57
I++FS FSA + +I++SPSL S PA AIRSS + P D +RF F +P F
Sbjct: 4 IMRFSGLFSAALVIIVLSPSLQSFP-PAEAIRSSHLDAYLRFPSSDPPPHRFSFRKAPVF 62
Query: 58 RNADQCEPISREIGACHPSLVHVAITLDVEYLRGSIAAVHSILYHASCPENVFFHFLVTD 117
RNA C + G C+PSLVHVAITLD EYLRGSIAAVHSIL H+SCPE+VFFHFLV++
Sbjct: 63 RNAADCAAADIDSGVCNPSLVHVAITLDFEYLRGSIAAVHSILKHSSCPESVFFHFLVSE 122
Query: 118 TDLETLVRTTFPQLRFKVYYFDRNIVKNLISTSVRQALEQPLNYARNYLADLLESCVKRV 177
TDLE+L+R+TFP+L+ KVYYFD IV+ LISTSVRQALEQPLNYARNYLADLLE CV+RV
Sbjct: 123 TDLESLIRSTFPELKLKVYYFDPEIVRTLISTSVRQALEQPLNYARNYLADLLEPCVRRV 182
Query: 178 IYLDSDLVLQDDIAKLWNTDLGLNTIGAPQYCHANFTKYFTAAFWSDPVFSTTFEKRKAC 237
IYLDSDL++ DDIAKLW T LG TIGAP+YCHANFTKYFT AFWSD FS F RK C
Sbjct: 183 IYLDSDLIVVDDIAKLWMTKLGSKTIGAPEYCHANFTKYFTPAFWSDERFSGAFSGRKPC 242
Query: 238 YFNTGVMVMDLVKWRKKGYTERIERWMEIQKVERIYELGSLPPFLLVFAGHVAAIEHRWN 297
YFNTGVMVMDL +WR+ GYTE IE+WMEIQK +RIYELGSLPPFLLVFAG VA IEHRWN
Sbjct: 243 YFNTGVMVMDLERWRRVGYTEVIEKWMEIQKSDRIYELGSLPPFLLVFAGEVAPIEHRWN 302
Query: 298 QHGLGGDNVKGSCRDLHPGPVSLLHWSGSGKPWRRLDESKPCPLDALWEPFDLYGHRRR 356
QHGLGGDNV+GSCRDLHPGPVSLLHWSGSGKPW RLD +PCPLD LW P+DLYGH R
Sbjct: 303 QHGLGGDNVRGSCRDLHPGPVSLLHWSGSGKPWFRLDSRRPCPLDTLWAPYDLYGHYSR 361
>At1g02720 unknown protein
Length = 361
Score = 497 bits (1279), Expect = e-141
Identities = 246/361 (68%), Positives = 285/361 (78%), Gaps = 10/361 (2%)
Query: 1 MSSIIKFSRFFSAVMTVIIISPSLHSSHYPALAIRSSIIHHIP-----HHDYRFYFHTSP 55
M I +FS FFSA + +I++SPSL S PA AIRSS + + DY F SP
Sbjct: 1 MHWITRFSAFFSAALAMILLSPSLQSFS-PAAAIRSSHPYADEFKPQQNSDYSS-FRESP 58
Query: 56 FFRNADQCEPISREIGACHPSLVHVAITLDVEYLRGSIAAVHSILYHASCPENVFFHFLV 115
FRNA+QC + G C+P+LVHVAITLD++YLRGSIAAV+SIL H+ CP++VFFHFLV
Sbjct: 59 MFRNAEQCRSSGEDSGVCNPNLVHVAITLDIDYLRGSIAAVNSILQHSMCPQSVFFHFLV 118
Query: 116 TDT--DLETLVRTTFPQL-RFKVYYFDRNIVKNLISTSVRQALEQPLNYARNYLADLLES 172
+ +LE+L+R+TFP+L K+YYF V++LIS+SVRQALEQPLNYARNYLADLLE
Sbjct: 119 SSESQNLESLIRSTFPKLTNLKIYYFAPETVQSLISSSVRQALEQPLNYARNYLADLLEP 178
Query: 173 CVKRVIYLDSDLVLQDDIAKLWNTDLGLNTIGAPQYCHANFTKYFTAAFWSDPVFSTTFE 232
CVKRVIYLDSDLV+ DDI KLW T LG TIGAP+YCHANFTKYFT FWSD F+ TF+
Sbjct: 179 CVKRVIYLDSDLVVVDDIVKLWKTGLGQRTIGAPEYCHANFTKYFTGGFWSDKRFNGTFK 238
Query: 233 KRKACYFNTGVMVMDLVKWRKKGYTERIERWMEIQKVERIYELGSLPPFLLVFAGHVAAI 292
R CYFNTGVMV+DL KWR+ +T+RIE+WMEIQK+ERIYELGSLPPFLLVFAGHVA I
Sbjct: 239 GRNPCYFNTGVMVIDLKKWRQFRFTKRIEKWMEIQKIERIYELGSLPPFLLVFAGHVAPI 298
Query: 293 EHRWNQHGLGGDNVKGSCRDLHPGPVSLLHWSGSGKPWRRLDESKPCPLDALWEPFDLYG 352
HRWNQHGLGGDNV+GSCRDLH GPVSLLHWSGSGKPW RLD PCPLD LW P+DLY
Sbjct: 299 SHRWNQHGLGGDNVRGSCRDLHSGPVSLLHWSGSGKPWLRLDSKLPCPLDTLWAPYDLYK 358
Query: 353 H 353
H
Sbjct: 359 H 359
>At4g02130 glycosyl transferase like protein
Length = 346
Score = 485 bits (1248), Expect = e-137
Identities = 242/353 (68%), Positives = 274/353 (77%), Gaps = 15/353 (4%)
Query: 4 IIKFSRFFSAVMTVIIISPSLHSSHYPALAIRSSIIHHIPHHDYRFYFHTSPFFRNADQC 63
I +F+ FSA M VI++SPSL S PA AIRSS P +R F N D+C
Sbjct: 4 ITRFAGLFSAAMAVIVLSPSLQSFP-PAAAIRSS-----PSPIFR---KAPAVFNNGDEC 54
Query: 64 EPISREIGACHPSLVHVAITLDVEYLRGSIAAVHSILYHASCPENVFFHFLVTDTD---L 120
G C+PSLVHVAITLDVEYLRGSIAAV+SIL H+ CPE+VFFHF+ + L
Sbjct: 55 LSSG---GVCNPSLVHVAITLDVEYLRGSIAAVNSILQHSVCPESVFFHFIAVSEETNLL 111
Query: 121 ETLVRTTFPQLRFKVYYFDRNIVKNLISTSVRQALEQPLNYARNYLADLLESCVKRVIYL 180
E+LVR+ FP+L+F +Y F V+ LIS+SVRQALEQPLNYAR+YLADLLE CV RVIYL
Sbjct: 112 ESLVRSVFPRLKFNIYDFAPETVRGLISSSVRQALEQPLNYARSYLADLLEPCVNRVIYL 171
Query: 181 DSDLVLQDDIAKLWNTDLGLNTIGAPQYCHANFTKYFTAAFWSDPVFSTTFEKRKACYFN 240
DSDLV+ DDIAKLW T LG IGAP+YCHANFTKYFT FWS+ FS TF RK CYFN
Sbjct: 172 DSDLVVVDDIAKLWKTSLGSRIIGAPEYCHANFTKYFTGGFWSEERFSGTFRGRKPCYFN 231
Query: 241 TGVMVMDLVKWRKKGYTERIERWMEIQKVERIYELGSLPPFLLVFAGHVAAIEHRWNQHG 300
TGVMV+DL KWR+ GYT+RIE+WMEIQ+ ERIYELGSLPPFLLVF+GHVA I HRWNQHG
Sbjct: 232 TGVMVIDLKKWRRGGYTKRIEKWMEIQRRERIYELGSLPPFLLVFSGHVAPISHRWNQHG 291
Query: 301 LGGDNVKGSCRDLHPGPVSLLHWSGSGKPWRRLDESKPCPLDALWEPFDLYGH 353
LGGDNV+GSCRDLHPGPVSLLHWSGSGKPW RLD +PCPLDALW P+DLY H
Sbjct: 292 LGGDNVRGSCRDLHPGPVSLLHWSGSGKPWIRLDSKRPCPLDALWTPYDLYRH 344
>At1g70090 unknown protein
Length = 390
Score = 446 bits (1147), Expect = e-126
Identities = 212/315 (67%), Positives = 248/315 (78%), Gaps = 11/315 (3%)
Query: 51 FHTSPFFRNADQC--EPISRE--IGACHPSLVHVAITLDVEYLRGSIAAVHSILYHASCP 106
F +P +RN +C + ++RE + +C SLVHVA+TLD EYLRGSIAAVHS+L HASCP
Sbjct: 51 FVEAPEYRNGKECVSQSLNRENFVSSCDASLVHVAMTLDSEYLRGSIAAVHSMLRHASCP 110
Query: 107 ENVFFHFLVTDTD------LETLVRTTFPQLRFKVYYFDRNIVKNLISTSVRQALEQPLN 160
ENVFFH + + D L LVR+TFP L FKVY F + V NLIS+S+RQALE PLN
Sbjct: 111 ENVFFHLIAAEFDPASPRVLSQLVRSTFPSLNFKVYIFREDTVINLISSSIRQALENPLN 170
Query: 161 YARNYLADLLESCVKRVIYLDSDLVLQDDIAKLWNTDL-GLNTIGAPQYCHANFTKYFTA 219
YARNYL D+L+ CV RVIYLDSD+++ DDI KLWNT L G IGAP+YCHANFTKYFT+
Sbjct: 171 YARNYLGDILDPCVDRVIYLDSDIIVVDDITKLWNTSLTGSRIIGAPEYCHANFTKYFTS 230
Query: 220 AFWSDPVFSTTFEKRKACYFNTGVMVMDLVKWRKKGYTERIERWMEIQKVERIYELGSLP 279
FWSDP F RK CYFNTGVMVMDLV+WR+ Y E++E WM+IQK +RIY+LGSLP
Sbjct: 231 GFWSDPALPGFFSGRKPCYFNTGVMVMDLVRWREGNYREKLETWMQIQKKKRIYDLGSLP 290
Query: 280 PFLLVFAGHVAAIEHRWNQHGLGGDNVKGSCRDLHPGPVSLLHWSGSGKPWRRLDESKPC 339
PFLLVFAG+V AI+HRWNQHGLGGDNV+GSCR LH GPVSLLHWSG GKPW RLDE +PC
Sbjct: 291 PFLLVFAGNVEAIDHRWNQHGLGGDNVRGSCRSLHKGPVSLLHWSGKGKPWVRLDEKRPC 350
Query: 340 PLDALWEPFDLYGHR 354
PLD LWEP+DLY H+
Sbjct: 351 PLDHLWEPYDLYEHK 365
>At1g24170 glycosyl transferase like protein
Length = 393
Score = 433 bits (1113), Expect = e-122
Identities = 205/319 (64%), Positives = 243/319 (75%), Gaps = 15/319 (4%)
Query: 51 FHTSPFFRNADQCEP--------ISREIGACHPSLVHVAITLDVEYLRGSIAAVHSILYH 102
F +P +RN +C +S + PSLVH+A+TLD EYLRGSIAAVHS+L H
Sbjct: 50 FMEAPEYRNGKECVSSSVNRENFVSSSSSSNDPSLVHIAMTLDSEYLRGSIAAVHSVLRH 109
Query: 103 ASCPENVFFHFLVTDTD------LETLVRTTFPQLRFKVYYFDRNIVKNLISTSVRQALE 156
ASCPENVFFHF+ + D L LVR+TFP L FKVY F + V NLIS+S+R ALE
Sbjct: 110 ASCPENVFFHFIAAEFDSASPRVLSQLVRSTFPSLNFKVYIFREDTVINLISSSIRLALE 169
Query: 157 QPLNYARNYLADLLESCVKRVIYLDSDLVLQDDIAKLWNTDL-GLNTIGAPQYCHANFTK 215
PLNYARNYL D+L+ V+RVIYLDSD++ DDI KLWNT L G IGAP+YCHANFT+
Sbjct: 170 NPLNYARNYLGDILDRSVERVIYLDSDVITVDDITKLWNTVLTGSRVIGAPEYCHANFTQ 229
Query: 216 YFTAAFWSDPVFSTTFEKRKACYFNTGVMVMDLVKWRKKGYTERIERWMEIQKVERIYEL 275
YFT+ FWSDP +K CYFNTGVMVMDLV+WR+ Y E++E+WM++QK RIY+L
Sbjct: 230 YFTSGFWSDPALPGLISGQKPCYFNTGVMVMDLVRWREGNYREKLEQWMQLQKKMRIYDL 289
Query: 276 GSLPPFLLVFAGHVAAIEHRWNQHGLGGDNVKGSCRDLHPGPVSLLHWSGSGKPWRRLDE 335
GSLPPFLLVFAG+V AI+HRWNQHGLGGDN++GSCR LHPGPVSLLHWSG GKPW RLDE
Sbjct: 290 GSLPPFLLVFAGNVEAIDHRWNQHGLGGDNIRGSCRSLHPGPVSLLHWSGKGKPWVRLDE 349
Query: 336 SKPCPLDALWEPFDLYGHR 354
+PCPLD LWEP+DLY H+
Sbjct: 350 KRPCPLDHLWEPYDLYKHK 368
>At3g28340 unknown protein
Length = 365
Score = 414 bits (1063), Expect = e-116
Identities = 190/311 (61%), Positives = 240/311 (77%), Gaps = 7/311 (2%)
Query: 54 SPFFRNADQCEPISRE--IGACHPSLVHVAITLDVEYLRGSIAAVHSILYHASCPENVFF 111
+P ++N C +++ + AC PS VH+A+TLD YLRG+++AVHSIL H SCPEN+FF
Sbjct: 45 APAYQNGLDCSVLAKNRLLLACDPSAVHIAMTLDPAYLRGTVSAVHSILKHTSCPENIFF 104
Query: 112 HFLVTDTDLETLVRT---TFPQLRFKVYYFDRNIVKNLISTSVRQALEQPLNYARNYLAD 168
HF+ + T +L +T FP L FKVY F+ VKNLIS+S+RQAL+ PLNYAR+YL++
Sbjct: 105 HFIASGTSQGSLAKTLSSVFPSLSFKVYTFEETTVKNLISSSIRQALDSPLNYARSYLSE 164
Query: 169 LLESCVKRVIYLDSDLVLQDDIAKLWNTDL-GLNTIGAPQYCHANFTKYFTAAFWSDPVF 227
+L SCV RVIYLDSD+++ DDI KLW L G TIGAP+YCHANFTKYFT +FWSD
Sbjct: 165 ILSSCVSRVIYLDSDVIVVDDIQKLWKISLSGSRTIGAPEYCHANFTKYFTDSFWSDQKL 224
Query: 228 STTFEKRKACYFNTGVMVMDLVKWRKKGYTERIERWMEIQKVE-RIYELGSLPPFLLVFA 286
S+ F+ + CYFNTGVMV+DL +WR+ YT +IE WM+IQK + RIYELGSLPPFLLVF
Sbjct: 225 SSVFDSKTPCYFNTGVMVIDLERWREGDYTRKIENWMKIQKEDKRIYELGSLPPFLLVFG 284
Query: 287 GHVAAIEHRWNQHGLGGDNVKGSCRDLHPGPVSLLHWSGSGKPWRRLDESKPCPLDALWE 346
G + AI+H+WNQHGLGGDN+ SCR LHPGPVSL+HWSG GKPW RLD+ KPCP+D LW
Sbjct: 285 GDIEAIDHQWNQHGLGGDNIVSSCRSLHPGPVSLIHWSGKGKPWVRLDDGKPCPIDYLWA 344
Query: 347 PFDLYGHRRRY 357
P+DL+ +R+Y
Sbjct: 345 PYDLHKSQRQY 355
>At1g13250 unknown protein
Length = 345
Score = 405 bits (1040), Expect = e-113
Identities = 186/307 (60%), Positives = 234/307 (75%), Gaps = 7/307 (2%)
Query: 51 FHTSPFFRNADQCEPISREIGACH---PSLVHVAITLDVEYLRGSIAAVHSILYHASCPE 107
F +P FRN +C + I + H PS++H+A+TLD YLRGS+A V S+L HASCPE
Sbjct: 31 FREAPAFRNGRECSKTTW-IPSDHEHNPSIIHIAMTLDAIYLRGSVAGVFSVLQHASCPE 89
Query: 108 NVFFHFLVT---DTDLETLVRTTFPQLRFKVYYFDRNIVKNLISTSVRQALEQPLNYARN 164
N+ FHF+ T DL ++ +TFP L + +Y+FD N+V++ IS+S+R+AL+QPLNYAR
Sbjct: 90 NIVFHFIATHRRSADLRRIISSTFPYLTYHIYHFDPNLVRSKISSSIRRALDQPLNYARI 149
Query: 165 YLADLLESCVKRVIYLDSDLVLQDDIAKLWNTDLGLNTIGAPQYCHANFTKYFTAAFWSD 224
YLADLL V+RVIY DSDLV+ DD+AKLW DL + +GAP+YCHANFT YFT+ FWS
Sbjct: 150 YLADLLPIAVRRVIYFDSDLVVVDDVAKLWRIDLRRHVVGAPEYCHANFTNYFTSRFWSS 209
Query: 225 PVFSTTFEKRKACYFNTGVMVMDLVKWRKKGYTERIERWMEIQKVERIYELGSLPPFLLV 284
+ + + RK CYFNTGVMV+DL KWR++ T ++E WM IQK RIYELGSLPPFLLV
Sbjct: 210 QGYKSALKDRKPCYFNTGVMVIDLGKWRERRVTVKLETWMRIQKRHRIYELGSLPPFLLV 269
Query: 285 FAGHVAAIEHRWNQHGLGGDNVKGSCRDLHPGPVSLLHWSGSGKPWRRLDESKPCPLDAL 344
FAG V +EHRWNQHGLGGDN++G CR+LHPGPVSLLHWSG GKPW RLD +PCPLD+L
Sbjct: 270 FAGDVEPVEHRWNQHGLGGDNLEGLCRNLHPGPVSLLHWSGKGKPWLRLDSRRPCPLDSL 329
Query: 345 WEPFDLY 351
W P+DL+
Sbjct: 330 WAPYDLF 336
>At3g06260 glycosyl transferase like protein
Length = 351
Score = 404 bits (1039), Expect = e-113
Identities = 188/327 (57%), Positives = 235/327 (71%), Gaps = 10/327 (3%)
Query: 32 LAIRSSIIHHIPHHDYRFYFHTSPFFRNADQCEPISREIGACHPSLVHVAITLDVEYLRG 91
+A+R +I H P F +P FRN DQC G +H+A+TLD YLRG
Sbjct: 27 MAVRVGVILHKPSAPTLPVFREAPAFRNGDQC-------GTREADQIHIAMTLDTNYLRG 79
Query: 92 SIAAVHSILYHASCPENVFFHFLVT---DTDLETLVRTTFPQLRFKVYYFDRNIVKNLIS 148
++AAV S+L H++CPEN+ FHFL + DL T +++TFP L FK+Y FD N+V++ IS
Sbjct: 80 TMAAVLSLLQHSTCPENLSFHFLSLPHFENDLFTSIKSTFPYLNFKIYQFDPNLVRSKIS 139
Query: 149 TSVRQALEQPLNYARNYLADLLESCVKRVIYLDSDLVLQDDIAKLWNTDLGLNTIGAPQY 208
S+RQAL+QPLNYAR YLAD++ S V R+IYLDSDLV+ DDI KLW+ ++ + AP+Y
Sbjct: 140 KSIRQALDQPLNYARIYLADIIPSSVDRIIYLDSDLVVVDDIEKLWHVEMEGKVVAAPEY 199
Query: 209 CHANFTKYFTAAFWSDPVFSTTFEKRKACYFNTGVMVMDLVKWRKKGYTERIERWMEIQK 268
CHANFT YFT FWSDPV E ++ CYFNTGVMV+D+ KWRK YT+++E WM IQK
Sbjct: 200 CHANFTHYFTRTFWSDPVLVKVLEGKRPCYFNTGVMVVDVNKWRKGMYTQKVEEWMTIQK 259
Query: 269 VERIYELGSLPPFLLVFAGHVAAIEHRWNQHGLGGDNVKGSCRDLHPGPVSLLHWSGSGK 328
+RIY LGSLPPFLL+FAG + A+ HRWNQHGLGGDN +G CR LHPGP+SLLHWSG GK
Sbjct: 260 QKRIYHLGSLPPFLLIFAGDIKAVNHRWNQHGLGGDNFEGRCRTLHPGPISLLHWSGKGK 319
Query: 329 PWRRLDESKPCPLDALWEPFDLYGHRR 355
PW RLD KPC +D LW P+DLY R
Sbjct: 320 PWLRLDSRKPCIVDHLWAPYDLYRSSR 346
>At1g19300 unknown protein
Length = 351
Score = 390 bits (1003), Expect = e-109
Identities = 194/342 (56%), Positives = 240/342 (69%), Gaps = 18/342 (5%)
Query: 26 SSHYPALAIRSSIIHHIPHHDYRFY--FHTSPFFRNADQC----------EPISREIGAC 73
S H L + S ++ H P F +P F N+ C + +++ I C
Sbjct: 2 SQHLLLLILLSLLLLHKPISATTIIQKFKEAPQFYNSADCPLIDDSESDDDVVAKPI-FC 60
Query: 74 HPSLVHVAITLDVEYLRGSIAAVHSILYHASCPENVFFHFLVTDT----DLETLVRTTFP 129
VHVA+TLD Y+RGS+AAV S+L H+SCPEN+ FHF+ + + L + ++FP
Sbjct: 61 SRRAVHVAMTLDAAYIRGSVAAVLSVLQHSSCPENIVFHFVASASADASSLRATISSSFP 120
Query: 130 QLRFKVYYFDRNIVKNLISTSVRQALEQPLNYARNYLADLLESCVKRVIYLDSDLVLQDD 189
L F VY F+ + V LIS+S+R AL+ PLNYAR+YLADLL CV+RV+YLDSDL+L DD
Sbjct: 121 YLDFTVYVFNVSSVSRLISSSIRSALDCPLNYARSYLADLLPPCVRRVVYLDSDLILVDD 180
Query: 190 IAKLWNTDLGLNTI-GAPQYCHANFTKYFTAAFWSDPVFSTTFEKRKACYFNTGVMVMDL 248
IAKL TDLG +++ AP+YC+ANFT YFT+ FWS+P S TF RKACYFNTGVMV+DL
Sbjct: 181 IAKLAATDLGRDSVLAAPEYCNANFTSYFTSTFWSNPTLSLTFADRKACYFNTGVMVIDL 240
Query: 249 VKWRKKGYTERIERWMEIQKVERIYELGSLPPFLLVFAGHVAAIEHRWNQHGLGGDNVKG 308
+WR+ YT RIE WM +QK RIYELGSLPPFLLVFAG + + HRWNQHGLGGDN +G
Sbjct: 241 SRWREGAYTSRIEEWMAMQKRMRIYELGSLPPFLLVFAGLIKPVNHRWNQHGLGGDNFRG 300
Query: 309 SCRDLHPGPVSLLHWSGSGKPWRRLDESKPCPLDALWEPFDL 350
CRDLHPGPVSLLHWSG GKPW RLD +PCPLDALW P+DL
Sbjct: 301 LCRDLHPGPVSLLHWSGKGKPWARLDAGRPCPLDALWAPYDL 342
>At3g50760 unknown protein
Length = 285
Score = 364 bits (934), Expect = e-101
Identities = 169/279 (60%), Positives = 215/279 (76%), Gaps = 6/279 (2%)
Query: 82 ITLDVEYLRGSIAAVHSILYHASCPENVFFHFLVTDTD--LETLVRTTFPQLRFKVYYFD 139
+TLD YLRGS+A + S+L H+SCP+N+ FHF+ + L+ V +FP L+F++Y +D
Sbjct: 1 MTLDTAYLRGSMAVILSVLQHSSCPQNIVFHFVTSKQSHRLQNYVVASFPYLKFRIYPYD 60
Query: 140 RNIVKNLISTSVRQALEQPLNYARNYLADLLESCVKRVIYLDSDLVLQDDIAKLWNTDLG 199
+ LISTS+R AL+ PLNYARNYLAD+L +C+ RV+YLDSDL+L DDI+KL++T +
Sbjct: 61 VAAISGLISTSIRSALDSPLNYARNYLADILPTCLSRVVYLDSDLILVDDISKLFSTHIP 120
Query: 200 LNTI-GAPQYCHANFTKYFTAAFWSDPVFSTTFE-KRKA--CYFNTGVMVMDLVKWRKKG 255
+ + AP+YC+ANFT YFT FWS+P S T R+A CYFNTGVMV++L KWR+
Sbjct: 121 TDVVLAAPEYCNANFTTYFTPTFWSNPSLSITLSLNRRATPCYFNTGVMVIELKKWREGD 180
Query: 256 YTERIERWMEIQKVERIYELGSLPPFLLVFAGHVAAIEHRWNQHGLGGDNVKGSCRDLHP 315
YT +I WME+QK RIYELGSLPPFLLVFAG++A ++HRWNQHGLGGDN +G CRDLHP
Sbjct: 181 YTRKIIEWMELQKRIRIYELGSLPPFLLVFAGNIAPVDHRWNQHGLGGDNFRGLCRDLHP 240
Query: 316 GPVSLLHWSGSGKPWRRLDESKPCPLDALWEPFDLYGHR 354
GPVSLLHWSG GKPW RLD+ +PCPLDALW P+DL R
Sbjct: 241 GPVSLLHWSGKGKPWVRLDDGRPCPLDALWVPYDLLESR 279
>At2g20807 unknown protein
Length = 536
Score = 112 bits (280), Expect = 3e-25
Identities = 68/190 (35%), Positives = 106/190 (55%), Gaps = 9/190 (4%)
Query: 159 LNYARNYLADLLESCVKRVIYLDSDLVLQDDIAKLWNTDLGLNTIGAPQYCHANFTKYFT 218
LN+ R Y+ ++ + +K+V++LD D+V+Q D++ L++ DL N GA + C F +Y
Sbjct: 342 LNHLRFYIPEVFPA-LKKVVFLDDDVVVQKDLSSLFSIDLNKNVNGAVETCMETFHRYHK 400
Query: 219 AAFWSDPVFSTTFEKRKACYFNTGVMVMDLVKWRKKGYTERIERWMEIQKVERIYELGSL 278
+S P+ + F+ AC + G+ V DLV+WRK+ T W E +++LG+L
Sbjct: 401 YLNYSHPLIRSHFDP-DACGWAFGMNVFDLVEWRKRNVTGIYHYWQEKNVDRTLWKLGTL 459
Query: 279 PPFLLVFAGHVAAIEHRWNQHGLGGDNVKGSCRDLHPGPVSLLHWSGSGKPWRRLDESKP 338
PP LL F G A+E W+ GLG NV R + G V LH++G+ KPW ++ K
Sbjct: 460 PPGLLTFYGLTEALEASWHILGLGYTNV--DARVIEKGAV--LHFNGNLKPWLKIGIEKY 515
Query: 339 CPLDALWEPF 348
P LWE +
Sbjct: 516 KP---LWERY 522
>At5g47780 unknown protein
Length = 616
Score = 106 bits (264), Expect = 2e-23
Identities = 57/172 (33%), Positives = 101/172 (58%), Gaps = 5/172 (2%)
Query: 159 LNYARNYLADLLESCVKRVIYLDSDLVLQDDIAKLWNTDLGLNTIGAPQYCHANFTKYFT 218
LN+ R YL ++ + +V++LD D+V+Q D++ LW+ DL N GA + C +F ++
Sbjct: 420 LNHLRFYLPEIFPK-LSKVLFLDDDIVVQKDLSGLWSVDLKGNVNGAVETCGESFHRFDR 478
Query: 219 AAFWSDPVFSTTFEKRKACYFNTGVMVMDLVKWRKKGYTERIERWMEIQKVERIYELGSL 278
+S+P+ S F+ R AC + G+ V DL +W+++ TE RW ++ + +++LG+L
Sbjct: 479 YLNFSNPLISKNFDPR-ACGWAYGMNVFDLDEWKRQNITEVYHRWQDLNQDRELWKLGTL 537
Query: 279 PPFLLVFAGHVAAIEHRWNQHGLGGDNVKGSCRDLHPGPVSLLHWSGSGKPW 330
PP L+ F ++ +W+ GL G N + RD+ V +H++G+ KPW
Sbjct: 538 PPGLITFWRRTYPLDRKWHILGL-GYNPSVNQRDIERAAV--IHYNGNLKPW 586
>At4g38270 unknown protein
Length = 680
Score = 102 bits (253), Expect = 4e-22
Identities = 63/223 (28%), Positives = 113/223 (50%), Gaps = 10/223 (4%)
Query: 132 RFKVYYFDRNIVKNLISTSVRQALEQP-----LNYARNYLADLLESCVKRVIYLDSDLVL 186
R K YYF N ++ + + P LN+ R YL ++ ++++++LD D+V+
Sbjct: 452 RLKEYYFKANHPSSISAGADNLKYRNPKYLSMLNHLRFYLPEVYPK-LEKILFLDDDIVV 510
Query: 187 QDDIAKLWNTDLGLNTIGAPQYCHANFTKYFTAAFWSDPVFSTTFEKRKACYFNTGVMVM 246
Q D+A LW D+ GA + C +F ++ +S+P S F+ AC + G+ +
Sbjct: 511 QKDLAPLWEIDMQGKVNGAVETCKESFHRFDKYLNFSNPKISENFDAG-ACGWAFGMNMF 569
Query: 247 DLVKWRKKGYTERIERWMEIQKVERIYELGSLPPFLLVFAGHVAAIEHRWNQHGLGGDNV 306
DL +WRK+ T W ++ + +++LGSLPP L+ F A++ W+ GLG D
Sbjct: 570 DLKEWRKRNITGIYHYWQDLNEDRTLWKLGSLPPGLITFYNLTYAMDRSWHVLGLGYDPA 629
Query: 307 KGSCRDLHPGPVSLLHWSGSGKPWRRLDESKPCPLDALWEPFD 349
+ +++H++G+ KPW L +K P + + +D
Sbjct: 630 LNQTAIEN---AAVVHYNGNYKPWLGLAFAKYKPYWSKYVEYD 669
>At3g58790 unknown protein
Length = 540
Score = 95.5 bits (236), Expect = 4e-20
Identities = 64/194 (32%), Positives = 102/194 (51%), Gaps = 14/194 (7%)
Query: 159 LNYARNYLADLLESCVKRVIYLDSDLVLQDDIAKLWNTDLGLNTIGA--PQYCHANF--- 213
LN+ R Y+ L + +++ LD D+V+Q D++ LW TDL +GA +C N
Sbjct: 336 LNHLRIYIPKLFPD-LNKIVLLDDDVVVQSDLSSLWETDLNGKVVGAVVDSWCGDNCCPG 394
Query: 214 TKYFTAAFWSDPVFSTTFEKRKACYFNTGVMVMDLVKWRKKGYTERIERW--MEIQKVER 271
KY +S P+ S+ ++ C + +G+ V DL WR+ TE W + ++ +
Sbjct: 395 RKYKDYFNFSHPLISSNL-VQEDCAWLSGMNVFDLKAWRQTNITEAYSTWLRLSVRSGLQ 453
Query: 272 IYELGSLPPFLLVFAGHVAAIEHRWNQHGLGGDNVKGSCRDLHPGPVSLLHWSGSGKPWR 331
+++ G+LPP LL F G ++E W+ GLG +VK L S+LH+SG KPW
Sbjct: 454 LWQPGALPPTLLAFKGLTQSLEPSWHVAGLGSRSVKSPQEILK--SASVLHFSGPAKPW- 510
Query: 332 RLDESKPCPLDALW 345
L+ S P + +LW
Sbjct: 511 -LEISNP-EVRSLW 522
>At3g25140 glycosyl transferase, putative
Length = 559
Score = 95.5 bits (236), Expect = 4e-20
Identities = 62/220 (28%), Positives = 107/220 (48%), Gaps = 11/220 (5%)
Query: 136 YYFDRNIVKNLISTSVRQALEQP-----LNYARNYLADLLESCVKRVIYLDSDLVLQDDI 190
+YF+ N ++N + P LN+ R YL ++ + R+++LD D+V+Q D+
Sbjct: 336 FYFE-NKLENATKDTTNMKFRNPKYLSILNHLRFYLPEMYPK-LHRILFLDDDVVVQKDL 393
Query: 191 AKLWNTDLGLNTIGAPQYCHANFTKYFTAAFWSDPVFSTTFEKRKACYFNTGVMVMDLVK 250
LW D+ GA + C +F +Y +S P+ F KAC + G+ DL
Sbjct: 394 TGLWEIDMDGKVNGAVETCFGSFHRYAQYMNFSHPLIKEKFNP-KACAWAYGMNFFDLDA 452
Query: 251 WRKKGYTERIERWMEIQKVERIYELGSLPPFLLVFAGHVAAIEHRWNQHGLGGDNVKGSC 310
WR++ TE W + + +++LG+LPP L+ F ++ W+ GL G N S
Sbjct: 453 WRREKCTEEYHYWQNLNENRALWKLGTLPPGLITFYSTTKPLDKSWHVLGL-GYNPSISM 511
Query: 311 RDLHPGPVSLLHWSGSGKPWRRLDESKPCPLDALWEPFDL 350
++ V +H++G+ KPW + ++ PL +DL
Sbjct: 512 DEIRNAAV--VHFNGNMKPWLDIAMNQFRPLWTKHVDYDL 549
>At3g61130 unknown protein
Length = 673
Score = 93.2 bits (230), Expect = 2e-19
Identities = 55/191 (28%), Positives = 98/191 (50%), Gaps = 5/191 (2%)
Query: 159 LNYARNYLADLLESCVKRVIYLDSDLVLQDDIAKLWNTDLGLNTIGAPQYCHANFTKYFT 218
LN+ R YL ++ + ++++LD D+++Q D+ LW +L GA + C +F ++
Sbjct: 477 LNHLRFYLPEVYPK-LNKILFLDDDIIVQKDLTPLWEVNLNGKVNGAVETCGESFHRFDK 535
Query: 219 AAFWSDPVFSTTFEKRKACYFNTGVMVMDLVKWRKKGYTERIERWMEIQKVERIYELGSL 278
+S+P + F AC + G+ + DL +W+K+ T +W + + +++LG+L
Sbjct: 536 YLNFSNPHIARNFNPN-ACGWAYGMNMFDLKEWKKRDITGIYHKWQNMNENRTLWKLGTL 594
Query: 279 PPFLLVFAGHVAAIEHRWNQHGLGGDNVKGSCRDLHPGPVSLLHWSGSGKPWRRLDESKP 338
PP L+ F G + W+ GL G N +D+ V +H++G+ KPW L SK
Sbjct: 595 PPGLITFYGLTHPLNKAWHVLGL-GYNPSIDKKDIENAAV--VHYNGNMKPWLELAMSKY 651
Query: 339 CPLDALWEPFD 349
P + FD
Sbjct: 652 RPYWTKYIKFD 662
>At3g02350 unknown protein
Length = 561
Score = 92.8 bits (229), Expect = 3e-19
Identities = 56/204 (27%), Positives = 103/204 (50%), Gaps = 11/204 (5%)
Query: 132 RFKVYYFDRNIVKNLISTSVRQALEQP-----LNYARNYLADLLESCVKRVIYLDSDLVL 186
+ + +YF+ N +N S + P LN+ R YL ++ + ++++LD D+V+
Sbjct: 334 KLQKFYFE-NQAENATKDSHNLKFKNPKYLSMLNHLRFYLPEMYPK-LNKILFLDDDVVV 391
Query: 187 QDDIAKLWNTDLGLNTIGAPQYCHANFTKYFTAAFWSDPVFSTTFEKRKACYFNTGVMVM 246
Q D+ LW +L GA + C +F +Y +S P+ F AC + G+ +
Sbjct: 392 QKDVTGLWKINLDGKVNGAVETCFGSFHRYGQYLNFSHPLIKENFNP-SACAWAFGMNIF 450
Query: 247 DLVKWRKKGYTERIERWMEIQKVERIYELGSLPPFLLVFAGHVAAIEHRWNQHGLGGDNV 306
DL WR++ T++ W + + +++LG+LPP L+ F +++ W+ GL G N
Sbjct: 451 DLNAWRREKCTDQYHYWQNLNEDRTLWKLGTLPPGLITFYSKTKSLDKSWHVLGL-GYNP 509
Query: 307 KGSCRDLHPGPVSLLHWSGSGKPW 330
S ++ V +H++G+ KPW
Sbjct: 510 GVSMDEIRNAGV--IHYNGNMKPW 531
>At5g15470 unknown protein
Length = 532
Score = 90.9 bits (224), Expect = 1e-18
Identities = 55/182 (30%), Positives = 93/182 (50%), Gaps = 18/182 (9%)
Query: 159 LNYARNYLADLLESCVKRVIYLDSDLVLQDDIAKLWNTDLGLNTIGAPQYCHA------- 211
LN+ R Y+ +L + + +V++LD D+V+Q D+ LW+ DLG GA + C
Sbjct: 329 LNHLRIYIPELFPN-LDKVVFLDDDIVVQGDLTPLWDVDLGGKVNGAVETCRGEDEWVMS 387
Query: 212 -NFTKYFTAAFWSDPVFSTTFEKRKACYFNTGVMVMDLVKWRKKGYTERIERWM--EIQK 268
YF +S P+ + + + C + G+ + DL WRK E W+ ++
Sbjct: 388 KRLRNYFN---FSHPLIAKHLDPEE-CAWAYGMNIFDLQAWRKTNIRETYHSWLRENLKS 443
Query: 269 VERIYELGSLPPFLLVFAGHVAAIEHRWNQHGLGGDNVKGSCRDLHPGPVSLLHWSGSGK 328
+++LG+LPP L+ F GHV I+ W+ GLG + K + ++ V +H++G K
Sbjct: 444 NLTMWKLGTLPPALIAFKGHVHIIDSSWHMLGLGYQS-KTNIENVKKAAV--IHYNGQSK 500
Query: 329 PW 330
PW
Sbjct: 501 PW 502
>At5g54690 putative protein
Length = 535
Score = 89.7 bits (221), Expect = 2e-18
Identities = 59/197 (29%), Positives = 99/197 (49%), Gaps = 15/197 (7%)
Query: 159 LNYARNYLADLLESCVKRVIYLDSDLVLQDDIAKLWNTDLGLNTIGAPQYCHANFTKYFT 218
+N+ R +L +L S + +V++LD D+V+Q D++ LW+ D+ GA + C +
Sbjct: 331 MNHIRIHLPELFPS-LNKVVFLDDDIVIQTDLSPLWDIDMNGKVNGAVETCRGEDKFVMS 389
Query: 219 AAF-----WSDPVFSTTFEKRKACYFNTGVMVMDLVKWRKKGYTERIERWME--IQKVER 271
F +S+P + F + C + G+ V DL WR+ + W++ ++
Sbjct: 390 KKFKSYLNFSNPTIAKNFNPEE-CAWAYGMNVFDLAAWRRTNISSTYYHWLDENLKSDLS 448
Query: 272 IYELGSLPPFLLVFAGHVAAIEHRWNQHGLGGDNVKGSCRDLHPGPVSLLHWSGSGKPWR 331
+++LG+LPP L+ F GHV I+ W+ GLG S D V +H++G KPW
Sbjct: 449 LWQLGTLPPGLIAFHGHVQTIDPFWHMLGLGYQETT-SYADAESAAV--VHFNGRAKPW- 504
Query: 332 RLDESKPCPLDALWEPF 348
LD + P L LW +
Sbjct: 505 -LDIAFP-HLRPLWAKY 519
>At3g01040 unknown protein
Length = 510
Score = 89.4 bits (220), Expect = 3e-18
Identities = 53/182 (29%), Positives = 92/182 (50%), Gaps = 18/182 (9%)
Query: 159 LNYARNYLADLLESCVKRVIYLDSDLVLQDDIAKLWNTDLGLNTIGAPQYCHAN------ 212
LN+ R YL +L + + +V++LD D+V+Q D++ LW+ DL GA + C
Sbjct: 307 LNHLRIYLPELFPN-LDKVVFLDDDIVIQKDLSPLWDIDLNGKVNGAVETCRGEDVWVMS 365
Query: 213 --FTKYFTAAFWSDPVFSTTFEKRKACYFNTGVMVMDLVKWRKKGYTERIERWME--IQK 268
YF +S P+ + + + C + G+ + DL WRK E W++ ++
Sbjct: 366 KRLRNYFN---FSHPLIAKHLDPEE-CAWAYGMNIFDLRTWRKTNIRETYHSWLKENLKS 421
Query: 269 VERIYELGSLPPFLLVFAGHVAAIEHRWNQHGLGGDNVKGSCRDLHPGPVSLLHWSGSGK 328
+++LG+LPP L+ F GHV I+ W+ GLG + + +++H++G K
Sbjct: 422 NLTMWKLGTLPPALIAFKGHVQPIDSSWHMLGLG---YQSKTNLENAKKAAVIHYNGQSK 478
Query: 329 PW 330
PW
Sbjct: 479 PW 480
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.326 0.140 0.449
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,623,055
Number of Sequences: 26719
Number of extensions: 383952
Number of successful extensions: 1045
Number of sequences better than 10.0: 35
Number of HSP's better than 10.0 without gapping: 27
Number of HSP's successfully gapped in prelim test: 8
Number of HSP's that attempted gapping in prelim test: 964
Number of HSP's gapped (non-prelim): 39
length of query: 360
length of database: 11,318,596
effective HSP length: 101
effective length of query: 259
effective length of database: 8,619,977
effective search space: 2232574043
effective search space used: 2232574043
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 61 (28.1 bits)
Medicago: description of AC145329.11


